Back to Journals » The Application of Clinical Genetics » Volume 15
Long Non-Coding RNAs ASB16-AS1 and AFAP1-AS1: Diagnostic, Prognostic Impact and Survival Analysis in Colorectal Cancer
Authors Elabd NS , Soliman SE , Elhamouly MS, Gohar SF, Elgamal A, Alabassy MM, Soliman HA, Gadallah AA, Elbahr OD, Soliman G , Saleh AA
Received 11 April 2022
Accepted for publication 18 July 2022
Published 1 August 2022 Volume 2022:15 Pages 97—109
DOI https://doi.org/10.2147/TACG.S370242
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Martin Maurer
Naglaa S Elabd,1 Shimaa E Soliman,2 Moamena S Elhamouly,1 Suzy F Gohar,3 Ayman Elgamal,4 Mahmoud Magdy Alabassy,5 Haitham A Soliman,5 Abdelnaser A Gadallah,6 Osama D Elbahr,7 Ghada Soliman,3 Amany A Saleh2
1Tropical Medicine Department, Faculty of Medicine - Menoufia University, Menoufia, Egypt; 2Medical Biochemistry and Molecular Biology Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 3Clinical Oncology Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 4Fellow of Tropical Medicine Department, Faculty of Medicine - Menoufia University, Menoufia, Egypt; 5General Medicine Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 6Internal Medicine Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 7Hepatology and Gastroenterology Department, National Liver Institute, Menoufia University, Menoufia, Egypt
Correspondence: Naglaa S Elabd, Tropical Medicine, Faculty of Medicine - Menoufia University- Egypt, Cairo, Egypt, Tel +201092304322, Email [email protected]
Background: We aimed to evaluate the diagnostic roles of AFAP1-AS1 and ASB16-AS1 in colorectal cancer and highlight their roles in predicting colorectal cancer patients’ prognosis.
Methods: In this case–control study, 146 participants were involved. Group I included 47 patients with CRC. Group II composed of 49 patients with benign lesions in the colon, and Group III included 50 apparently normal subjects of coincided age and gender as controls. All participants were subjected to clinical and endoscopic evaluations, CA19-9, CEA, and quantification of relative expression of lncRNAs ASB16-AS1 and AFAP1-AS1.
Results: CRC patients had significantly elevated expression levels of both lncRNAs in tissue and plasma samples versus benign and control groups (p < 0.001). Despite the higher sensitivity of tissue samples results, the relative expression of both lncRNAs in plasma samples was very encouraging in the discrimination between patients with CRC versus control and benign groups. Furthermore, both lncRNAs could discriminate patients with early-stage CRC (stage I&II) from being colonic lesion and control groups with better sensitivity and specificity presented by ASB16-AS1 in tissue and plasma than results detailed by AFAP1-AS1. High expression levels of ASB16-AS1 in tissue and plasma and tissue lncRNA AFAP1-AS1 are significantly correlated with decreased overall survival (p < 0.001) and reduced progression-free (p < 0.001) compared to low expression in CRC patients.
Conclusion: We propose the utilization of lncRNA ASB16-AS1 and lncRNA AFAP1-AS1 as biomarkers in diagnosis and prognosis estimation for CRC patients. Moreover, their value in early CRC patients may affect the assortment of target therapy and treatment protocols.
