Back to Journals » Breast Cancer: Targets and Therapy » Volume 15
Guideline-Based, Multi-Gene Panel Germline Genetic Testing for at-Risk Patients with Breast Cancer
Authors Abdel-Razeq H , Abujamous L, Al-Azzam K, Abu-Fares H, Bani Hani H, Alkyam M, Sharaf B , Elemian S, Tamimi F , Abuhijla F , Edaily S , Salama O, Abdulelah H, Daoud R, Abubaker M, Al-Atary A
Received 31 October 2022
Accepted for publication 15 December 2022
Published 13 January 2023 Volume 2023:15 Pages 1—10
DOI https://doi.org/10.2147/BCTT.S394092
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pranela Rameshwar
Hikmat Abdel-Razeq,1,2 Lama Abujamous,3 Khansa Al-Azzam,1 Hala Abu-Fares,1 Hira Bani Hani,1 Mais Alkyam,1 Baha’ Sharaf,1 Shatha Elemian,1 Faris Tamimi,1 Fawzi Abuhijla,4 Sarah Edaily,1 Osama Salama,1 Hazem Abdulelah,1 Rand Daoud,1 Mohammad Abubaker,1 Areej Al-Atary5
1Department of Internal Medicine, King Hussein Cancer Center, Amman, Jordan; 2School of Medicine, The University of Jordan, Amman, Jordan; 3Department of Cell Therapy & Applied Genomic, King Hussein Cancer Center, Amman, Jordan; 4Department of Radiation Oncology, King Hussein Cancer Center, Amman, Jordan; 5Department of Nursing, King Hussein Cancer Center, Amman, Jordan
Correspondence: Hikmat Abdel-Razeq, Department of Internal Medicine, King Hussein Cancer Center, School of Medicine, University of Jordan, Queen Rania Al Abdullah Street, P.O. Box 1269, Amman, 11941, Jordan, Tel +962-6 5300460, Ext 1000, Email [email protected]
Background: Genetic testing for at-risk patients with breast cancer should be routinely offered. Knowledge generated may influence both treatment decisions and cancer prevention strategies among the patients themselves and their relatives. In this study, we report on the prevalence and patterns of germline mutations, using commercially available next-generation sequencing (NGS)-based multi-gene panels (MGP).
Patients and Methods: Consecutive at-risk breast cancer patients, as determined by international guidelines, were offered germline genetic testing using a 20-gene NGS-based panel at a reference lab. Samples of peripheral blood were obtained for DNA extraction and genetic variants were classified as benign/likely benign (negative), pathogenic/likely pathogenic (positive) or variants of uncertain significance (VUS).
Results: A total of 1310 patients, median age (range) 43 (19– 82) years, were enrolled. Age ≤ 45 years (n = 800, 61.1%) was the most common indication for testing. Positive family history of breast, ovarian, pancreatic or prostate cancers, and triple-negative disease were among the common indications. Among the whole group, 184 (14.0%) patients had pathogenic/likely pathogenic variants; only 90 (48.9%) were in BRCA1 or BRCA2, while 94 (51.9%) others had pathogenic variants in other genes; mostly in APC, TP53, CHEK2 and PALB2. Mutation rates were significantly higher among patients with positive family history (p = 0.009); especially if they were 50 years or younger at the time of breast cancer diagnosis (p < 0.001). Patients with triple-negative disease had relatively higher rate (17.5%), and mostly in BRCA1/2 genes (71.4%). Variants of uncertain significance (VUS) were reported in 559 (42.7%) patients; majority (90.7%) were in genes other than BRCA1 or BRCA2.
Conclusion: Pathogenic mutations in genes other than BRCA1/2 are relatively common and could have been missed if genetic testing was restricted to BRCA1/2. The significantly high rate of VUS associated with multi-gene panel testing can be disturbing.
