Back to Journals » Breast Cancer: Targets and Therapy » Volume 14
Chromobox Family Proteins as Putative Biomarkers for Breast Cancer Management: A Preliminary Study Based on Bioinformatics Analysis and qRT-PCR Validation
Authors Tian H, Zhao T, Li Y, Sun N, Ma D, Shi Q, Zhang G, Chen Q, Zhang K, Chen C, Zhang Y, Qi X
Received 15 July 2022
Accepted for publication 8 December 2022
Published 30 December 2022 Volume 2022:14 Pages 515—535
DOI https://doi.org/10.2147/BCTT.S381856
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Pranela Rameshwar
Hao Tian,1,* Tingting Zhao,1,* Yanling Li,1,* Na Sun,1 Dandan Ma,1 Qiyun Shi,1 Guozhi Zhang,1 Qingqiu Chen,1 Kongyong Zhang,1 Ceshi Chen,2,* Yi Zhang,1 Xiaowei Qi1
1Department of Breast and Thyroid Surgery, Southwest Hospital, Army Medical University, Chongqing, People’s Republic of China; 2Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaowei Qi; Yi Zhang, Department of Breast and Thyroid Surgery, Southwest Hospital, Army Medical University, Gaotanyan Street 29, Chongqing, 400038, People’s Republic of China, Tel/Fax +86-23-68754160, Email [email protected]; [email protected]
Background: Epigenetic modification of chromatin is an important step in the regulation of gene expression. The chromobox family proteins (CBXs), as epigenetic modifier, may play a vital role in tumorigenesis and cancer progression. Herein we explored the correlation between CBXs and breast cancer (BC) via the bioinformatics approach and qRT-PCR validation.
Methods: Several databases, including GEPIA, TCGA, GEO, K-M plotter, STRING, DAVID, cBioPortal, CIBERSORT, and HPA were employed to analyze the expression levels of CBXs and the correlations between CBXs and prognosis (overall and recurrence-free survival) in BC. We analyzed molecular functions, genetic variations, transcription factors of CBXs, and immune cell infiltration status. ROC curve analysis was performed to determine the predictive value of CBXs. RNA extracted from 11 human BC and paired adjacent normal tissues were subjected to qRT-PCR.
Results: The mRNA expression level of CBX1– 5 was significantly upregulated, while that of CBX7 was significantly downregulated in BC; no expression disparities were observed in CBX6/8 expression. Further, high mRNA expression of CBX1/2/3/4/8 correlated with advanced BC, whereas high mRNA expression of CBX6/7 correlated with early BC. High mRNA expressions of CBX1/2/3/5 predict poor OS and RFS, while higher mRNA expressions of CBX6/7 predict better OS and RFS in patients with BC. ROC curve analysis revealed that CBX3 showed excellent discriminatory ability. Gene ontology enrichment analysis showed that CBXs primarily participated in SUMOylation and post-/transcriptional regulation. Moreover, they presented varying degrees of amplification in BC tissues and were related to the infiltration of various immune cells.
Conclusion: CBXs can serve as putative biomarkers for BC. Further studies are warranted to determine the exact molecular mechanisms underlying the action of CBXs in BC, particularly CBX1/2/3/5/7.
Keywords: chromobox family proteins, breast cancer, prognosis, bioinformatics, qRT-PCR
Introduction
Breast cancer (BC) is an increasingly common malignancy among women across the world. It shows the second highest mortality rate after lung cancer,1 and recently, its incidence was reported to show a substantial increase.2 Considering the high invasiveness and heterogeneity of BC, outcomes in case of many patients who undergo conventional treatment are not always satisfactory. Moreover, high-risk patients often experience recurrence or metastases post-treatment. There is a dearth of efficient prognostic markers to guide clinical practice, including treatment priority/decision, and prognosis prediction. Fortunately, various molecular assays and advanced sequencing technologies that are widely employed in clinical practice can help resolve, to a certain extent, treatment dilemmas related to tumor heterogeneity.
As an important part of epigenetic regulation, chromatin remodeling largely determines gene transcription, which accounts for the diversity and functional specificity of cells, organs, living organisms, as well as tumor formation and evolution.3 Chromobox family proteins (CBX2/4/6/7/8) are core components of the polycomb repressor complex 1 (PRC1), which is involved in epigenetic regulation and several biological processes, including embryonic stem cell differentiation and self-renewal.4 Besides, CBXs (CBX5/HP1α, CBX1/HP1β, and CBX3/HP1γ) constitute the heterochromatin protein 1 (HP1) family, which is also involved in the process of post-transcriptional regulation, similar to PRC1.5 CBXs act as methyl readers in PRC1 and HP1 and recognize the histone methyl marks H3K27me3 and H3K9m2/3 (Figure 1). Subsequently, chromatin compaction and target gene silencing are induced.6 The biological functions of CBXs include stem cell self-renewal, tissue development, cell cycle regulation, cell proliferation, and senescence. Notably, the aberrant expression of CBXs is closely related to various cancers.7
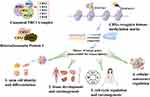 |
Figure 1 Biological functions of CBXs and involvement in carcinogenesis. |
CBX family proteins have attracted extensive attention as they play a key role in tumorigenesis and cancer progression.4,8–10 At present, eight CBXs (CBX1–8) have been identified in the human genome, and their main functions are related to heterochromatin, gene expression, apoptosis, and developmental regulation.7 CBX4 is involved in regulating hypoxia-inducible factor-1-alpha expression and is an important regulator of tumor angiogenesis.11 Further, AKT-related pathways and the tumor suppressor protein P16 maintain the stemness of gastric cancer cells by regulating the function of CBX7.12 However, in clear cell renal cell carcinoma, CBX7 is involved in TRAIL pathway-induced apoptosis and is correlated with prognosis.13 Besides, CBX8 overexpression in hepatocellular carcinoma has been reported to be associated with poor overall survival (OS).14 Epigenetic regulation is known to play a key role in the occurrence and development of BC, but the functions of CBXs in BC have been rarely reported.15 Mao et al recently explored the correlations between all CBXs and BC.16 While all of these studies are lack of any experimental validation. Therefore, it is of high clinical significance to elucidate this correlation to identify novel and efficient biomarkers for BC.