Keywords: CRC, expression, lncRNA, overall survival, progression-free survival
Introduction
Colorectal cancer (CRC) is the third most prevalent cancer in males, while it is the second most commonly reported cancer in females. CRC is considered the second most common cause of cancer-associated death, accounting for almost 935,000 cancer deaths yearly.1 The total number of rectal and colon cancer deaths is expected to increase by 60% and 71.5%, respectively, by 2035.2
The development of CRC has been linked to many factors that can be classified into lifestyle or behavioral variables and genetically based variables.3 Individuals with a personal or family history of cancer, colon polyps, inflammatory bowel disease (IBD), or diabetes mellitus were found to be at an elevated risk for CRC. Furthermore, the gut microbiome, race, gender, age, and socioeconomic state are all recognized to impact CRC risk.4 Lifestyle or behavioral variables such as physical inactivity, obesity, alcohol consumption, cigarette smoking, and improper dietary patterns (a diet high in red and a diet low in fiber and calcium) are all implicated as considerable risk factors for CRC.5 Recently, there was significant progress in understanding biogenesis of CRC with the identification of potential genetic biomarkers devising aptitudes in estimating disease prognosis.6
The survival of CRC is affected by the stage at the time of diagnosis, with later-stage diagnoses having a worse survival rate. Early diagnosed have a 90% five-year survival rate, compared to 13% for those discovered late.7
Long non-coding RNAs (lncRNAs) are a category of RNAs with more than 200 nucleotides but no protein products. In multiple cancers, lncRNAs organize cell proliferation and act as tumor suppressors or tumor oncogenes.8,9 LncRNAs regulate gene expression in the vicinity of the transcription site (cis) or leave the site of transcription to moderate cellular functions in trans.10
The ASB16 gene-coded protein is a member of the ankyrin repeat motif protein family. This protein family affects cancer progression and the epithelial-mesenchymal transition (EMT).11 The lncRNA ASB16-AS1 is transcribed from the antisense strand of the ASB16 gene. The expression level of ASB16-AS1 may influence the expression of ASB16 and other related family genes.12
Formerly, it has been elucidated that actin filament-associated protein 1 antisense RNA1 (AFAP1-AS1) plays a role as an oncogene in various cancers. It is a lncRNA derived from the antisense DNA strand in the actin filament-associated protein 1 gene.13 The lncRNA AFAP1-AS1 organizes the actin filament integrity and acts as an adaptor protein linking Src family members.14
Efforts to overcome the diagnostic restrictions in CRC should be the fundamental purpose of modern medicine. A simple circulatory noninvasive test that has adequate sensitivity and specificity would be idealistic, enabling early detection of CRC at a time where curative management is still convenient. Thus, in this study, we aimed to evaluate the diagnostic roles of AFAP1-AS1 and ASB16-AS1 in colorectal cancer and highlight their roles in predicting CRC patients’ prognosis.
Subjects and Methods
Subjects: In this case–control study, 146 participants were involved. This study was carried out in the Departments of Tropical Medicine, Internal Medicine, General surgery, and Oncology in partnership with the Medical Biochemistry and Molecular Biology department, Faculty of Medicine. The participants were chosen between March 2017 and August 2018 and categorized into one of three groups: Group I included 47 patients with colorectal cancer. Group II composed of 49 patients with benign lesions in the colon including benign polyps or patients with inflammatory bowel diseases (IBD) Group III included 50 apparently normal subjects of coincided age and gender as controls.
Every participant included in this study was subjected to history appraisal and clinical evaluation. Laboratory investigations (involving complete blood count, liver and kidney function tests, ESR, Carbohydrate Antigen 19–9 (CA19-9), Carcinoembryonic Antigen (CEA), and quantification of relative expression of lncRNA ASB16-AS1 and AFAP1-AS1) in addition to abdominal pelvic ultrasonography were performed for each subject. Patients having other cancer or previously treated CRC patients were excluded from the study. Control group was chosen from patients attending endoscopy unit for colonoscopy (bleeding per rectum or change bowel habits) while no benign or malignant lesions detected in colonoscopy assessment with tissue biopsies obtained for measuring the studied lncRNAs in normal tissues. Patients with benign or malignant colorectal lesions were diagnosed by clinical assessment together with colonoscopy evaluation and tissue biopsies were taken for histopathological examination as well as assessment of studied lncRNAs in tissues. Patients with confirmed CRC underwent radiological evaluation in the form of baseline-computed tomography (CT scan) of abdomen, chest, and pelvis as well as a bone scan to detect distant metastases. TNM staging was done for all colorectal cancer patients; moreover, grading was determined based on the WHO criteria and performance status (PS) was estimated based on ECOG classification.15 For CRC patients’ colectomy was performed General Surgery Department and follow-up was done in the Clinical Oncology Department for 36 months (until the end of September 2021).
Ethical Approval
Before being enrolled in this study, all participants were given a description of the study and given the opportunity to give their informed consent. The study was carried out after approval from the ethical committee, Faculty of Medicine, Menoufia University (number; 2/2022TROP) and per the Declaration of Helsinki.
Blood and Tissue Sampling
All tissue specimens including fresh parts of the tumor mass or of the benign and normal tissues were delivered for RNA extraction. Eight milliliters of peripheral venous blood were withdrawn from every participant (before starting any treatment protocol for patients): 2 mL of fresh blood was collected into EDTA tubes, and the samples were centrifuged (at 4℃) and the attained plasma was used for RNA extraction. Another EDTA tube with 2mL of blood was used for complete blood count (CBC) using a Sysmex XN-1000 (Japan, 19,723; B.M, Egypt). Sera were separated from the remaining 4mL blood for measurement of CA19-9 and CEA concentrations by enzyme-linked immunosorbent assay (ELISA) with kits from Chemux BioScience, Inc. (USA), Alanine Aminotransferase (ALT) and Aspartate Aminotransferase (AST) (LTEC Kit, England) by a kinetic UV optimized method (International Federation of Clinical Chemistry and Laboratory Medicine (IFCC)).