Keywords: breast cancer, BRCA1, BRCA2, multigene panel, hereditary breast cancer, next generation sequencing
Introduction
Breast cancer continues to be the most common cancer among women worldwide.1 Regionally, it constitutes one-fifth of all cancer cases and almost 40% of all female cancers.2 With a median age of 51 years, breast cancer in the Arab world is diagnosed at much younger age compared to the West. Additionally, more than 30% of patients present late with locally-advanced or metastatic disease.3,4
Recent data had shown that 5–10% of breast cancers are related to inherited germline mutations, mostly in BRCA1 or BRCA2.5,6 Carriers of these mutations are at higher risk for both breast and ovarian cancers. Professional societies had published genetic testing guidelines for patients with breast cancer based on their family or personal history of cancer or tumor molecular subtypes.7,8
The risk of breast and ovarian cancers among individuals with pathogenic variants of BRCA1 and BRCA2 is well known.9 In one prospective study that included a cohort of 978 BRCA1 and 909 BRCA2 carriers from the United Kingdom, the average cumulative risks by age 70 years for BRCA1 carriers were estimated to be 60% for breast cancer and 59% for ovarian cancer. Women with BRCA2 pathogenic variants had a corresponding risk of 55% and 16% for breast and ovarian cancers, respectively.10 A meta-analysis of studies looked at the penetrance rates of BRCA1 and BRCA2 reached similar conclusions.11 Risk-reduction interventions including bilateral mastectomies12,13 and oophorectomies14,15 are highly recommended in such patients.16
We recently reported our experience on BRCA1 and BRCA2 mutations among a total of 517 at-risk patients tested as per the National Comprehensive Cancer Network (NCCN) guidelines.7 Among the whole group, 72 (13.9%) patients had pathogenic or likely pathogenic BRCA1 or BRCA2 mutations, while 53 (10.3%) others had VUS. Higher mutation rates were observed among patients with bilateral or second primary breast cancer, those with positive family history of breast and/or ovarian cancers, and patients with triple-negative disease (negative for estrogen receptors (ER), progesterone receptors (PR), and human epidermal growth factor (HER2) receptors).17
About a third of patients with strong family history of breast cancers remain negative for BRCA1 or BRCA2 mutation.18 Several non-BRCA genes have been recently identified and were introduced as part of multigene breast cancer gene panels. Such genes include ATM, CHEK2, PALB2, PTEN, TP53, and several others.
Multi-gene panels testing, utilizing next-generation sequencing (NGS) is widely used and had become more affordable. Some of these multi-gene panels are offered directly to customers and not necessarily through a controlled health-care provider setting. As such, more genes associated with breast, ovarian and other cancers were recently identified.19,20
Regional data on frequency of BRCA1 and BRCA2 pathogenic variants are scarce and data on newly identified genes does not exist.21–23 Knowledge about pattern and prevalence of such mutations can add to our efforts to improve preventive and treatment strategies of breast and other cancers, too.
In this study, we evaluate the contribution of germline mutations in genes other than BRCA1 and BRCA2 to breast cancer among our local population with selected high-risk profile as recommended by the NCCN guidelines.
Methods
Breast cancer patients with selected high-risk profile as recommended by the NCCN guidelines were invited for multi-gene panel (MGP) testing. All patients had their diagnosis, treatment and follow up at our center.
Since November 2019, we introduced and included in our clinical practice guidelines, multi-gene panel testing (20 genes) for at-risk breast cancer patients. The 20 genes are: ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, NBN, NF1, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11 and TP53. Eligible patients were identified at their first encounter by their primary medical or surgical oncologists during their routine clinic visit, or during the weekly breast multidisciplinary team (MDT) meeting. Patients who consented to MGP testing were then referred to a genetic counseling clinic where a detailed pre-testing counseling is carried out by trained counselors. Clinical and psychosocial consequences of positive test results were discussed at length with the patients, and when requested, with the spouse and/or close family members.
The study was conducted in accordance with the local and international guidelines and regulations on human research including the 1964 Helsinki declaration and its later amendments. The study was approved by Institutional Review Board (IRB) at King Hussein Cancer Center (approval number: 20-KHCC-198), and all patients signed informed consent for genetic testing. Testing was done at no-cost to all patients. Cascade family screening of positive patients was also offered, almost free of charge, to all at-risk close relatives.
Samples of peripheral blood (10 milliliters) were obtained for DNA extraction utilizing methods previously detailed,24 and MGP testing was performed at Invitae, San Francisco, USA. Mutations in breast cancer predisposing genes were classified as pathogenic/likely pathogenic (positive), no pathogenic mutations (negative) and variant of uncertain significance (VUS). Clinical and pathological data were obtained from patients’ medical records, and a detailed 3-generation family history was also obtained by a genetic counselor or one of the investigators.