Herein we used several databases, such as Gene Expression Profiling Interactive Analysis (GEPIA), The Cancer Genome Atlas (TCGA), Kaplan–Meier Plotter, Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), Database for Annotation, Visualization, and Integrated Discovery (DAVID), cBioPortal, and cell-type identification by estimating relative subsets of RNA transcript (CIBERSORT), to investigate the expression levels of CBXs in BC and assessed their diagnostic/prognostic value, molecular functions, genetic variations, and immune cell infiltration status. Clinical specimens were assessed to ensure robust data validation after performing comprehensive in silico analyses. Our objective was to assess the potential function of CBXs in BC to assess their prognostic value and to identify novel biomarkers/strategies for BC management.
Materials and Methods
GEPIA, TCGA and GEO Database
The Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/) contains RNA sequencing expression data of 9736 tumors and 8587 normal samples from The Cancer Genome Atlas (TCGA) and Genotype–Tissue Expression (GTEx) projects.17 Herein we used GEPIA to analyze differences in the mRNA expression levels of CBXs in normal (n = 291) and BC tissues (n = 1085) and to assess the relationship between CBXs and clinicopathological grades. The significance threshold was set to log2FC > 0.5 and p < 0.05. Considering data heterogeneity, another two microarray datasets, GSE65194 (153 BC and 11 normal tissues) and GSE42568 (104 BC and 17 normal tissues), from the GEO database were selected for validation. GEO is a public functional genomics data repository supporting MIAME-compliant data submissions. (https://www.ncbi.nlm.nih.gov/geo/). Correlation results were visualized using the R package “ggpubr”. TCGA was employed for survival analysis in immune cell setting.
Kaplan–Meier Plotter
Kaplan–Meier plotter (K-Mplotter) (http://kmplot.com/analysis/) is an online database that can be used to assess the effect of 54,000 genes on survival in 21 different types of cancers.18 We herein utilized the GeneChip of Kaplan–Meier plotter to analyze the prognostic value of CBXs on the OS (n = 1402) and recurrence-free survival (RFS, n = 3955) of patients with BC. Subsequently, we explored the prognostic value of CBXs on OS (n=2976) using the RNAseq dataset. The median value of gene expression was used as the grouping basis, and p < 0.05 indicated statistical significance.
STRING & DAVID
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (https://string-db.org/) is commonly used for analyzing protein interactions.19 In this study, we employed STRING for protein–protein interaction (PPI) analyses involving eight CBXs and 10 proteins interacting with CBXs, and medium confidence (0.4) was selected. The results were visualized using Cytoscape v3.7.2. To further explore the function of pertinent genes, the results were uploaded to Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) for gene ontology (GO) enrichment analysis.20 False discovery rate < 0.05 indicated statistical significance.
cBioPortal and the Signaling Pathways Project
cBioPortal (www.cbioportal.org) provides a web resource to explore, analyze, and visualize multidimensional cancer genomics data.21 We used cBioPortal to analyze genetic variations in CBXs in patients with BC, and visualization was achieved by combining heat maps depicting CBX expression. SPP (The Signaling Pathways Project) is a multi-omics knowledge mine based upon public, manually curated transcriptomic and cistromic (ChIP-Seq) datasets involving genetic and small molecule manipulations of cellular receptors, enzymes, and transcription factors. It was employed by using the package “ggalluvial” of R software to investigate transcription factors of CBXs.
CIBERSORT
CIBERSORT is a deconvolution method that characterizes the cell composition of complex tissue from their gene expression profiles.22 Herein we employed CIBERSORT to analysis the differences of immune cell infiltration in BC. More than half of the immune cells with zero abundance in the sample were removed (https://cibersortx.stanford.edu/), and R package “ggplot2” was used for plotting in combination with TCGA data.
The Human Protein Atlas (HPA) Database
The HPA database (www.proteinatlas.org) provides free access to immunohistochemistry results for many cancer tissues. Depending on the intensity of staining and percentage of stained cells, the immunohistochemistry score is primarily classified into strong, medium, weak, and negative.23 In this study, protein expression levels of CBXs in BC and adjacent normal tissues were analyzed using the “Pathology Atlas” and “Tissue Atlas” modules on www.proteinatlas.org.
Receiver Operating Characteristic (ROC) Curve Analysis
CBX1–8 expression levels from TCGA were discriminated by ROC curve analysis. TCGA BC dataset, containing information for 1104 BC and 113 normal tissues, was downloaded from UCSC (https://xenabrowser.net/datapages/), and ROC curves were plotted using the R package “pROC.”