RNA Isolation and Reverse Transcription
The total RNA was extracted from both fresh plasma samples and fresh tissue specimens using the miRNeasy® Mini kit (QIAGEN, Germany) that contains QIAzol reagent consistent with the manufacturer’s procedure. The isolated RNA was kept at −80 ℃.
The cDNA was produced by reverse transcription using RevertAid First Strand cDNA Synthesis Kit, Thermo Scientific, USA. A two-step reaction was conducted on ice with a net 20-µL volume: primary, 10 µL of extracted RNA were added to 1 µL of random primers and 1 µL of nuclease-free water, and the 12 volume was incubated for 5 min at 65 ℃, followed by chilling on ice. Finally, we added to the previous mixture 4 µL of 5 × reaction buffer, 1 µL of Ribolock RNase inhibitor, 2 µL of 10 mM dNTPs and 1 µL of Revertaid RT. The final 20 µL volume was incubated using the 2720 thermal cycler (ABO systems, Singapore) for one cycle carried out as: 25 ℃ for 5 min, 42 ℃ for 60 min, and finally, 5 min at 70 ℃ f.16 The obtained cDNA was preserved at −20 ℃ for real-time PCR step.
Quantification of lncRNAs Expression
The SensiFASTTM SYBR Lo-ROX Kit USA was used for conduction of quantitative real-time PCR. The reaction volume contained 6 µL of the preserved cDNA, 10 µL of SYBR green, 1 µL of nuclease-free water, and 1.5 µL of each primer. The primers’ sequences were as following: ASB16-AS1 gene: (forward) 5ʹ-GACAACAGAATTGGAAGGTCC-3ʹ and (reverse) 5ʹ- CTGTCTGAGGCAGTGAGTAC −3ʹ; AFAP1‑AS1 gene (forward) 5ʹ‑CGTTCACTTCAATAGCCG C‑3ʹ and (reverse) 5ʹ‑GGAGAAGGGATCGTCCCA‑3’; and GAPDH gene as an internal control or reference gene (forward) 5ʹ-GAAGGTGAAGGTCGGAGTC-3ʹ and (reverse) 5ʹ-GAAGATGGTGATGGGATTTC-3ʹ. The specificity of the used primers’ sequences was affirmed by the National Center for Biotechnology Information (NCBI). The amplification conditions composed of: a primary stage at 95 ℃ for 5 min followed by 50 cycles at (95 ℃ for 15s, 60 ℃ for 60s, and 72 ℃ for 60s), then 10 min at 72 ℃ as a terminal extension stage. The relative expressions of both lncRNAs were evaluated by the 2−ΔΔCt method normalized to the internal reference gene (GAPDH) and relative to the control17.
Statistical Analysis of the Data
Data of this study were coded and analyzed by the IBM SPSS software package version 20.0. (Armonk, NY: IBM Corp). Shapiro–Wilk test was used to verify the normality of distribution of different variables. Categorical variables were compared by the Chi-square test or Fisher’s Exact and Monte Carlo correction when more than 20% of the cells have an expected count less than 5. To compare more than two groups, we used F-test (ANOVA) for normally distributed quantitative variables with the Post Hoc test (Tukey). Kruskal Wallis test for abnormally distributed quantitative variables with Post Hoc (Dunn’s multiple comparisons test). Mann Whitney test was used to compare between two groups for abnormally distributed quantitative variables. The receiver operating characteristic curve (ROC) was used to determine the diagnostic performance of the markers. Spearman coefficient was used to correlate between quantitative variables. Kaplan–Meier survival curve and Cox regression were illustrated for the significant relationship between progression-free and overall survival. The significance of the obtained results was refereed at a 5% level.
Results
Analysis of our data displayed that age and gender did not vary significantly among our groups. The CRC and benign groups had significantly higher body mass index (BMI) and lower hemoglobin concentration (Hb%) than controls. Regarding other laboratory investigations, patients with CRC had significantly higher levels of ALT, AST, CEA, and CA19-9 compared to subjects with benign colonic lesions and controls. Evaluating the expression level of lncRNA ASB16-AS1 and AFAP1-AS1 in both tissue and plasma revealed a stepwise increase pattern from control to the benign lesion with the highest level recorded in the CRC group (p < 0.001). The CRC patients had significantly elevated expression levels of both lncRNAs in tissue and plasma samples versus benign and control groups (p < 0.001). Moreover, lncRNA ASB16-AS1 in tissue and plasma (p < 0.001 and p = 0.020, respectively) and tissue lncRNA AFAP1-AS1 (p < 0.001) were upregulated in subjects with benign colonic lesion compared to controls (Table 1). Additionally, patients with early-stage CRC (stage I&II) showed higher significant levels of both tissue and plasma lncRNAs compared to benign colonic lesion and controls (Figure 1A and B).