Analysis was performed using an Agilent SureSelect custom design reagent to screen for germline pathogenic variants. Genomic DNA regions including coding exons and intron/exon boundaries are targeted by hybridization capture and sequenced on the Illumina platform with a sensitivity of at least 95%. The target region of selected transcripts is covered to a minimum read depth of 30x. Analysis for large deletion and duplication is performed using comparative depth of coverage of NGS data and - or MLPA analysis using P087, P045, P260.
Statistical Analysis
Both clinical and pathological characteristics were tabulated and then described by percentages (%), medians and range. Analyses included all patients tested during the study period; however, close relatives diagnosed with breast cancer and tested following the identification of the index case in the family were all excluded. Proportion of patients with pathogenic/likely pathogenic variants were calculated and compared according to age at diagnosis, family history of breast, ovarian, pancreatic and prostate cancers and triple-negative status. Analysis was performed utilizing version 9.4 of SAS software (SAS Institute Inc., Cary, NC).
Results
Patients’ Characteristics
Between November 2019 and October 2021, a total of 1310 patients were enrolled, median age (range) was 43 (19–82) years and 496 (37.9%) patients were 40 years or younger. All, but 24 (1.8%) were females and majority (n = 1213, 92.6%) were Jordanians. Non-Jordanians were mostly from Iraq, Libya, Palestine and Syria. Majority of the patients had hormone receptor positive disease (ER: 74.3%, PR: 73.7%), while human epidermal growth factor receptor-2 (HER2) was positive in 285 (21.8%) by immunohistochemistry (IHC) and/or Fluorescent in Situ Hybridization (FISH), and 166 (12.7%) had triple-negative disease; 160 (96.4%) of them were 60 years or younger at time of breast cancer diagnosis. Positive family history (first-, second- or third-degree) of breast, ovarian, pancreatic or high-grade prostate cancer was found in majority of the patients (n = 972, 74.2%), Table 1.
 |
Table 1 Patients’ Characteristics (n = 1310) |
Multi-Gene Panel Testing
Patients were tested according to latest NCCN guidelines. Age ≤45 years (n = 800, 61.1%) was the most common indication for genetic testing and counseling. Other common indications include family history of breast, ovarian, pancreatic or prostate cancers, and triple-negative disease.
Mutation rates were significantly higher among a group of 372 women (diagnosed at any age), with one or more close relatives with breast cancer, diagnosed at age 50 years or younger (18.0% compared to 12.5%, p = 0.009), and among 541 younger patients (diagnosed at age ≤50 years) with one or more close relatives with breast, ovarian, pancreatic, or high-grade prostate cancer (Gleason score ≥7); 18.1% vs 11.2%, p < 0.001 (Figure 1, Table 2).
 |
Table 2 Rates of Positive Mutation Across Different Indications |
 |
Figure 1 Rates of pathogenic/likely pathogenic variants. |
Triple-Negative Disease
A total of 28 (17.5%) of the 160 patients tested because of triple-negative disease had pathogenic/likely pathogenic mutations; 20 (12.5%) of them were in BRCA1 or BRCA2, while only 8 (5.0%) patients had pathogenic mutations in genes other than BRCA1 or BRCA2 (RAD51D, NF1 and APC Exon 16 c.3920T>A). On the other hand, a total of 65 (40.6%) patients had VUS; 54 (33.8%) were in genes other than BRCA1 or BRCA2. An additional 10 (6.3%) had both VUS and pathogenic mutations.
Mutations in Genes Other Than BRCA1/2
Rate of mutations in non-BRCA1/2 genes is relatively common and represents 51.1% (n = 94) of all detected mutations. APC (n = 19, 10.3%), TP53 (n = 14, 7.6%), CHEK2 (n = 12, 6.5%) and PALB2 (n = 10, 5.4%) were the most encountered mutations (Figure 2). Mutation rate in non-BRCA1/2 genes was lowest (5.0%) among patients with TN disease, and highest (12.6%) among patients diagnosed (at any age) with one or more close relatives with epithelial ovarian or pancreatic cancers (Figure 1). A list of all detected pathogenic/likely pathogenic variants is detailed in Supplementary Table.