Specimens for Experimental Validation, RNA Extraction, and Quantitative Reverse-Transcriptase Polymerase Chain Reaction (qRT-PCR)
All 11 BC and paired adjacent normal tissue samples were stored at −80°C immediately after resection (Table 1). Total RNA was extracted using TRIzol (ServiceBio, Wuhan, China). Approximately 1 μg total RNA was then reverse transcribed into cDNA using PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time), according to manufacturer instructions (Takara, Dalian, China). qRT-PCR was performed using TB Green™ Premix Ex Taq II (Takara) on a CFX Connect™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). ACTB (forward: CTCCTACCTGGCCTCGCTGT, reverse: GCTGTCACCTTCACCGTTCC) served as the internal control. Data were analyzed using the comparative Ct method (ΔΔCt). Supplementary Table S lists all sense/antisense primers used in this study. This study was approved by the Human Ethics Committee of the First Affiliated Hospital of Army Medical University.
 |
Table 1 Clinicopathological Characteristics of Patients Whose Primary Tumor Specimens Were Subjected to qRT-PCR |
Statistical Analysis
An unpaired t-test was used to analyze differences in the transcription level in BC. Survival analysis was performed using Log rank test or multivariate cox regression analysis. qRT-PCR data were analyzed with Wilcoxon signed-rank test. R programming language was used for statistical analysis and plotting charts and graphs. p < 0.05 indicated statistical significance.
Results
Expression of CBXs Considerably Varies Between BC and Adjacent Normal Tissues
The expression levels of CBX1–5 and CBX8 were significantly upregulated in BC tissues (Figure 2, p < 0.05). In contrast, the CBX7 expression level was significantly downregulated in BC tissues (p < 0.05). Further, CBX6 expression levels did not significantly differ between normal and BC tissues.
 |
Figure 2 Gene expression level of CBXs in BC and normal tissues (GEPIA). |
Similarly, in GSE65194 and GSE42568, the expression level of most CBXs was comparable with that in the GEPIA database (Figure 3). The expression level of CBX1–5 was significantly upregulated (p < 0.001), and that of CBX7 was significantly downregulated in BC tissues in both the datasets (p1 < 0.001 and p2 < 0.05); these data were in accordance with those in GEPIA. In contrast, CBX6 expression was significantly upregulated in BC tissues in GSE42568 (p < 0.05) but downregulated in GSE65194 (p < 0.01), while CBX8 expression was upregulated in BC tissues in GSE42568 but showed no significant differences in GSE65194. CBXs expressions were furtherly evaluated in PAM50-based intrinsic subtype of BC. All expression signatures were consistent with the above results, except that there is no difference in the expression of CBX5 in BC and normal tissues (Figure 4).
 |
Figure 3 mRNA expression levels of CBXs in BC were validated in GSE42568 (A) and GSE65194 (B) (NS > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001). |
 |
Figure 4 mRNA expression levels of CBXs in PAM-50 based intrinsic subtype. |
Protein expression levels of CBXs in BC were evaluated using the HPA database (Figure 5). The protein expression levels of CBX3/4/5 in BC tissues were found to be higher than in adjacent normal tissues, and those of CBX2/7 in adjacent normal tissues were higher than in BC tissues. However, the protein expression levels of CBX1/8 did not show any differences between BC and adjacent normal tissues. CBX6 expression was not detected in either tissue specimen.
 |
Figure 5 Immunohistochemical staining of CBX protein in BC and normal breast tissues summarized from the HPA database. Abbreviations: ND, not detected; Ab, antibody. |
Experimental Validation of the Expression of CBXs
Based on qRT-PCR results, BC tissues showed a significant upregulation of CBX3 (p < 0.01) and CBX5 (p < 0.01) expression levels, while CBX7 (p < 0.01) expression level was downregulated. No significant differences were found in the expression level of CBX1/2/8 between BC and adjacent normal tissues (Figure 6). Unfortunately, qRT-PCR did not generate reliable expression data for CBX4 and CBX6 (Ct values were too high, >40), despite repeating the experiment with different primers. This could be primarily attributed to insufficient sample size or low-quality specimens, given that little credible data are available. Table 2 summarizes the mRNA/protein expression levels of CBXs in BC and adjacent normal tissues.
 |
Table 2 CBX mRNA/Protein Expression Levels in BC and Normal Tissues |
 |
Figure 6 mRNA expression levels of CBX1–8, as determined by qRT-PCR. |
Relationship Between CBX Expression Levels and Clinicopathological Grade
According to GEPIA, the expression levels of eight CBXs were correlated with clinicopathological grade (Figure 7). Among them, the expression levels of CBX1 (P = 0.034) and CBX2 (p < 0.001) were positively correlated with clinicopathological grade and those of CBX4 (P = 0.445) and CBX8 (P = 0.066) tended to increase with an increase in clinicopathological grade. In contrast, CBX7 (P = 0.089) expression level was negatively correlated with clinicopathological grade. However, the expression levels of CBX3 (P = 0.405), CBX5 (P = 0.531), and CBX6 (P = 0.084) were not significantly different in tumor tissues of different clinicopathological grades. To be further aligned with clinical practice, we further classified the cases into high- and low-stage cohorts (Figure 8). The expression levels of CBX1/2 (p < 0.05) were higher in advanced BC (high stage), which is consistent with previous results, and those of CBX3/4/8 (p < 0.05) were higher in advanced BC, which contradicts previous results. Further, the expression levels of CBX6/7 (p < 0.05) were higher in early BC (low stage).
