Back to Journals » Journal of Inflammation Research » Volume 16
Association of Enolase-1 with Prognosis and Immune Infiltration in Breast Cancer by Clinical Stage
Authors Shi YY , Chen XL, Chen QX, Yang YZ, Zhou M, Ren YX, Tang LY, Ren ZF
Received 5 November 2022
Accepted for publication 10 January 2023
Published 7 February 2023 Volume 2023:16 Pages 493—503
DOI https://doi.org/10.2147/JIR.S396321
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Yue-Yu Shi,1 Xing-Lei Chen,1 Qian-Xin Chen,1 Yuan-Zhong Yang,2 Meng Zhou,1 Yue-Xiang Ren,3 Lu-Ying Tang,3 Ze-Fang Ren1
1School of Public Health, Sun Yat-sen University, Guangzhou, People’s Republic of China; 2The Sun Yat-sen University Cancer Center, Guangzhou, People’s Republic of China; 3The Third Affiliated Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China
Correspondence: Ze-Fang Ren, School of Public Health, Sun Yat-sen University, 74 Zhongshan 2nd Road, Guangzhou, Guangdong, People’s Republic of China, Tel/Fax +86-20-87332577, Email [email protected] Lu-Ying Tang, The Third Affiliated Hospital, Sun Yat-sen University, 600 Tianhe Road, Guangzhou, Guangdong, People’s Republic of China, Tel +86-20-85253000, Fax +86-20-85253336, Email [email protected]
Purpose: Enolase-1 (ENO1) plays a key role in malignancies. Previous studies on the association between ENO1 expression and breast cancer prognosis had yielded inconsistent results. In the present study, we assessed the prognostic effect of ENO1 in breast cancer using Guangzhou Breast Cancer Study (GZBCS) cohort with full consideration of the potential confounders and the modification effects. The results were further validated in the TCGA-BRCA cohort and explained by tumor immunity.
Methods: ENO1 protein expressions were evaluated by immunohistochemistry in tissue microarrays from 961 patients with primary invasive breast cancer. Chi-square tests were used to assess the association of ENO1 levels with the patient’s characteristics. Cox regression models were applied to assess the prognostic effects. The TCGA-BRCA cohort was utilized to validate the results and explore the potential mechanisms. The immune infiltration was determined using the CIBERSORT and ssGSEA algorithms; the correlation between ENO1 expression and the abundance of tumor-infiltrating immune cells (TIICs) and scores of immune-related functions was evaluated by Wilcoxon signed-rank tests and Spearman’s rank test.
Results: ENO1 protein expression exerted a protective effect on OS in stage I/II patients (HR=0.58, 95% CI: 0.35– 0.96) but not in stage III patients (HR=1.42, 95% CI: 0.81– 2.49, P interaction=0.04) in GZBCS; consistent results were obtained at mRNA levels in TCGA cohort. Immune infiltration analyses revealed that ENO1 was positively correlated with multiple antitumor TIICs (including M1 macrophages, B cells, CD8 T cells, T helper 2 cells, and NK cells) only in stage I/II but not stage III patients.
Conclusion: A higher expression of ENO1 was associated with a better prognosis only in early-stage breast cancer, which may be related to the different effects of ENO1 on immune infiltration, suggesting that ENO1 may be a promising target for precision immunotherapy in breast cancer.
Keywords: ENO1, breast cancer, prognosis, tissue microarray, tumor-infiltrating immune cells
Introduction
Breast cancer is the most prevalent malignancy and the leading cause of cancer death in women worldwide.1 After its diagnosis, the most immediate challenge is to tailor treatment strategies and predict the prognosis; traditional clinicopathologic features, including estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), etc, provided relatively effective biomarkers.2 However, it is widely believed that these factors alone are not sufficient for optimal patient management; patients with the same characters (such as ER+) would differently respond to the treatment and have various prognosis.3–5 Therefore, more biomarkers are needed to precisely determine treatment strategy and predict prognosis for breast cancer.