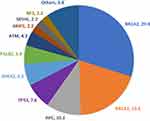 |
Figure 2 Positive/Likely positive variants (n=184) in percentage*. *Numbers next to gene involved represent percentage from the 184 variants detected. |
VUS Rates
Variants of uncertain significance (VUS) were observed among 559 (42.7%) patients. Because many patients (n = 287, 21.9%) had more than variants, a total of 846 VUS were observed and majority (n = 767, 90.7%) were found in genes other than BRCA1 or BRCA2 as illustrate in Figure 3. Another 78 (6.0%) had VUS in addition to another pathogenic mutation. Ratio of VUS to positive variants was significantly higher in non-BRCA1/2; 8.2 versus 0.9, p =< 0 0.001. We also analyzed VUS to pathogenic/likely pathogenic ratio for the 10 most common genes as illustrated in Figure 4.
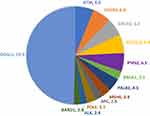 |
Figure 3 Rates of Variants of Uncertain Significance (VUS) in percentage*. *Numbers next to gene involved represent percentage from all VUS. |
 |
Figure 4 Ratio of variants of uncertain significance (VUS) to pathogenic/likely pathogenic variants. |
Discussion
To our knowledge, this is the first study from the region addressing the use of multi-gene panel testing in patients with breast cancer. It is clear from our study that pathogenic/likely pathogenic mutations in non-BRCA1/2 genes are not uncommon and represent at least 50% of all encountered positive mutations. Prior to 2020, the NCCN guidelines focused largely on testing for BRCA1 and BRCA2 and its associated risk management interventions. However, the latest updated version recognized several other genes and endorsed its testing.7 Cancer risk management interventions are recommended when the absolute cancer risk for mutation carriers exceed that of average non-carrier population, which is estimated at 12–13%. The most common mutations identified in our current study are actionable and testing may be helpful. However, no clear data or recommendations exist regarding pathogenic/likely pathogenic mutations in many of such genes.25 To complicate the issue further, variants in the same gene may be associated with different risk levels, like what we see with the ATM different mutation variants; some are associated with early onset and even bilateral disease while other variants are not.26,27
It is also clear that with the expansion of gene tested, more VUS will be encountered. Rates of VUS among non-BRCA1/2 genes are relatively high. In our current study, at least one in three tested patients had a VUS; 90% of them were in genes other than BRCA1 or BRCA2. Additionally, many of the selected genes in the panel have a high VUS to pathogenic/likely pathogenic ratio (Figure 4). Furthermore, many of the gene tested in our cohort, like PMS2, RAD51C, MSH2, STK11 and MLH1, had only VUS and never pathogenic mutations. The generated anxiety for patients, families and even the treating physicians might outweigh the anticipated benefit.
The added value of extended testing might be limited to special group of at-risk breast cancer patients. Our data clearly illustrated that testing patients with TN disease beyond the usual BRCA1/2 is associated with little added value; only 8 (5.0%) cases of non-BRCA1/2 mutations were identified in 160 patients with TN-disease.
Family involvement in preventive decisions is inheritably limited.28,29 Most breast cancer predisposing genes are inherited in an autosomal dominant fashion, thus the risk of carrying the same mutation is 50% among first-degree relatives. But such numbers might not necessarily be taken seriously by family members. Add to this, the fact that patients themselves are occasionally not willing to share such information with their relatives. We are in the process of collecting data on these issues which is somewhat more important in smaller communities and cultures, like ours. Our findings might add to the national efforts exercised to prevent cancer in general, and breast in particular. Identifying inherited cancer predisposing genes in a patient should reflect positively in preventing the occurrence of cancer in close relatives.