 |
Figure 7 Relationship between mRNA expression of CBXs and clinicopathological grade in BC (GEPIA). |
 |
Figure 8 Relationship between mRNA expression of CBXs and clinicopathological grade in BC (high/low stage). |
Relationship Between CBXs and OS
To determine the prognostic value of CBXs, Kaplan–Meier plotter was used to perform survival analysis (genechip) (Figure 9). The highly expressed CBX2 (HR = 1.5, 95% CI = 1.1–2.06, p = 0.011), CBX3 (HR = 1.43, 95% CI = 1.15–1.77, p = 0.001), and CBX5 (HR = 1.35, 95% CI = 1.09–1.67, p = 0.006) were related to poorer OS in BC. In contrast, the highly expressed CBX4 (HR = 0.73, 95% CI = 0.53–1, p = 0.049), CBX6 (HR = 0.78, 95% CI = 0.63–0.96, p = 0.021), and CBX7 (HR = 0.55, 95% CI = 0.44–0.69, p < 0.001) were associated with better OS. CBX1 (HR = 1.04, 95% CI = 0.84–1.29, p = 0.690) or CBX8 (HR = 1.05, 95% CI = 0.85–1.3, p = 0.670) was not associated with OS.
 |
Figure 9 Relationship between mRNA expression of CBXs and OS in BC (KM plotter, cut-off = 50%). |
However, different results were obtained when using RNAseq data (Figure 10). High expressions of CBX1 and CBX2 were related to poor OS, respectively, while high expressions of CBX3, CBX5, and CBX6 indicated better OS in BC patients. CBX4, CBX7, and CBX8 were not related to OS in BC patients. However, multivariate cox regression analysis indicated that all CBXs were not independent prognostic factors in BC patients (Figure 11).
 |
Figure 10 Relationship between mRNA expression of CBXs and OS in BC (RNAseq) (KM plotter, cut-off = 50%). |
 |
Figure 11 Multivariate cox regression analysis of CBXs and OS in BC. |
Relationship Between CBXs and RFS
As evident from Figure 12, the highly expressed CBX1 (HR = 1.26, 95% CI = 1.13–1.41, p < 0.001), CBX2 (HR = 1.79, 95% CI = 1.53–2.1, p < 0.001), CBX3 (HR = 1.46, 95% CI = 1.31–1.63, p < 0.001), and CBX5 (HR = 1.34, 95% CI = 1.2–1.5, p < 0.001) were significantly negatively correlated with RFS. In contrast, the highly expressed CBX4 (HR = 0.69, 95% CI = 0.59–0.81, p < 0.001), CBX6 (HR = 0.73, 95% CI = 0.66–0.82, p < 0.001), and CBX7 (HR = 0.53, 95% CI = 0.47–0.59, p< 0.001) were significantly positively correlated with RFS, while CBX8 (HR = 1.04, 95% CI = 0.93–1.16, p = 0.480) showed no significant relationship with RFS. Table 3. summarizes the correlation between the expression of CBX mRNA and clinicopathological grade, OS and RFS.
 |
Table 3 Correlation Between mRNA Expression of CBXs and Clinicopathological Grade, OS, and RFS |
 |
Figure 12 Relationship between mRNA expression of CBXs and RFS in BC (KM plotter, cut-off = 50%). |
Functional Enrichment Analysis
To assess the involvement of CBXs in biological processes, we analyzed the STRING database to shortlist 10 genes that interact with CBXs, and a PPI network was constructed (Figure 13A). DAVID was then utilized for GO enrichment analysis of the aforementioned 18 genes (Figure 13B). Consequently, it became evident that CBXs were principally involved in protein SUMOylation, covalent chromatin modification, negative regulation of transcription from RNA polymerase II promoter, DNA-templated transcriptional regulation, histone H2A monoubiquitination, and DNA-templated negative transcriptional regulation. False discovery rate < 0.05 indicated statistical significance.
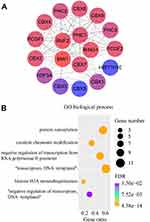 |
Figure 13 Functional analysis of CBXs: the PPI network of CBXs and interacting proteins (A): GO enrichment analysis, (B): STRING and DAVID databases. |
Genetic Variations and Transcription Factors of CBXs
In general, >2 gene mutations were detected in BC, and amplification was the most common form (Figure 14). Further, 292 CBX gene mutations were identified in 1108 patients with BC; the mutation rate was 30%. Various CBXs showed the following mutation rates: CBX1, 8%; CBX2, 10%; CBX3, 5%; CBX4, 9%; CBX5, 4%; CBX6, 4%; CBX7, 4%; and CBX8, 11%. All transcription factors of CBXs were presented in an alluvial plot (Figure 15). Notably, FOXA1, ELK1, and TCF7L2 were involved in the transcription of multiple CBXs.
 |
Figure 14 Genetic variations in CBXs in breast cancer (cBioPortal database). |
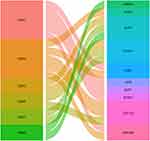 |
Figure 15 Alluvial plot of transcription factors regulating CBXs: the left column represents genes, the right represents transcription factors, and the edge represents the relationship between them. |
Correlation Between CBXs and Immune Cell Infiltration, and Immune Cell Related OS
Immune cell infiltration is closely related to tumor initiation and progression, which also guide immunotherapy in BC. In this study, we explored the correlation between CBXs and immune cell infiltration using CIBERSORT (Figures 16 and17). The expressions of CBX5/6/7 were positively related to the infiltration of B cells, and CBX2 expression was negatively related to the infiltration of B cells. The expressions of CBX7/8 were positively related to the infiltration of CD8+ T cells, and CBX1/5 expression was negatively related to the infiltration of CD8+ T cells. The expressions of CBX5/6/7 were positively related to the infiltration of CD4+ T cells, and CBX2/3/8 expressions were negatively related to the infiltration of CD4+ T cells. The expressions of CBX2/4/8 were positively related to the infiltration of Tregs, and CBX5/7 expressions were negatively related to the infiltration of Tregs cells.