Enolase-1 (ENO1) is generally recognized to be a glycolytic enzyme catalyzing the conversion of 2-phospho-
For breast cancer, two previous studies applied public databases to analyze the association between ENO1 expression levels in the tissues and the prognosis. Cancemi et al22 found in the Kaplan–Meier plotter (http://kmplot.com/analysis/) that a high ENO1 expression level was associated with a poor survival; Xu et al23 reported a null association between the expression and the prognosis in the GEPIA database (http://gepia.cancer-pku.cn/). Due to limited information in the databases, both studies were unable to consider the confounding of clinicopathological characteristics which may distort the results.
In the present study, we performed an immunohistochemical analysis of tissue microarrays from patients in Guangzhou Breast Cancer Study (GZBCS) cohort to assess the prognostic effect of ENO1 in breast cancer with full consideration of the potential confounders and the modification effects. The results were then validated in the TCGA-BRCA cohort. Furthermore, we performed immune infiltration analyses to explore the potential mechanisms.
Materials and Methods
Subjects
The subjects were a subgroup from the GZBCS cohort, as described elsewhere.24 Briefly, a total of 1063 females with pathologically diagnosed primary invasive breast cancer and >1 cm of tumor size in diameter were recruited between January 2008 and December 2015 from the Cancer Center of Sun Yat-sen University in Guangzhou, China. The study was approved by the Ethics Committee of the School of Public Health, Sun Yat-sen University. All participants provided written informed consent. Patients with stage I–III breast cancer and those with available ENO1 expression information were eligible for this study (N=975). A total of 961 cases of the included patients were successfully followed up until December 31, 2021.
Baseline Information Collection and Follow-Up
Baseline demographic characteristics were collected by trained investigators in face-to-face interviews using structured questionnaires. Body mass index (BMI) and clinicopathologic characteristics at baseline were collected from medical records. Patients were followed up by phone calls or outpatient visits every 3 months in the first year, every 6 months in the second and third year after diagnosis, and annually thereafter. Overall survival (OS) was defined as the time from diagnosis to death and progression-free survival (PFS) was defined as the time from diagnosis to disease progression including recurrence, metastasis, and death. Survival status was censored at the latest follow-up date.
Construction of Tissue Microarray (TMA) and Immunohistochemistry (IHC)
TMA construction and IHC staining were carried out according to standard procedure, the details of which we have previously described.25 Briefly, tissue cylinders with a diameter of 1 mm were punched out of the corresponding paraffin block as donor block and placed into the TMA paraffin block. After dewaxing, hydration, and antigen retrieval (Target Retrieval; Dako pH 6.0), slides were incubated in rabbit monoclonal (EPR 19758) to ENO1 (ab 227978, diluted 1:3000, Abcam) and labeled with the EnVision Detection System (Peroxidase/Diaminobenzidine, Rabbit/Mouse) (Dako K5007).
IHC stained sections were digitally imaged using Pannoramic Scanner and CaseViewer software. IHC staining was analyzed by an experienced pathologist and scored for staining intensity (0 – no staining, 1 – weak, 2 – moderate, and 3 – strong, as shown in Figure 1A–D) and percentage of tumor cell staining (0–100). Multiplying staining intensity with percentages yields an H-score ranging from 0 to 300. The mean value of the H-score from duplicate cores was taken.
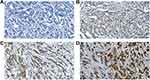 |
Figure 1 Representative images were shown for IHC staining intensity of ENO1 in breast cancer tissues. (A) No staining, (B) weak, (C) moderate, and (D) strong. |
Validation of the ENO1’prognostic Effect in TCGA-BRCA Cohort
To validate the prognostic effect of ENO1 in breast cancer, we downloaded publicly available gene expression profiles (workflow type: HTSeq-FPKM) and corresponding clinical information from The Cancer Genome Atlas Breast Invasive Carcinoma (TCGA-BRCA, https://portal.gdc.cancer.gov/, accessed in February 2022). Patients with stage I–III breast cancer with complete gene expression profiles and survival information were included in the study (N=1034). Then, FPKM values were transformed into transcripts per kilobase million (TPM) values for subsequent analysis.