In addition to breast and ovarian cancer prevention, the utilization of our knowledge about mutations in BRCA1 and BRCA2, and even in non-BRCA mutations, in treatment decisions is increasing.30 Patients with TN-breast cancer and deleterious BRCA1/2 mutations have better response rate and progression-free survival (PFS) when treated with carboplatin compared to docetaxel.31 PARP (poly (ADP)-ribose polymerase) inhibitors were also tried in patients with pathogenic BRCA1/2 advanced-stage breast cancer. The randomized phase-3 trial (OlympiAD) showed that olaparib, when compared to palliative chemotherapy, in a cohort of 302 HER2-negative metastatic breast cancer patients with pathogenic germline BRCA1/2 mutation, was associated with better PFS.32 Talazoparib, another PARP inhibitor, had shown similar results in another phase-3 randomized trial (EMBRACA).33 More recently, PARP inhibitors were also tried in the setting of high-risk early-stage breast cancer with germline pathogenic BRCA1/2 mutations. Following the completion neoadjuvant or adjuvant therapy and standard local treatment, adjuvant olaparib for one year was associated with significant improvement in invasive (iDFS), distant (dDFS) disease-free survivals and possibly overall survival (OS), when compared to placebo, in a randomized phase-3 trial (OlympiA).34,35 In addition to BRCA1 and BRCA2, meaningful responses were seen in patients with germline PALB2, but not those with ATM or CHEK2 mutations alone.36
Given the increasing percentage of women with VUS and pathogenic/likely pathogenic variants in genes with no much data on its associated risk, pre- and post-test genetic counseling are highly needed. Studies had shown that satisfaction is significantly higher among women who had undergone genetic counseling.37 It is also important that preventive and therapeutic decisions in relation to genetic testing made in a multidisciplinary setting with active participation of oncologists, surgeons and geneticists with high level of psychosocial support.
Conclusions
Pathogenic mutations in genes other than BRCA1 or BRCA2 are relatively common and could have been missed if genetic testing was restricted to BRCA1 and BRCA2. MGP testing results in a significantly higher rate of VUS, a finding that may increase the anxiety of patients and physicians, alike. Germline genetic testing had gone beyond cancer prevention and currently is incorporated in treatment decisions of both early and advanced stage breast cancer.
Abbreviations
CI, Confidence Intervals; ER, Estrogen Receptors; FISH, Fluorescent in Situ Hybridization; HER2, Human Epidermal Growth Factor Receptor; IHC, Immunohistochemistry; IRB, Institutional Review Board; MDT, Multidisciplinary Team; MGP, Multi-gene Panel; NCCN, National Comprehensive Cancer Network; NGS, Next-Generation Sequencing; PFS, Progression-Free Survival; PR, Progesterone Receptors; VUS, Variant of Uncertain Significance.
Data Sharing Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The study was approved by Institutional Review Board (IRB) at King Hussein Cancer Center (approval number, 20-KHCC-198), and all patients signed informed consent for genetic testing.
Consent for Publication
Data submitted are entirely unidentifiable, and there are no details on individuals reported within the manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The study was funded by unrestricted competitive grant from Pfizer International. The sponsor has no access to the data or the manuscript.
Disclosure
First author HA received an institutional grant from Pfizer International to study genetic mapping of breast cancer. All other authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript, this includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties. The abstract of this paper was presented at the 44th annual Breast Cancer Symposium (7–10 Dec 2021) as a poster presentation with interim findings. The poster’s abstract was published in “Poster Abstracts”, Cancer Res 2022; 82(4 Suppl): Abstract number P3-07-06.
Link, https://doi.org/10.1158/1538-7445.SABCS21-P3-07-06
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020, GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Jordan Cancer Registry. Cancer Incidence in Jordan. Non-Communicable Diseases Directorate Ministry of Health, Jordan; 2015. Available from: https://moh.gov.jo/EN/Pages/Health_Statistics_and_Indicators.
3. Chouchane L, Boussen H, Sastry KSR. Breast cancer in Arab populations: molecular characteristics and disease management implications. Lancet Oncol. 2013;14(10):e417–e424. doi:10.1016/S1470-2045(13)70165-7
4. El Saghir NS, Khalil MK, Eid T, et al. Trends in epidemiology and management of breast cancer in developing Arab countries, a literature and registry analysis. Int J Surg. 2007;5(4):225–233. doi:10.1016/j.ijsu.2006.06.015
5. Larsen MJ, Thomassen M, Gerdes AM, Kruse TA. Hereditary breast cancer, clinical, pathological and molecular characteristics. Breast Cancer (Auckl). 2014;8:145–155. doi:10.4137/BCBCR.S18715
6. Drohan B, Roche CA, Cusack JC, Hughes KS. Hereditary breast and ovarian cancer and other hereditary syndromes, using technology to identify carriers. Ann Surg Oncol. 2012;19(6):1732–1737. doi:10.1245/s10434-012-2257-y
7. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines). Genetic/familial high-risk assessment, breast, ovarian, and pancreatic. Version 2; 2021. Available from: https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf.