 |
Figure 16 Correlation between CBXs and immune cell infiltration: B cells, CD4+/CD8+ T cells and Tregs. |
 |
Figure 17 Correlation between CBXs and immune cell infiltration: M1/M2 macrophages, NK cells and Dendritic cells. |
The expressions of CBX1/2/3/8 were positively related to the infiltration of M1 macrophages, and CBX6/7 expressions were negatively related to the infiltration of M1 macrophages. The expressions of CBX7 were positively related to the infiltration of M2 macrophages, and CBX1/2/3/4/8 expressions were negatively related to the infiltration of M2 macrophages. The expressions of CBX2/7/8 were positively related to the infiltration of NK cells, and CBX1/3/5 expressions were negatively related to the infiltration of NK cells. Dendritic cells account for a smaller part of immune cells. The expressions of CBX1/2/3 were positively related to the infiltration of dendritic cells, and CBX4/6/7 expressions were negatively related to the infiltration of dendritic cells. Survival analysis showed that M2 macrophages and dendritic cells in BC correlate with poor prognosis. While CD8+ T cells in BC indicated better OS (Figure 18).
 |
Figure 18 Relationship between infiltrating immune cell count and OS in BC (KM plotter, cut-off = 50%). |
Diagnostic Ability of CBXs in BC
ROC curve analysis was performed according to mRNA expression of CBXs in BC and normal tissues (obtained from TCGA). CBX1 (AUC = 0.783, p < 0.001), CBX2 (AUC = 0.790, p < 0.001), CBX3 (AUC = 0.939, p < 0.001), CBX4 (AUC = 0.924, p < 0.001), and CBX8 (AUC = 0.892, p < 0.001), particularly CBX3/4/8, showed good predictive value. Furthermore, the combination of all CBXs (AUC = 0.993, p < 0.001) was associated with an excellent predictive value (Figure 19).
 |
Figure 19 Sensitivity and specificity for ROC curve analysis of CBXs in predicting a diagnostic value in patients with BC. Abbreviation: AUC, area under the curve. |
Discussion
As the most common malignancy in female around the world, BC is associated with high invasiveness and heterogeneity, hence treatment strategies and clinical outcomes considerably vary depending on different subtypes. In addition to transcriptome abnormalities, epigenetic dysregulation plays a vital role in BC occurrence and progression.24 As core components of PRC1 and HP1, CBXs are one of the main executors of epigenetic regulation in vivo.7 In this study, we systematically explored the correlation between CBX1–8 and BC, and the possibility of CBXs serving as diagnostic and prognosis markers for BC is also estimated.
Functional enrichment analyses indicated that CBXs were involved in protein SUMOylation, covalent chromatin modification, negative regulation of transcription from RNA polymerase II promoter, DNA-templated negative/transcriptional regulation, and histone H2A monoubiquitination. Besides being a member of PRC1, human CBX4 presents the small ubiquitin-related modifier (SUMO) E3 ligase activity, owing to its structural properties.25 Moreover, Li et al reported that CBX4 contributes to the angiogenesis of hepatocellular carcinoma by regulating HIF-1 transactivation with its SUMO E3 ligase activity.11 Other functions, such as chromatin modification and transcriptional regulation, are well understood as CBXs are critical components of PRC1 (CBX2/4/6/7/8) as well as HP1 (CBX1/3/5), whose main function is to mediate epigenetic regulation.6,7 What interests us the most is whether the SUMOylation of CBX4 and other CBXs plays a role in BC tumorigenesis and progression. Further studies on this topic are warranted.
In the present study, multidataset-based bioinformatics analyses provided us with consistent results, that there are significant differences in CBXs expression between BC and adjacent normal tissues. With regard to mRNA expression of CBXs in BC and adjacent normal tissues, different datasets/approaches showed acceptable concordance. And the results were furtherly confirmed by human specimen-based qRT-PCR. Protein expression level were almost consistent with transcriptional level data of CBXs. However, CBXs protein expressions in this study used as a reference to some extent since we obtained them from HPA instead of human specimen-based experiments like Western blotting. The mRNA expression levels of CBX6/8 in BC and adjacent normal tissues remain unknown because our analysis showed considerably different trends (Table 2).
In silico analysis results indicated that the mRNA expression levels of CBX1/2/3/4/8 were significantly higher in advanced BC, while those of CBX6/7 were significantly higher in early BC. No difference in the mRNA expression of CBX5 was observed between advanced and early BC. Similar trends were observed in the correlations between CBX and OS/RFS. When the mRNA expression levels of CBX2/3/5 and CBX4/6/7 were markedly upregulated, patients with BC showed shorter and longer OS and RFS, respectively.
As for CBX4/6/8, some studies have suggested their role in various cancers, including BC. Jiang et al reported that CBX4 transcriptionally suppresses the tumor suppressor KLF6 by interacting with HDAC1 to exert oncogenic activities in clear cell renal cell carcinoma.26 In BC, however, the analysis revealed that both CBX4 mRNA and protein were highly expressed in advanced BC, and high expression of CBX4 mRNA was related to better OS and RFS. Obviously, it is inconsistent with the biological behavior of BC that advanced BC is usually related to poor OS or RFS. Further, as per two previous studies, CBX8 plays a pivotal role in BC; one of them reported that CBX8 acts non-canonically with WDR5 to promote mammary tumorigenesis.27,28 In this study, however, CBX8 were neither significantly differentially expressed in BC and adjacent normal tissues nor related to prognosis in patients with BC. Further, CBX6 overexpression evidently promotes the growth of hepatocellular carcinoma cells and accelerates the progression of hepatocellular carcinoma by regulating the S100A9/NF-κB/MAPK pathway.29 In this study, analysis about CBX6 presented with much bias, and besides, we did not obtain reliable data for the expression of both CBX4/6 mRNA in BC. Altogether, based on our comprehensive results (Table 2), CBX1/2/3/5/7 appears to be novel prognostic biomarkers for BC.