 |
Table 1 Comparison Between the Three Studied Groups According to Demographic and Laboratory Parameters |
This analysis included 47 CRC patients; the clinical characteristics of the selected patients (Table 2) specified that the family history of CRC presented in only 3 (6.4%) patients. The CRC lesions were in left (53.2%), right (38.3%) colon, while rectal cancer was detected in only (8.5%) of patients. Regarding the pathological analysis, 72.3% of the patients identified adenocarcinoma, while mucinous and signet ring tumors were detected in only 19.1% and 8.5% of patients. Only 18 (38.3%) patients showed initial good performance (PS0). Based on tumor characters, 36.2% of the patients had grade II (early-stage), and more than half (63.8%) of our patients revealed late stage. Distance metastasis was present in 17% of the patients with the liver and lung as the main metastatic sites. The main clinical presentations of patients with CRC were bleeding per rectum (29.8%), anemia (25.5%), abdominal pain (19.1%) and less frequently change bowel habits, intestinal obstruction or jaundice. These clinical presentations were comparable with those of benign group. By the end of the follow-up period, 28 (59.6%) patients had progressive disease, and 21 (44.7%) died.
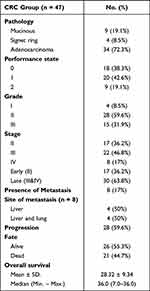 |
Table 2 Distribution of the Studied CRC Cases According to Pathological Finding, Staging and Survival (n = 47) |
The ROC curve was applied to identify the sensitivity of each biomarker in differentiating between our groups. Despite the higher sensitivity of tissue samples results, the relative expression of both lncRNAs in plasma samples was very encouraging and analogous to results detailed by tissues in the discrimination between patients with CRC versus controls and subjects with benign colonic lesions with better performance than CEA and CA19-19 in the discrimination between CRC and benign colonic lesions. Furthermore, both lncRNAs could discriminate between patients with early-stage CRC and being colonic lesion and controls better sensitivity and specificity presented by ASB16-AS1 in tissue and plasma than results detailed by AFAP1-AS1 (Table 3, Figure 1C–F).
 |
Table 3 Validity (AUC, Sensitivity, Specificity) of the lncRNAs to Discriminate Between Different Groups |
The elevated expression levels of ASB16-AS1 and AFAP1-AS1 in tissues and plasma were significantly more prevalent in the advanced stage. Likewise, the expression levels of ASB16-AS1 in tissue and plasma and tissue lncRNA AFAP1-AS1 were related to advanced grade tumors than low-grade tumors and in tumors that metastasized to distant organs and were elevated in right colonic lesion compared to left site lesion and rectal one. All indicate higher expressions in aggressive tumors (Table 4). Moreover, the expression level of ASB16-AS1 in both tissue and plasma showed a significant positive correlation with serum levels of the tumor markers CA19-9 and CEA in the CRC group. In contrast, only AFAP1-AS1expression in cancerous tissue revealed a significant correlation with serum CA19-9 levels (Figure 2).
 |
Table 4 Relation Between Studied Markers with Different Parameters in CRC Patients (n = 47) |
The relation between lncRNAs expression was evaluated by applying Kaplan–Meier survival curve and a log-rank (Mantel-Cox) analysis. High expression levels of ASB16-AS1 in tissue and plasma and tissue lncRNA AFAP1-AS1 are significantly correlated with decreased overall survival (p < 0.001) and reduced progression-free (p < 0.001) compared to low expression in CRC patients (Figure 3).