Immune Infiltration Analysis
To explore the potential role of ENO1 in the tumor immunity of breast cancer patients, we evaluated the association between ENO1 expression and immune infiltration in the TCGA-BRCA cohort. Cell-type identification by estimating relative subsets of RNA transcripts (CIBERSORT)26 algorithm can accurately determine the composition of 22 types of tumor-infiltrating immune cells (TIICs) in tissues based on a “signature matrix” of 547 genes. First, we uploaded the standardized processed expression data to the CIBERSORT website (https://cibersortx.stanford.edu/index.php), selected LM22, set the number of iterations to 1000, and analyzed the results. Then using R package GSVA, single-sample gene set enrichment analysis (ssGSEA)27 algorithm was performed to calculate the scores of infiltrating immune cells and to evaluate the activity of immune-related pathways based on 29 immune gene sets.
Statistical Analysis
The median was used as the cut-off value to divide ENO1 H-score and mRNA expression into high and low levels. Chi-square tests were used to analyze the association between ENO1 level and patient characteristics. Cox regression models were used to estimate the hazard ratios (HR) and 95% confidence interval (CI). Stratified analysis was further performed to assess the modification effects of clinicopathological characteristics on the association of ENO1 expression with breast cancer prognosis. Wilcoxon signed-rank tests and Spearman’s rank correlation tests were used to examine the correlation between immune infiltration and ENO1 expression. All analyses were conducted using R 3.6.2 and a two-sided P-value below 0.05 was considered statistical significance.
This study followed the reporting recommendations for tumor marker prognostic studies (REMARK) criteria.28
Results
Demographic and Clinicopathological Characteristics of the GZBCS Cohort
More than half of the women included were between 36 and 50 years of age at diagnosis (65.9%), premenopausal (58.0%), and between 18.5 and 23.9 kg/m2 of BMI (56.4%). The majority of the women were diagnosed with low histological grade (grade I/II: 73.3%), early clinical stage (stage I/II: 72.2%), ER-positive (73.2%), PR-positive (72.3%), or HER2-negative (66.7%) (Table 1).
 |
Table 1 Demographic and Clinicopathological Characteristics and the Associations with ENO1 Protein Expression in Breast Cancer Tissue |
ENO1 Protein Expression in Breast Cancer Tissues and the Association with Demographic and Clinicopathological Characteristics
As shown in Table 1, there were 470 patients with low ENO1 expression and 491 patients with high ENO1 expression. Patients with high-level ENO1 expression were more likely to have grade III, ER-negative, PR-negative, and HER2-positive tumors than the subjects with low ENO1.
Prognostic Effect of ENO1 Protein Expression on Breast Cancer and the Modification Effects of Clinical Stage
Of all the 961 women included in the survival analysis, 137 died and 215 experienced disease progression with a median follow-up time of 80.98 months. ENO1 expression was not statistically significantly associated with breast cancer prognosis in the whole subjects. When stratified by clinical stage, however, we observed that a high ENO1 expression was significantly associated with a lower risk of death among stage I/II patients (HR=0.58, 95% CI: 0.35–0.96) while there was an inverse association among stage III patients (HR=1.42, 95% CI: 0.81–2.49); the interaction was significant (P interaction=0.04). Similar results were observed for PFS (Table 2). We further conducted the stratified analyses by histological grade, ER, and HER-2 status, and there were no differential effects between the strata (Supplemental Tables 1–3).
 |
Table 2 Association of ENO1 Protein Expression in Breast Cancer Tissue and Prognosis Stratified by Clinical Stage |
Association of ENO1 Expression at the mRNA Level with Breast Cancer Prognosis Varied by Clinical Stage
To validate our results, we examined the association between ENO1 mRNA expression level and breast cancer prognosis in the TCGA-BRCA cohort. Clinicopathological characteristics and the association with ENO1 expression in the TCGA-BRCA cohort were overall consistent with that in our cohort (Supplemental Table 4). Similarly, the stratified analysis showed that the clinical stage significantly modified the association between ENO1 expression and OS (P interaction <0.01); as shown in Table 3, a high mRNA level of ENO1 was marginally significantly associated with a better OS among stage I/II patients (HR=0.48, 95% CI: 0.21–1.09) but not among stage III patients (HR=1.40, 95% CI: 0.41–4.41). No modification effects were observed for ER or HER2 status (Supplemental Tables 5 and 6).