CBXs are closely related to various cancers. A study reported CBX2 mRNA and protein levels to be significantly higher in gastric cancer tissues than in normal tissues.30 CBX2 regulates proliferation and apoptosis via YAP phosphorylation in hepatocellular carcinoma.31 Many other studies have elucidated the oncogenic role of CBX2 and reported robust data.32,33 Mohammad et al reported that CBX2 participates in metabolic reprogramming, which ultimately promotes BC growth; CBX7 was also reported to be involved in metabolic reprogramming, and compared to CBX2, it plays an antagonistic role in aerobic glycolysis in BC.34 In this study, even we obtained contradictory expression data for CBX2 and CBX7 in BC. The differential expression of CBX2 mRNA and its prognostic value in BC were identified on analyzing multiple datasets.
Similar to CBX2, CBX3 also regulates glycolysis by inhibiting fructose-1, 6-bisphosphatase 1, and eventually promotes the proliferation of pancreatic cancer cells.35 A study reported CBX3 expression level to be significantly upregulated in brain glioma tissues; further, CBX3 stimulated the growth of glioma U87 cells by targeting CDKN1A, leading to unfavorable prognosis.36 Similar biological effects were reported for hepatocellular carcinoma.37 Yu et al38 found that CBX5 promoted the proliferation and metastasis of lung cancer cells, leading to poor prognosis in patients with lung cancer.
CBX7 is a unique member of the CBX family acting as a tumor suppressor in most situations, including in cases of BC.39,40 In ovarian cancer, CBX7 inhibits the function of TWIST-1 by combining with E-box.41 Moreover, it inhibits pancreatic cancer progression by negatively regulating the PTEN/AKT signaling pathway;42 CBX7 also reportedly suppresses urinary bladder cancer progression.43 Kim et al found that CBX7 can serve as a BC suppressor because it inhibits BC tumorigenesis by mediating the suppression of the Wnt/β-catenin pathway.44 As a possible suppressor, the effects of CBX7 mutation on BC occurrence and progression remain to be systematically reported. Interestingly, the transcription of two CBXs (CBX5/7) are regulated by transcription factor FOXA1. A recent study by Scaltriti and their colleagues indicated that FOXA1 mutations perturb its function to dictate cancer progression and lower response to aromatase inhibitors in BC.45 It’s reasonable to infer that CBX7 may be one of the executors of this process.
ROC curve analysis indicated that CBX1/2/3/4/8 presented promising predictive values. The calculated AUC was 0.993 (p < 0.001) when all CBXs were included. However, considering cost and feasibility, it is hard to expand all or combined CBXs to clinical practice. CBX3 (AUC = 0.939) was the most efficient at discriminating between BC and normal tissues; thus, it can serve as an independent diagnostic biomarker.
CBXs were found to be involved in cell differentiation, tissue development, cell cycle regulation, and cell differentiation. Therefore, CBX mutations are more likely to impact cell/organ development and cancer induction. Unfortunately, studies on the correlation between CBX mutations and cancer development are limited. CBX7 knockout mice were found to be more prone to liver and lung cancer.46,47
Previous survival analysis revealed that expressions of CBX1/2/3/4/8.
The tumor microenvironment is essential for tumor progression and recurrence. Various cues in the environment support therapeutic decision-making, particularly in the case of immunotherapy. Immune cells in the tumor microenvironment promote or inhibit tumor activity, and they are generally considered to be important determinants of clinical outcomes and immunotherapy response.48 CBXs in BC is related to immune cell infiltration including B cells, CD4/8+ T cell, Tregs, macrophage, NK cells and Dendritic cell.
M1 and M2 macrophages are widely recognized as anti-cancer and pro-cancer cells respectively.49 In this study, CBX1/2/3 is highly expressed in BC and related to poor outcomes, and survival analysis revealed that high M2 macrophage or dendritic cell count predicts poor OS in BC. However, CBX1/2/3 is negatively related to M2 macrophage count and positively related to M1 macrophage count. The same phenomenon has been observed in CBX7. On the contrary, High dendritic cell count indicated poor OS in BC, and CBX1/2/3 is positively related to dendritic cell count. CBXs in BC may involve in a complex immune process and cannot be directly summarized and correlated.
The role of CBX7 in inhibiting tumorigenesis of BC has been detailed. Therefore, CBX-targeted therapy appears promising to treat BC. In fact, Simhadri et al have already reported the first chromodomain antagonists that target CBX7.50 Moreover, Wang et al proposed pyrvinium pamoate to be an indirect inhibitor of CBX4, to inhibit osteosarcoma metastasis via the CK1α/CBX4 axis.51 It’s important to note that CBXs cannot help to choose the preferred regimens for patients among chemotherapy, anti-HER2 therapy, endocrine therapy and immunotherapy, since no reliable data is available.