Additionally, according to Cox regression analysis, the presence of metastasis was the only independent predictor for patients’ survival in multivariate analysis (Table 5).
 |
Table 5 Univariate and Multivariate Cox Regression Analysis for the Parameters Affecting Patients’ Survival |
Discussion
Expanding patterns in the incidence and mortality rates of CRC in younger ages have been markedly recognized in the last years.18 Despite the fact that CRC is one of the preventable malignancies due to utilizing of effective screening programs and elimination of the precancerous lesions, the incidence of CRC is predicted to elevate by sixty percent by the end of the current decade.19,20 Various risk factors are involved in the pathogenesis of CRC, such as behavioral and genetically based variables.3 LncRNAs are rolled up in various cellular processes, and dysregulation of lncRNAs expression has been related to the development and progression of diverse malignancies.21 Recognition of circulating lncRNAs is evolving as novel noninvasive biomarkers in different cancers and diseases.16,22,23 In this study, we aimed to evaluate the potential roles of lncRNA ASB16-AS1 and lncRNA AFAP1-AS1 expression levels in plasma and tissue specimens’ in predicting the diagnosis and progression of CRC. Our findings revealed that both lncRNA ASB16-AS1 and lncRNA AFAP1-AS1 expression levels in plasma and tissues were upregulated in patients with CRC compared to healthy control and to subjects with benign colonic lesions. They reported good sensitivity to discriminate between these groups and even distinguish early-stage CRC from benign lesion and controls as revealed by ROC curve analysis. Higher expressions were identified in advanced tumors (grade III and stage III&IV) and in patients with metastasis. Moreover, lncRNA ASB16-AS1 in tissue and plasma and tissue lncRNA AFAP1-AS1 were upregulated in subjects with benign colonic lesions compared to controls. Their high expressions were associated with lower survival and a poor prognosis in patients with CRC.
Come online with our findings, a recent study by Jia et al,24 in esophageal cancer reported an increased lncRNA ASB16-AS1 expression level in esophageal cancer tissues compared to neighboring normal tissues, and they added that ASB16-AS1 stimulates the propagation and migration of malignant cells by affecting miR-1258.
The ASB16-AS1 expression was evaluated in patients with gastric cancer and found to be upregulated in malignant tissues compared to normal tissues and in advanced tumor stages compared to stage I and II cancers. Also, Fu et al added that higher expression was associated with cisplatin resistance of gastric cancer cells.25 In hepatocellular carcinoma (HCC), ASB16-AS1 expression was elevated in cancerous tissues with marked elevation in advanced stage tumors. Additionally, ASB16-AS1 higher expressions in HCC tissues were associated with a low survival rate, which align with our results.26 The ASB16-AS1 was suggested to enhance gastric cell proliferation by activating the nuclear factor kappa B (NF-κB).25 The NF-κB pathway is a mutual oncogenic mechanism in different malignancies. Ubiquitination and degradation of Inhibitors-of-kappaB (IκBα) enable p65 nuclear translocation, consequently activating the NF-κB pathway and enhancing epithelial-mesenchymal transition (EMT) in CRC cells.27 Additionally, ASB16-AS1 was found to promote HCC development by moderating miR-1827/FZD4/Wnt/β-catenin pathways.26 The Wnt/β-catenin pathway activation was reported as an oncogenic mechanism in CRC cells.28 NF-κB acts as a crucial link between inflammation and malignancy via direct regulation of cellular proliferation. Moreover, NF-κB has a positive effect on the Wnt/β-catenin pathway.29 The interaction between NF-κB and β-catenin could modify Wnt genes expression in cancer colon.30
The ASB16-AS1 expression was upregulated in cervical cancer and supposed to act as an oncogene by enhancing Wnt/β-catenin signals.31 Likewise, the expression of ASB16-AS1 ASB16-AS1 was significantly elevated in human glioma tissues versus normal tissues and high-grade glioma versus low-grade glioma. Furthermore, the silencing of ASB16-AS1expression suppresses cellular proliferation as reduced ASB16-AS1 expression inhibits the cell cycle progression and hinders cells in the G2/M phase,12 which may explain our finding of elevated ASB16-AS1 expression in patients with benign proliferative lesions compared to controls as well.