 |
Table 3 Association of ENO1 mRNA Expression with OS in TCGA-BRCA Cohort Stratified by Clinical Stage |
Association of ENO1 Expression with Immune Infiltration
The CIBERSORT and ssGSEA algorithms were employed within stage I/II and stage III samples to investigate the correlation of ENO1 expression with immune cells and immune functions. CIBERSORT analysis demonstrated that 14 types of TIICs were associated with ENO1 expression within stage I/II patients (Figure 2A). To be specific, seven types of TIICs (ie, CD4 memory-activated T cells, T follicular helper cells, regulatory T cells, macrophage M0, macrophage M1, activated dendritic cells, and resting NK cells) were positively related to ENO1 expression; seven TIICs types were negatively correlated with ENO1 expression, including plasma cells, CD4 memory resting T cells, naive B cells, resting dendritic cells, macrophage M2, resting mast cells and monocytes. In stage III patients (Figure 2B), those with elevated ENO1 levels had more CD4 memory-activated T cells, T follicular helper cells, and macrophage M0, but fewer plasma cells, CD4 memory resting T cells, naive B cells, resting mast cells, and monocytes compared with those with ENO1 low-expressing. The results of ssGSEA analysis (Table 4) showed that ENO1 expression was associated with almost all immune cells and immune functions in stage I/II but not stage III patients. In particular, several antitumor immune-related terms, including B cells, CD8 T cells, T helper 2 (Th2) cells, NK cells, cytolytic activity, human leukocyte antigen (HLA), major histocompatibility complex (MHC) class I, parainflammation, and type I IFN responses were positively correlated with ENO1 expression only in stage I/II patients (All Spearman’s rho >0, P<0.05).
 |
Table 4 Association Between ENO1 Expression with the Enrichment Scores of 29 Immune-Related Terms Evaluated Based on ssGSEA in Different Stage Patients |
 |
Figure 2 Correlation between ENO1 expression and immune cell infiltration in stage I/II (A) and stage III (B) breast cancer patients. *P<0.05, **P<0.01, ****P<0.0001. |
Discussion
In the present study, we found that ENO1 protein expression exerted a protective effect on survival in stage I/II breast cancer patients, while this protective effect disappeared in stage III patients. The results were subsequently confirmed in mRNA levels within the TCGA-BRCA cohort. Furthermore, immune infiltration analyses showed that the correlation of ENO1 expression with immune-related functions and immune infiltration differed in patients of different stages.
For the relationship of ENO1 expression in breast cancer with the prognosis, Xu et al23 revealed a null association and Cancemi et al22 found a negative association, which were not consistent with our result. One possible reason was that the proportions of patients’ clinical stages varied in different studies, as we found that the association between ENO1 expression and breast cancer prognosis varied by clinical stage. Unfortunately, we could not confirm this assumption due to the lack of information on the clinical stage of the subjects in the previous two studies. Nevertheless, our results, a beneficial marker of ENO1 only for early breast cancer patients, were mutually verified at a protein level in our cohort and an mRNA level in the TCGA-BRCA cohort.