Conclusions
To summarize, we herein conducted comprehensive bioinformatics analyses and qRT-PCR validation to assess the correlation between CBXs and BC. Our data revealed differential expression of CBX mRNAs between BC and adjacent normal tissues. CBX1/2/3/5/7 were significantly related to prognosis and thus can serve as potential prognostic biomarkers for BC. CBX3 can also serve as a diagnostic marker. Moreover, most CBXs are related with immune cell infiltration. Our results provide a framework for further in-depth research.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information Files).
Ethics Approval and Consent to Participate
The trial protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of the First Affiliated Hospital of Army Medical University (No: KY2020055). Written informed consent was obtained from individual participants, including consent to have identifying details (containing age) published.
Acknowledgments
We thank all the patients who have kindly given their time to participate in our research.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Project of National Key Clinical Specialty Construction (413F1Z113), Chongqing Basic Research and Frontier Exploration Project (No. cstc2018jcyjA0137), and the Military Medical Staff Innovation Plan of Southwest Hospital (No. SWH2018BJLC-04) as well as Army Medical University (No. XZ-2019-505-042).
Disclosure
The authors declare that they have no competing interests.
References
1. Siegel RL, Miller KDJemal A, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi:10.3322/caac.21590
2. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
3. Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers, and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17(5):284–299. doi:10.1038/nrg.2016.13
4. Klauke K, Radulovic V, Broekhuis M, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol. 2013;15:353–362. doi:10.1038/ncb2701
5. Machida S, Takizawa Y, Ishimaru M, et al. Structural basis of heterochromatin formation by human HP1. Mol Cell. 2018;69(3):385–397.e8. doi:10.1016/j.molcel.2017.12.011
6. van Wijnen AJ, Bagheri L, Badreldin AA, et al. Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer, and development. Bone. 2021;143:115659. doi:10.1016/j.bone.2020.115659
7. Ma RG, Zhang Y, Sun TT, Cheng B. Epigenetic regulation by polycomb group complexes: focus on roles of CBX proteins. J Zhejiang Univ Sci B. 2014;15(5):412–428. doi:10.1631/jzus.B1400077
8. Chen WY, Zhang XY, Liu T, Liu Y, Zhao YSPang D, Pang D. Chromobox homolog 2 protein: a novel biomarker for predicting prognosis and Taxol sensitivity in patients with breast cancer. Oncol Lett. 2017;13:1149–1156. doi:10.3892/ol.2016.5529
9. Lee SH, Um SJ, Kim EJ. CBX8 antagonizes the effect of Sirtinol on premature senescence through the AKT-RB-E2F1 pathway in K562 leukemia cells. Biochem Biophys Res Commun. 2016;469:884–890. doi:10.1016/j.bbrc.2015.12.070
10. Pallante P, Forzati F, Federico A, Arra CFusco A, Fusco A. Polycomb protein family member CBX7 plays a critical role in cancer progression. Am J Cancer Res. 2015;5:1594–1601.
11. Li J, Xu Y, Long XD, et al. Cbx4 governs HIF-1alpha to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 2014;25:118–131. doi:10.1016/j.ccr.2013.12.008
12. Ni SJ, Zhao LQ, Wang XF, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-kappaB-miR-21 pathways. J Hematol Oncol. 2018;11:17. doi:10.1186/s13045-018-0562-z
13. Shinjo K, Yamashita Y, Yamamoto E, et al. Expression of chromobox homolog 7 (CBX7) is associated with poor prognosis in ovarian clear cell adenocarcinoma via TRAIL-induced apoptotic pathway regulation. Int J Cancer. 2014;135:308–318. doi:10.1002/ijc.28692
14. Gao SB, Sun SL, Zheng QL, et al. Genetic alteration and misexpression of Polycomb group genes in hepatocellular carcinoma. Am J Cancer Res. 2015;5:2969–2979.
15. Li X, Gou J, Li H, Yang X. Bioinformatic analysis of the expression and prognostic value of chromobox family proteins in human breast cancer. Sci Rep. 2020;10:17739. doi:10.1038/s41598-020-74792-5
16. Mao G, Zheng Y, Lin S, Ma L, Zhou Z, Zhang S. Bioinformatic analysis of prognostic value, genetic interaction, and immune infiltration of chromobox family proteins in breast cancer. Int J Gen Med. 2021;14:9181–9191. doi:10.2147/IJGM.S343948
17. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi:10.1093/nar/gkx247
18. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi:10.1007/s10549-009-0674-9
19. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi:10.1093/nar/gky1131
20. Dennis G
21. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi:10.1158/2159-8290.CD-12-0095
22. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
23. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi:10.1126/science.1260419
24. Karsli-Ceppioglu S, Dagdemir A, Judes G, et al. Epigenetic mechanisms of breast cancer: an update of the current knowledge. Epigenomics. 2014;6:651–664. doi:10.2217/epi.14.59
25. Luis NM, Morey L, Mejetta S, et al. Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell. 2011;9(3):233–246. doi:10.1016/j.stem.2011.07.013