Regarding lncRNA AFAP1-AS1 expression and consistency with our findings, AFAP1-AS1 expression was significantly elevated in CRC.32 Also, Tang et al identified a significant increase in AFAP1-AS1 expression in CRC tissue samples compared to adjacent tissue and normal colon specimens’. Besides, they have linked this elevated expression to poor prognosis and decreased survival in patients with CRC and suggested a possible effect of AFP1-AS1 on cell cycle progression.33 Also, Wang et al added that AFAP1-AS1 might act as an independent prognostic factor for CRC patients and that AFAP1-AS1 repression led to cell cycle arrest in G0/G1 phase.34 Parallel to our findings, the AFAP1-AS1expression was related to cell proliferation and metastasis of CRC cells.35
The AFAP1-AS1 expression was upregulated in tongue squamous cell carcinoma tissues, and patients with higher AFAP1-AS1 expression were risky for shorter overall survival. Inhibition of AFAP1-AS1expression repressed cancer cell proliferation and inhibited the expression of EMT-related genes as Wnt/β-catenin pathways.36
AFAP1-AS1 relative expression was also elevated in nasopharyngeal carcinoma tissues,37 melanoma cell lines,38 and non–small cell lung cancer tissues and cell lines.39
Our lncRNAs showed better performance than the usual tumor markers CEA and CA19-9 in ROC analysis with CEA had better sensitivity and specificity than CA19-9. Tayel et al also reported that CEA was superior than CA19-19 in CRC according to ROC curve analysis.40
To the best of our knowledge, our current study is the first one that evaluated the lncRNA ASB16-AS1 in patients with CRC. Additionally, the lncRNA ASB16-AS1 and lncRNA AFAP1-AS1 were assessed for the first time in both plasma and tissue specimens of CRC patients. Despite the higher sensitivity of tissue samples results, the relative expression in plasma samples was very promising and parallel to results reported by tissues in the discrimination of the groups and with tumor stage, grade, and the presence of metastasis. Also, LncRNA ASB16-AS1 expression in plasma reported similar results to tissues in predicting overall survival and progression-free survival.
The lncRNAs expression is commonly tissue-specific. However, analogous to proteins and microRNAs, lncRNAs had been considered as potential biomarkers in circulation as well as in malignant tissues.41 Circulatory RNAs are released by cancer tissues and cells.42 Noticeably, lncRNAs can be identified and can resist degradation by ribonucleases in body fluids.22
Conclusion
In conclusion, we propose the utilization of lncRNA ASB16-AS1 and lncRNA AFAP1-AS1 as biomarkers in diagnosis and prognosis estimation for patients with CRC. Both have good value in predicting metastasis and survival. Specifically, we suggest the evaluation of circulating ASB16-AS1 and AFAP1-AS1 expression levels as a practical noninvasive method for predicting patients with CRC, as practical noninvasive biomarkers alerting physicians to perform colonoscopy, the gold standard in CRC screening and diagnosis, to improve the early discovery of CRC which may affect the assortment of target therapy and treatment protocols.
Abbreviations
CRC, Colorectal cancer; LncRNAs, Long non-coding RNAs; EMT, epithelial-mesenchymal transition; AFAP1-AS1, actin filament-associated protein 1 antisense RNA1; BMI, Body Mass Index; Hb, Hemoglobin concentration; ALT, Alanine Aminotransferase; AST, Aspartate Aminotransferase; CEA, Carcinoembryonic Antigen; CA19-9, Carbohydrate Antigen 19-9.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval and Consent to Participate
Before being enrolled in this study, all participants were given a description of the study and given the opportunity to give their informed consent. The study was carried out after approval from the ethical committee, Faculty of Medicine, Menoufia University and per the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The study was fully non-funded from any organization.
Disclosure
The authors declare that they have no competing interests.
References
1. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi:10.1136/GUTJNL-2015-310912
2. Douaiher J, Ravipati A, Grams B, Chowdhury S, Alatise O, Are C. Colorectal cancer-global burden, trends, and geographical variations. J Surg Oncol. 2017;115(5):619–630. doi:10.1002/JSO.24578
3. Ding X, Duan H, Luo H. Identification of core gene expression signature and key pathways in colorectal cancer. Front Genet. 2020;11:45. doi:10.3389/FGENE.2020.00045/BIBTEX
4. Sawicki T, Ruszkowska M, Danielewicz A, Niedźwiedzka E, Arłukowicz T, Przybyłowicz KE. A review of colorectal cancer in terms of epidemiology, risk factors, development, symptoms and diagnosis. Cancers. 2021;13(9):2025. doi:10.3390/CANCERS13092025
5. Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet. 2016;7(3):105. doi:10.3109/9781420016307-2
6. Pan F, Chen T, Sun X, et al. Prognosis prediction of colorectal cancer using gene expression profiles. Front Oncol. 2019;9(APR):252. doi:10.3389/FONC.2019.00252/BIBTEX