As previously mentioned, given the close relationship between ENO1 and tumor immunity, we performed immune analyses (ssGSEA and CIBERSORT) to explore differences in the immune infiltration between the high- and low-ENO1 expression groups in patients with different stages. CIBERSORT analysis showed that ENO1 expression was positively correlated with infiltration of M1 macrophages and negatively correlated with infiltration of resting dendritic cells and M2 macrophages in stage I/II patients, while these correlations disappeared in stage III patients. Resting dendritic cells can down-regulate T cell immune response by upregulating immune checkpoints including PD-L1 and CTLA-4,29,30 and M2 macrophages can produce immunosuppressive cytokines, both of which possess pro-tumor activity, whereas M1 macrophages have antitumor activity.31,32 Furthermore, ssGSEA analysis also showed that some cells that play an important role in antitumor immune response, such as B cells,33 CD8 T cells,34 Th2 cells,35 and NK cells,36 were positively associated with ENO1 only in early-stage patients. These results were supported by previous findings that ENO1, as a tumor antigen, can exert an antitumor effect by eliciting integrated humoral and cellular immune responses and forming a specific tumor immune microenvironment.14,37,38 Interestingly, some antigenic peptide presentation-related molecules,39,40 such as HLA and MHC class I, were shown in our study to be positively associated with ENO1 only in early-stage patients. Thus, in patients with early-stage breast cancer, a high expression of ENO1 was associated with a favorable prognosis, which may be mediated by activating an antitumor immune response and affecting immune infiltration. However, with the progression of breast tumors, tumor immunity is inhibited by multiple mechanisms, such as the upregulation of immune checkpoints,41 and the downregulation of antigenic peptide presentation-related molecules,42 especially in patients with advanced-stage breast cancer,43,44 such that ENO1 alone is unable to mobilize antitumor immunity and may need to be combined with immune checkpoint inhibitors.45,46
We found that a higher ENO1 level was associated with some of the traditionally more aggressive characteristics of breast cancer, such as a higher histological grade, ER and PR negativity, and HER2 positivity, which was consistent with the results of other studies.47–49 However, this finding seemed contradictory to our subsequent result that ENO1 expression exerted a protective effect on survival. One possible reason is that these clinicopathological features (ER, PR, and HER2) have a limited impact on prognosis. ER-negative tumors have a worse prognosis than ER-positive tumors, due in large part to the latter receiving hormonal therapy. Previous studies have shown that ER was not an independent prognostic factor, but more a predictor of endocrine therapy.50,51 In addition, a recent study showed that HER2 positivity did not show a negative prognostic impact in the era of trastuzumab, a drug that targets HER2.52 The other possible reason might be that patients with a higher ENO1 expression level were more likely to be in the early stage of breast cancer, and clinical stage played a greater impact on the prognosis of breast cancer than ER, PR, and HER2.53,54
There were some limitations in this study. First, only patients with tumors >1 cm were included, which may lead to selective bias. However, ENO1 expression was independent of tumor size in this study and the selection may not affect our findings. Second, we did not collect the information on treatment. However, since the treatment was determined according to the clinicopathological characteristics, adjustment of these characteristics in the analysis was able to largely control the confounding effects of the treatment. Finally, we assessed the association of ENO1 expression with immune infiltration using the bioinformatics methods only, which provided initial clues to the role of ENO1 in breast tumor immunity, and further experimental validation is still needed.
Conclusions
In summary, we found that a high expression of ENO1 was associated with a favorable prognosis in patients with early-stage breast cancer but not in advanced-stage patients; there was a similar differential association between ENO1 expression and the infiltration of immune cells by clinical stages. These findings provided an insight into the roles of ENO1 as a prognostic marker associated with immune infiltration and suggested that induction of ENO1-specific antitumor immunity may represent a novel strategy for precision immunotherapy in breast cancer.
Data Sharing Statement
The public datasets analyzed during the present study are available from TCGA (https://portal.gdc.cancer.gov/) data portal. The IHC data generated during and analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Statement
The public datasets analyzed during the present study are available from TCGA (https://portal.gdc.cancer.gov/) data portal, which allowed researchers to download and analyze datasets for scientific purposes free of charge and without ethical issues. The protocol of the GZBCS cohort was approved by the Ethics Committee of School of Public Health, Sun Yat-sen University. All participants provided written informed consent.
Acknowledgments
We sincerely thank the patients who participated in this study, the staff who conducted the baseline and the follow-up data collection, and the medical staff in the breast departments of the First Affiliated Hospital, and the Cancer Center of Sun Yat-sen University.