26. Jiang N, Niu G, Pan YH, et al. CBX4 transcriptionally suppresses KLF6 via interaction with HDAC1 to exert oncogenic activities in clear cell renal cell carcinoma. EBioMedicine. 2020;53:102692. doi:10.1016/j.ebiom.2020.102692
27. Chung C-Y, Sun Z, Mullokandov G, et al. Cbx8 acts non-canonically with Wdr5 to promote mammary tumorigenesis. Cell Rep. 2016;16:472–486. doi:10.1016/j.celrep.2016.06.002
28. Shi D, Ao L, Yu H, et al. Chromobox homolog 8 (CBX8) in human tumor carcinogenesis and prognosis: a pancancer analysis using multiple databases. Front Genet. 2021;12:745277. doi:10.3389/fgene.2021.745277
29. Zheng H, Jiang WH, Tian T, et al. CBX6 overexpression contributes to tumor progression and is predictive of a poor prognosis in hepatocellular carcinoma. Oncotarget. 2017;8:18872–18884. doi:10.18632/oncotarget.14770
30. Ma T, Ma N, Chen JL, et al. Expression and prognostic value of Chromobox family members in gastric cancer. J Gastrointest Oncol. 2020;11:983–998. doi:10.21037/jgo-20-223
31. Mao J, Tian Y, Wang C, et al. CBX2 regulates proliferation and apoptosis via the phosphorylation of YAP in hepatocellular carcinoma. J Cancer. 2019;10:2706–2719. doi:10.7150/jca.31845
32. Clermont P-L, Sun L, Crea F, et al. Genotranscriptomic meta-analysis of the Polycomb gene CBX2 in human cancers: initial evidence of an oncogenic role. Br J Cancer. 2014;111:1663–1672. doi:10.1038/bjc.2014.474
33. Piqué DG, Montagna C, Greally JM, et al. A novel approach to modelling transcriptional heterogeneity identifies the oncogene candidate CBX2 in invasive breast carcinoma. Br J Cancer. 2019;120:746–753. doi:10.1038/s41416-019-0387-8
34. Iqbal MA, Siddiqui S, Ur Rehman A, et al. Multiomics integrative analysis reveals antagonistic roles of CBX2 and CBX7 in metabolic reprogramming of breast cancer. Mol Oncol. 2021;15:1450–1465. doi:10.1002/1878-0261.12894
35. Chen LY, Cheng CS, Qu C, et al. CBX3 promotes proliferation and regulates glycolysis via suppressing FBP1 in pancreatic cancer. Biochem Biophys Res Commun. 2018;500:691–697. doi:10.1016/j.bbrc.2018.04.137
36. Zhao SP, Wang F, Yang M, et al. CBX3 promotes glioma U87 cell proliferation and predicts an unfavorable prognosis. J Neurooncol. 2019;145:35–48. doi:10.1007/s11060-019-03286-w
37. Zhong X, Kan A, Zhang W, et al. CBX3/HP1gamma promotes tumor proliferation and predicts poor survival in hepatocellular carcinoma. Aging (Albany NY). 2019;11:5483–5497. doi:10.18632/aging.102132
38. Yu YH, Chiou GY, Huang PI, et al. Network biology of tumor stem-like cells identified a regulatory role of CBX5 in lung cancer. Sci Rep. 2012;2:584. doi:10.1038/srep00584
39. Forzati F, Federico A, Pallante P, et al. CBX7 is a tumor suppressor in mice and humans. J Clin Invest. 2012;122(2):612–623. doi:10.1172/JCI58620
40. Mohammad HP, Cai Y, McGarvey KM, et al. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Res. 2009;69(15):6322–6330. doi:10.1158/0008-5472.CAN-09-0065
41. Li J, Alvero AB, Nuti S, et al. CBX7 binds the E-box to inhibit TWIST-1 function and inhibit tumorigenicity and metastatic potential. Oncogene. 2020;39:3965–3979. doi:10.1038/s41388-020-1269-5
42. Ni S, Wang H, Zhu X, et al. CBX7 suppresses cell proliferation, migration, and invasion through the inhibition of PTEN/Akt signaling in pancreatic cancer. Oncotarget. 2017;8:8010–8021. doi:10.18632/oncotarget.14037
43. Huang Z, Yan Y, Zhu Z, et al. CBX7 suppresses urinary bladder cancer progression via modulating AKR1B10-ERK signaling. Cell Death Dis. 2021;12(6):537. doi:10.1038/s41419-021-03819-0
44. Kim HY, Park JH, Won HY, Lee JY, Kong G. CBX7 inhibits breast tumorigenicity through DKK-1-mediated suppression of the Wnt/beta-catenin pathway. FASEB J. 2015;29:300–313. doi:10.1096/fj.14-253997
45. Arruabarrena-Aristorena A, Maag JLV, Kittane S, et al. FOXA1 mutations reveal distinct chromatin profiles and influence therapeutic response in breast cancer. Cancer Cell. 2020;38(4):534–550. doi:10.1016/j.ccell.2020.08.003
46. Forzati F, Federico A, Pallante P, et al. CBX7 gene expression plays a negative role in adipocyte cell growth and differentiation. Biol Open. 2014;3(9):871–879. doi:10.1242/bio.20147872
47. Forzati F, Federico A, Pallante P, Fedele M, Fusco A. Tumor suppressor activity of CBX7 in lung carcinogenesis. Cell Cycle. 2012;11(10):1888–1891. doi:10.4161/cc.20022
48. Sun J, Zhang Z, Bao S, et al. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J Immunother Cancer. 2020;8:e000110. doi:10.1136/jitc-2019-000110
49. Locati M, Curtale G, Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev Pathol. 2020;15:123–147. doi:10.1146/annurev-pathmechdis-012418-012718
50. Simhadri C, Daze KD, Douglas SF, et al. Chromodomain antagonists that target the polycomb-group methyllysine reader protein chromobox homolog 7 (CBX7). J Med Chem. 2014;57(7):2874–2883. doi:10.1021/jm401487x
51. Wang X, Qin G, Liang X, et al. Targeting the CK1α/CBX4 axis for metastasis in osteosarcoma. Nat Commun. 2020;11(1):1141. doi:10.1038/s41467-020-14870-4
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