7. Wong MCS, Huang J, Lok V, et al. Differences in incidence and mortality trends of colorectal cancer worldwide based on sex, age, and anatomic location. Clin Gastroenterol Hepatol. 2021;19(5):955–966.e61. doi:10.1016/J.CGH.2020.02.026
8. Sabry HS, Tayel SI, Enar ME, Elabd NS. Differential expression of long noncoding RNAin hepatocellular carcinoma on top of chronic HCVand HBV infections. Clin Exp Hepatol. 2021;7(4):337–350. doi:10.5114/CEH.2021.111060
9. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393. doi:10.1016/J.CELL.2018.01.011
10. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18(1):1–13. doi:10.1186/S13059-017-1348-2
11. Liu S, Iaria J, Simpson RJ, Zhu HJ. Ras enhances TGF-β signaling by decreasing cellular protein levels of its type II receptor negative regulator SPSB1. Cell Commun Signal. 2018;16(1):1–15. doi:10.1186/S12964-018-0223-4/FIGURES/9
12. Zhang D, Zhou H, Liu J, Mao J. Long noncoding RNA ASB16-AS1 promotes proliferation, migration, and invasion in glioma cells. Biomed Res Int. 2019;2019. doi:10.1155/2019/5437531
13. Abdul S, Majid A, Wang J, Liu Q, Sun MZ, Liu S. Bidirectional interaction of lncRNA AFAP1-AS1 and CRKL accelerates the proliferative and metastatic abilities of hepatocarcinoma cells. J Adv Res. 2020;24:121. doi:10.1016/J.JARE.2020.03.010
14. Baisden JM, Qian Y, Zot HM, Flynn DC. The actin filament-associated protein AFAP-110 is an adaptor protein that modulates changes in actin filament integrity. Oncogene. 2001;20(44):6435–6447. doi:10.1038/sj.onc.1204784
15. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. doi:10.1016/j.ejca.2008.10.026
16. Saleh AA, Kasem HE, Zahran E, El-Hefnawy SM. Dysregulation of cell-free long non-coding RNAs (NEAT2, CTC-471J1.2 and lnc-DC) in Egyptian systemic lupus and lupus nephritis patients. Meta Gene. 2020;24:100665. doi:10.1016/j.mgene.2020.100665
17. Dorak MT. Real-time PCR. Real-Time PCR. 2007;1–333. doi:10.4324/9780203967317
18. Burnett-Hartman AN, Lee JK, Demb J, Gupta S. An update on the epidemiology, molecular characterization, diagnosis, and screening strategies for early-onset colorectal cancer. Gastroenterology. 2021;160(4):1041–1049. doi:10.1053/J.GASTRO.2020.12.068
19. Thomas M, Sakoda LC, Hoffmeister M, et al. Genome-wide modeling of polygenic risk score in colorectal cancer risk. Am J Hum Genet. 2020;107(3):432. doi:10.1016/J.AJHG.2020.07.006
20. Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Przeglad Gastroenterol. 2019;14(2):89. doi:10.5114/PG.2018.81072
21. Chakraborty S, Andrieux G, Hasan AMM, et al. Harnessing the tissue and plasma lncRNA-peptidome to discover peptide-based cancer biomarkers. Sci Rep. 2019;9(1):1–17. doi:10.1038/s41598-019-48774-1
22. Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers. 2016;2016:1–10. doi:10.1155/2016/9085195
23. Saleh AA, Kasem HE, Zahran ES, El-Hefnawy SM. Cell-free long non-coding RNAs (LY86-AS1 & HCG27_201and GAS5) as biomarkers for pre-diabetes and type 2 DM in Egypt. Biochem Biophys Rep. 2020;23(May):100770. doi:10.1016/j.bbrep.2020.100770
24. Jia Z, Wang P, Yang Y, Zhu D, Wang Z, Wang W. LncRNA ASB16-AS1 regulates the proliferation, migration and invasion of esophageal cancer cells by targeting miR-1258. Zhonghua Zhong Liu Za Zhi. 2021;43(7):762–768. doi:10.3760/CMA.J.CN112152-20200509-00430
25. Fu T, Ji K, Jin L, et al. ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-κB pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric Cancer. 2021;24(1):45–59. doi:10.1007/S10120-020-01096-Y/FIGURES/6
26. Yao X, You G, Zhou C, Zhang D. LncRNA ASB16-AS1 promotes growth and invasion of hepatocellular carcinoma through regulating miR-1827/FZD4 axis and activating Wnt/β-catenin pathway. Cancer Manag Res. 2019;11:9371. doi:10.2147/CMAR.S220434
27. Xiao C, Wu G, Zhou Z, et al. RBBP6, a RING finger-domain E3 ubiquitin ligase, induces epithelial–mesenchymal transition and promotes metastasis of colorectal cancer. Cell Death Dis. 2019;10(11):1–17. doi:10.1038/s41419-019-2070-7
28. Fang G, Ye BL, Hu BR, Ruan XJ, Shi YX. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2018;504(1):184–189. doi:10.1016/J.BBRC.2018.08.152
29. Ma B, Hottiger MO. Crosstalk between wnt/β-catenin and NF-κB signaling pathway during inflammation. Front Immunol. 2016;7(SEP):378. doi:10.3389/FIMMU.2016.00378/BIBTEX
30. Schwitalla S, Fingerle AA, Cammareri P, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152(1–2):25–38. doi:10.1016/J.CELL.2012.12.012
31. Liu W, Zhuang R, Feng S, et al. Long non-coding RNA ASB16-AS1 enhances cell proliferation, migration and invasion via functioning as a ceRNA through miR-1305/Wnt/β-catenin axis in cervical cancer. Biomed Pharmacother. 2020;125:109965. doi:10.1016/J.BIOPHA.2020.109965
32. Li Y, Zhu Z, Hou X, Sun Y. LncRNA AFAP1-AS1 promotes the progression of colorectal cancer through miR-195-5p and WISP1. J Oncol. 2021;2021. doi:10.1155/2021/6242798
33. Tang J, Zhong G, Wu J, Chen H, Jia Y. Long noncoding RNA AFAP1-AS1 facilitates tumor growth through enhancer of zeste homolog 2 in colorectal cancer. Am J Cancer Res. 2018;8(5):892.