Funding
This research was supported by the National Natural Science Foundation of China (81973115) and Science and Technology Planning Project of Guangdong Province, China (2019B030316002). The founders have no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin Cancer Biol. 2018;52(Pt 1):56–73. doi:10.1016/j.semcancer.2017.08.010
3. Early Breast Cancer Trialists’ Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. doi:10.1016/S0140-6736(05)66544-0
4. Byar DP, Sears ME, McGuire WL. Relationship between estrogen receptor values and clinical data in predicting the response to endocrine therapy for patients with advanced breast cancer. Eur J Cancer. 1979;15(3):299–310. doi:10.1016/0014-2964(79)90041-0
5. Patani N, Martin LA. Understanding response and resistance to oestrogen deprivation in ER-positive breast cancer. Mol Cell Endocrinol. 2014;382(1):683–694. doi:10.1016/j.mce.2013.09.038
6. Diaz-Ramos A, Roig-Borrellas A, Garcia-Melero A, Lopez-Alemany R. alpha-Enolase, a multifunctional protein: its role on pathophysiological situations. J Biomed Biotechnol. 2012;2012:156795. doi:10.1155/2012/156795
7. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi:10.1126/science.1160809
8. Niccolai E, Cappello P, Taddei A, et al. Peripheral ENO1-specific T cells mirror the intratumoral immune response and their presence is a potential prognostic factor for pancreatic adenocarcinoma. Int J Oncol. 2016;49(1):393–401. doi:10.3892/ijo.2016.3524
9. Mandili G, Curcio C, Bulfamante S, et al. In pancreatic cancer, chemotherapy increases antitumor responses to tumor-associated antigens and potentiates DNA vaccination. J Immunother Cancer. 2020;8(2):e001071. doi:10.1136/jitc-2020-001071
10. Zhang C, Zhang K, Gu J, Ge D. ENO1 promotes antitumor immunity by destabilizing PD-L1 in NSCLC. Cell Mol Immunol. 2021;18(8):2045–2047. doi:10.1038/s41423-021-00710-y
11. Liu S, Sun X, Li K, et al. Generation of the tumor-suppressive secretome from tumor cells. Theranostics. 2021;11(17):8517–8534. doi:10.7150/thno.61006
12. Li K, Sun X, Zha R, et al. Counterintuitive production of tumor-suppressive secretomes from Oct4- and c-Myc-overexpressing tumor cells and MSCs. Theranostics. 2022;12(7):3084–3103. doi:10.7150/thno.70549
13. Mojtahedi Z, Safaei A, Yousefi Z, Ghaderi A. Immunoproteomics of HER2-positive and HER2-negative breast cancer patients with positive lymph nodes. OMICS. 2011;15(6):409–418. doi:10.1089/omi.2010.0131
14. Cappello P, Principe M, Bulfamante S, Novelli F. Alpha-Enolase (ENO1), a potential target in novel immunotherapies. Front Biosci. 2017;22:944–959. doi:10.2741/4526
15. Cappello P, Rolla S, Chiarle R, et al. Vaccination with ENO1 DNA prolongs survival of genetically engineered mice with pancreatic cancer. Gastroenterology. 2013;144(5):1098–1106. doi:10.1053/j.gastro.2013.01.020
16. Qiao H, Wang Y, Zhu B, et al. Enolase1 overexpression regulates the growth of gastric cancer cells and predicts poor survival. J Cell Biochem. 2019;120(11):18714–18723. doi:10.1002/jcb.29179
17. Cheng Z, Shao X, Xu M, Zhou C, Wang J. ENO1 acts as a prognostic biomarker candidate and promotes tumor growth and migration ability through the regulation of Rab1A in colorectal cancer. Cancer Manag Res. 2019;11:9969–9978. doi:10.2147/CMAR.S226429
18. Zhu W, Li H, Yu Y, et al. Enolase-1 serves as a biomarker of diagnosis and prognosis in hepatocellular carcinoma patients. Cancer Manag Res. 2018;10:5735–5745. doi:10.2147/CMAR.S182183
19. Lomnytska MI, Becker S, Gemoll T, et al. Impact of genomic stability on protein expression in endometrioid endometrial cancer. Br J Cancer. 2012;106(7):1297–1305. doi:10.1038/bjc.2012.67
20. Chang YS, Wu W, Walsh G, Hong WK, Mao L. Enolase-alpha is frequently down-regulated in non-small cell lung cancer and predicts aggressive biological behavior. Clin Cancer Res. 2003;9(10 Pt 1):3641–3644.