8. National Collaborating Centre for Cancer (UK). Familial breast cancer, classification and care of people at risk of familial breast cancer and management of breast cancer and related risks in people with a family history of breast cancer. Cardiff (UK): National Collaborating Centre for Cancer (UK); 2013. Available from: https://www.ncbi.nlm.nih.gov/books/NBK247567/.
9. Maxwell KN, Domchek SM. Cancer treatment according to BRCA1 and BRCA2 mutations. Nat Rev Clin Oncol. 2012;9(9):520–528. doi:10.1038/nrclinonc.2012.123
10. Mavaddat N, Peock S, Frost D, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers, results from prospective analysis of EMBRACE. J Natl Cancer Inst. 2013;105(11):812–822. doi:10.1093/jnci/djt095
11. Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25(11):1329–1333. doi:10.1200/JCO.2006.09.1066
12. Chai X, Friebel TM, Singer CF, et al. Use of risk-reducing surgeries in a prospective cohort of 1499 BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat. 2014;148(2):397–406. doi:10.1007/s10549-014-3134-0
13. Rebbeck TR, Friebel T, Lynch HT, et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers, the PROSE Study Group. J Clin Oncol. 2004;22(6):1055–1062. doi:10.1200/JCO.2004.04.188
14. Olopade OI, Artioli G. Efficacy of risk-reducing salpingo-oophorectomy in women with BRCA-1 and BRCA-2 mutations. Breast J. 2004;10:S5–S9. doi:10.1111/j.1524-4741.2004.101s3.x
15. Eleje GU, Eke AC, Ezebialu IU, Ikechebelu JI, Ugwu EO, Okonkwo OO. Risk-reducing bilateral salpingo-oophorectomy in women with BRCA1 or BRCA2 mutations. Cochrane Database Syst Rev. 2018;8(8):CD012464. doi:10.1002/14651858.CD012464.pub2
16. Domchek SM, Friebel TM, Singer CF, et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA. 2010;304(9):967–975. doi:10.1001/jama.2010.1237
17. Abdel-Razeq H, Abujamous L, Jadaan D. Patterns and prevalence of germline brca1 and brca2 mutations among high-risk breast cancer patients in Jordan, A study of 500 Patients. J Oncol. 2020;2020:8362179. doi:10.1155/2020/8362179
18. Coppa A, Nicolussi A, D’Inzeo S, et al. Optimizing the identification of risk-relevant mutations by multigene panel testing in selected hereditary breast/ovarian cancer families. Cancer Med. 2018;7(1):46–55. doi:10.1002/cam4.1251
19. Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Intern Med. 2014;174(2):275–280. doi:10.1001/jamainternmed.2013.12048
20. Kurian AW, Hare EE, Mills MA, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32(19):2001–2009. doi:10.1200/JCO.2013.53.6607
21. Jalkh N, Nassar-Slaba J, Chouery E, et al. Prevalance of BRCA1 and BRCA2 mutations in familial breast cancer patients in Lebanon. Hered Cancer Clin Pract. 2012;10(1):7. doi:10.1186/1897-4287-10-7
22. Laraqui A, Uhrhammer N, Lahlou-Amine I, et al. Mutation screening of the BRCA1 gene in early onset and familial breast/ovarian cancer in Moroccan population. Int J Med Sci. 2013;10(1):60–67. doi:10.7150/ijms.5014
23. Abdel-Razeq H, Al-Omari A, Zahran F, Arun B. Germline BRCA1/BRCA2 mutations among high risk breast cancer patients in Jordan. BMC Cancer. 2018;18(1):152. doi:10.1186/s12885-018-4079-1
24. Abdel-Razeq H, Tamimi F, Abujamous L, et al. Patterns and prevalence of BRCA1 and BRCA2 germline mutations among patients with triple-negative breast cancer, regional perspectives. Cancer Manag Res. 2021;13:4597–4604. doi:10.2147/CMAR.S316470
25. Powers B, Pal T, Laronga C. Considerations in Testing for Inherited Breast Cancer Predisposition in the Era of Personalized Medicine. Surg Oncol Clin N Am. 2018;27(1):1–22. doi:10.1016/j.soc.2017.08.003