34. Wang F, Ni H, Sun F, Li M, Chen L. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomed Pharmacother. 2016;81:152–159. doi:10.1016/J.BIOPHA.2016.04.009
35. Han X, Wang L, Ning Y, Li S, Wang Z. Long non-coding RNA AFAP1-AS1 facilitates tumor growth and promotes metastasis in colorectal cancer. Biol Res. 2016;49(1):1–7. doi:10.1186/S40659-016-0094-3/FIGURES/7
36. Wang ZY, Hu M, Dai MH, et al. Upregulation of the long non-coding RNA AFAP1-AS1 affects the proliferation, invasion and survival of tongue squamous cell carcinoma via the Wnt/β-catenin signaling pathway. Mol Cancer. 2018;17(1). doi:10.1186/S12943-017-0752-2
37. Fang M, Zhang M, Wang Y, et al. Long noncoding RNA AFAP1-AS1 is a critical regulator of nasopharyngeal carcinoma tumorigenicity. Front Oncol. 2020;10:2510. doi:10.3389/FONC.2020.601055/BIBTEX
38. Liu F, Hu L, Pei Y, et al. Long non-coding RNA AFAP1-AS1 accelerates the progression of melanoma by targeting miR-653-5p/RAI14 axis. BMC Cancer. 2020;20(1):1–11. doi:10.1186/S12885-020-6665-2/FIGURES/4
39. Yin D, Lu X, Su J, et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer. 2018;17(1):1–12. doi:10.1186/S12943-018-0836-7/FIGURES/6
40. Tayel SI, Fouda EAM, Gohar SF, Elshayeb EI, El-sayed EH, El-kousy SM. Potential role of MicroRNA 200c gene expression in assessment of colorectal cancer. Arch Biochem Biophys. 2018;647:41–46. doi:10.1016/J.ABB.2018.04.009
41. Yao Y, Chen X, Lu S, et al. Circulating long noncoding RNAs as biomarkers for predicting head and neck squamous cell carcinoma. Cell Physiol Biochem. 2018;50(4):1429–1440. doi:10.1159/000494605
42. Ren S, Wang F, Shen J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49(13):2949–2959. doi:10.1016/J.EJCA.2013.04.026
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Efficacy of Bevacizumab in High-Grade Meningiomas: A Retrospective Clinical Study
Bai X, Liu X, Wen J
Neuropsychiatric Disease and Treatment 2022, 18:1619-1627
Published Date: 6 August 2022
Real-World Outcomes and Prognostic Factors Among Patients with Advanced Non-Small Cell Lung Cancer and High PD-L1 Expression Treated with Immune Checkpoint Inhibitors as First-Line Therapy
Ge W, Wu N, Jalbert JJ, Quek RGW, Liu J, Rietschel P, Pouliot JF, Harnett J, Hsu ML, Feliciano JL
Cancer Management and Research 2022, 14:3191-3202
Published Date: 16 November 2022
The Systemic Inflammation Score is Associated with the Survival of Patients with Prostate Cancer
Xie J, Xiao X, Dong Z, Wang Q
Journal of Inflammation Research 2023, 16:963-975
Published Date: 7 March 2023
Prognostic Value of Ki67 in Epithelial Ovarian Cancer: Post-Neoadjuvant Chemotherapy Ki67 Combined with CA125 Predicting Recurrence
Liu Y, Gu Q, Xiao Y, Wei X, Wang J, Huang X, Linghu H
Cancer Management and Research 2024, 16:761-769
Published Date: 9 July 2024