21. White-Al Habeeb NM, Di Meo A, Scorilas A, et al. Alpha-enolase is a potential prognostic marker in clear cell renal cell carcinoma. Clin Exp Metastasis. 2015;32(6):531–541. doi:10.1007/s10585-015-9725-2
22. Cancemi P, Buttacavoli M, Roz E, Feo S. Expression of alpha-enolase (ENO1), myc promoter-binding protein-1 (MBP-1) and matrix metalloproteinases (MMP-2 and MMP-9) reflect the nature and aggressiveness of breast tumors. Int J Mol Sci. 2019;20:16. doi:10.3390/ijms20163952
23. Xu W, Yang W, Wu C, Ma X, Li H, Zheng J. Enolase 1 correlated with cancer progression and immune-infiltrating in multiple cancer types: a pan-cancer analysis. Front Oncol. 2020;10:593706. doi:10.3389/fonc.2020.593706
24. Zhou M, Chen QX, Yang YZ, et al. Prognostic value of glutaminase 1 in breast cancer depends on H3K27me3 expression and menopausal status. Virchows Archiv. 2022;480(2):259–267. doi:10.1007/s00428-021-03210-6
25. Chen QX, Yang YZ, Liang ZZ, et al. Time-varying effects of FOXA1 on breast cancer prognosis. Breast Cancer Res Treat. 2021;187(3):867–875. doi:10.1007/s10549-021-06125-7
26. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
27. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
28. Sauerbrei W, Taube SE, McShane LM, Cavenagh MM, Altman DG. Reporting recommendations for tumor marker prognostic studies (REMARK): an abridged explanation and elaboration. J Natl Cancer Inst. 2018;110(8):803–811. doi:10.1093/jnci/djy088
29. Probst HC, McCoy K, Okazaki T, Honjo T, van den Broek M. Resting dendritic cells induce peripheral CD8+ T cell tolerance through PD-1 and CTLA-4. Nat Immunol. 2005;6(3):280–286. doi:10.1038/ni1165
30. Adema GJ. Dendritic cells from bench to bedside and back. Immunol Lett. 2009;122(2):128–130. doi:10.1016/j.imlet.2008.11.017
31. Gunassekaran GR, Poongkavithai Vadevoo SM, Baek MC, Lee B. M1 macrophage exosomes engineered to foster M1 polarization and target the IL-4 receptor inhibit tumor growth by reprogramming tumor-associated macrophages into M1-like macrophages. Biomaterials. 2021;278:121137. doi:10.1016/j.biomaterials.2021.121137
32. Laria A, Lurati A, Marrazza M, Mazzocchi D, Re KA, Scarpellini M. The macrophages in rheumatic diseases. J Inflamm Res. 2016;9:1–11. doi:10.2147/JIR.S82320
33. Hollern DP, Xu N, Thennavan A, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 2019;179(5):1191–1206 e1121. doi:10.1016/j.cell.2019.10.028
34. Ali HR, Provenzano E, Dawson SJ, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25(8):1536–1543. doi:10.1093/annonc/mdu191
35. Schreiber S, Hammers CM, Kaasch AJ, Schraven B, Dudeck A, Kahlfuss S. Metabolic interdependency of Th2 cell-mediated type 2 immunity and the tumor microenvironment. Front Immunol. 2021;12:632581. doi:10.3389/fimmu.2021.632581
36. Shimasaki N, Jain A, Campana D. NK cells for cancer immunotherapy. Nat Rev Drug Discov. 2020;19(3):200–218. doi:10.1038/s41573-019-0052-1
37. Cappello P, Tomaino B, Chiarle R, et al. An integrated humoral and cellular response is elicited in pancreatic cancer by alpha-enolase, a novel pancreatic ductal adenocarcinoma-associated antigen. Int J Cancer. 2009;125(3):639–648. doi:10.1002/ijc.24355
38. Cook K, Daniels I, Symonds P, et al. Citrullinated alpha-enolase is an effective target for anti-cancer immunity. Oncoimmunology. 2018;7(2):e1390642. doi:10.1080/2162402X.2017.1390642