26. Jerzak KJ, Mancuso T, Eisen A. Ataxia-telangiectasia gene (ATM) mutation heterozygosity in breast cancer, a narrative review. Curr Oncol. 2018;25(2):e176–e180. doi:10.3747/co.25.3707
27. Brunet J, Gutiérrez-Enríquez S, Torres A, et al. ATM germline mutations in Spanish early-onset breast cancer patients negative for BRCA1/BRCA2 mutations. Clin Genet. 2008;73(5):465–473. doi:10.1111/j.1399-0004.2008.00987
28. Barsevick AM, Montgomery SV, Ruth K, et al. Intention to communicate BRCA1/BRCA2 genetic test results to the family. J Fam Psychol. 2008;22(2):303–312. doi:10.1037/0893-3200.22.2.303
29. MacDonald DJ, Sarna L, van Servellen G, Bastani R, Giger JN, Weitzel JN. Selection of family members for communication of cancer risk and barriers to this communication before and after genetic cancer risk assessment. Genet Med. 2007;9(5):275–282. doi:10.1097/gim.0b013e31804ec075
30. Edaily S, Abdel-Razeq H. Management strategies of breast cancer patients with BRCA1 and BRCA2 pathogenic germline variants. Onco Targets Ther. 2022;27(15):815–826. PMID, 35923470; PMCID, PMC9343017. doi:10.2147/OTT.S369844
31. Tutt A, Tovey H, Cheang MCU, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups, the TNT Trial. Nat Med. 2018;24(5):628–637. doi:10.1038/s41591-018-0009-7
32. Robson ME, Tung N, Conte P, et al. OlympiAD final overall survival and tolerability results, Olaparib versus chemotherapy treatment of physician’s choice in patients with a germline BRCA mutation and HER2-negative metastatic breast cancer. Ann Oncol. 2019;30(4):558–566. doi:10.1093/annonc/mdz012
33. Litton JK, Hurvitz SA, Mina LA, et al. Talazoparib versus chemotherapy in patients with germline BRCA1/2-mutated HER2-negative advanced breast cancer, final overall survival results from the EMBRACA trial. Ann Oncol. 2020;31(11):1526–1535. doi:10.1016/j.annonc.2020.08.2098
34. Tutt ANJ, Garber JE, Kaufman B, et al. Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer. N Engl J Med. 2021;384(25):2394–2405. doi:10.1056/NEJMoa2105215
35. Tutt A, Garber J, Gelber R, et al. VP1-2022, pre-specified event driven analysis of Overall Survival (OS) in the OlympiA Phase III trial of adjuvant olaparib (OL) in germline BRCA1/2 mutation (gBRCAm) associated breast cancer. ESMO Virtual Plenary; 2022. Available from: https://oncologypro.esmo.org/meeting-resources/esmo-virtual-plenary-resources/olympia-phase-iii-pre-specified-event-driven-analysis-of-overall-survival-of-olaparib-in-gbrcam-breast-cancer.
36. Tung NM, Robson ME, Ventz S, et al. TBCRC 048, Phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J Clin Oncol. 2020;38(36):4274–4282. doi:10.1200/JCO.20.02151
37. Cohen SA, Bradbury A, Henderson V, Hoskins K, Bednar E, Arun BK. Genetic counseling and testing in a community setting, quality, access, and efficiency. Am Soc Clin Oncol Educ Book. 2019;39:e34–e44. doi:10.1200/EDBK_238937
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Management Strategies of Breast Cancer Patients with BRCA1 and BRCA2 Pathogenic Germline Variants
Edaily S, Abdel-Razeq H
OncoTargets and Therapy 2022, 15:815-826
Published Date: 27 July 2022
Association Between Germline BRCA1/2 Gene Variants and Clinicopathological Features of Ovarian Cancer
Luo Y, Pan R, Rao H, Chen X, Yang H
International Journal of General Medicine 2024, 17:75-84
Published Date: 11 January 2024
Functional Analysis of BRCA1 3’UTR Variants Predisposing to Breast Cancer
Sierra-Díaz DC, Cabrera R, Gonzalez-Vasquez LA, Angulo-Aguado M, Llinás-Caballero K, Fonseca-Mendoza DJ, Contreras-Bravo NC, Restrepo CM, Ortega-Recalde O, Morel A
The Application of Clinical Genetics 2024, 17:57-62
Published Date: 23 May 2024