39. Chong C, Coukos G, Bassani-Sternberg M. Identification of tumor antigens with immunopeptidomics. Nat Biotechnol. 2022;40(2):175–188. doi:10.1038/s41587-021-01038-8
40. Yoshihama S, Vijayan S, Sidiq T, Kobayashi KS. NLRC5/CITA: a key player in cancer immune surveillance. Trends Cancer. 2017;3(1):28–38. doi:10.1016/j.trecan.2016.12.003
41. Gil Del Alcazar CR, Huh SJ, Ekram MB, et al. Immune escape in breast cancer during in situ to invasive carcinoma transition. Cancer Discov. 2017;7(10):1098–1115. doi:10.1158/2159-8290.CD-17-0222
42. Liu Y, Komohara Y, Domenick N, et al. Expression of antigen processing and presenting molecules in brain metastasis of breast cancer. Cancer Immunol Immunother. 2012;61(6):789–801. doi:10.1007/s00262-011-1137-9
43. Gil Del Alcazar CR, Aleckovic M, Polyak K. Immune escape during breast tumor progression. Cancer Immunol Res. 2020;8(4):422–427. doi:10.1158/2326-6066.CIR-19-0786
44. Khan M, Arooj S, Wang H. Soluble B7-CD28 family inhibitory immune checkpoint proteins and anti-cancer immunotherapy. Front Immunol. 2021;12:651634. doi:10.3389/fimmu.2021.651634
45. Lentz RW, Colton MD, Mitra SS, Messersmith WA. Innate immune checkpoint inhibitors: the next breakthrough in medical oncology? Mol Cancer Ther. 2021;20(6):961–974. doi:10.1158/1535-7163.MCT-21-0041
46. Criscitiello C, Esposito A, Gelao L, et al. Immune approaches to the treatment of breast cancer, around the corner? Breast Cancer Res. 2014;16(1):204. doi:10.1186/bcr3620
47. Kulkarni YM, Suarez V, Klinke DJ. Inferring predominant pathways in cellular models of breast cancer using limited sample proteomic profiling. BMC Cancer. 2010;10:291. doi:10.1186/1471-2407-10-291
48. Zhang D, Tai LK, Wong LL, Chiu LL, Sethi SK, Koay ES. Proteomic study reveals that proteins involved in metabolic and detoxification pathways are highly expressed in HER-2/neu-positive breast cancer. Mol Cell Proteomics. 2005;4(11):1686–1696. doi:10.1074/mcp.M400221-MCP200
49. Didiasova M, Zakrzewicz D, Magdolen V, et al. STIM1/ORAI1-mediated Ca2+ influx regulates enolase-1 exteriorization. J Biol Chem. 2015;290(19):11983–11999. doi:10.1074/jbc.M114.598425
50. Silvestrini R, Daidone MG, Luisi A, et al. Biologic and clinicopathologic factors as indicators of specific relapse types in node-negative breast cancer. J Clin Oncol. 1995;13(3):697–704. doi:10.1200/JCO.1995.13.3.697
51. Bundred NJ. Prognostic and predictive factors in breast cancer. Cancer Treat Rev. 2001;27(3):137–142. doi:10.1053/ctrv.2000.0207
52. Li S, Wu J, Huang O, et al. HER2 positivity is not associated with adverse prognosis in high-risk estrogen receptor-positive early breast cancer patients treated with chemotherapy and trastuzumab. Breast. 2020;54:235–241. doi:10.1016/j.breast.2020.10.002
53. Allison KH. Prognostic and predictive parameters in breast pathology: a pathologist’s primer. Mod Pathol. 2021;34(Suppl 1):94–106. doi:10.1038/s41379-020-00704-7
54. Fei F, Zhang K, Siegal GP, Wei S. A simplified breast cancer prognostic score: comparison with the AJCC clinical prognostic staging system. Mod Pathol. 2021;34(12):2141–2147. doi:10.1038/s41379-021-00890-y
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
