Back to Journals » Infection and Drug Resistance » Volume 16
The Biological and Regulatory Role of Type VI Secretion System of Klebsiella pneumoniae
Authors Liu W, Li M, Cao S, Ishaq HM, Zhao H, Yang F , Liu L
Received 27 June 2023
Accepted for publication 30 September 2023
Published 30 October 2023 Volume 2023:16 Pages 6911—6922
DOI https://doi.org/10.2147/IDR.S426657
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Wenke Liu,1,* Min Li,1,* Shiwen Cao,1 Hafiz Muhammad Ishaq,2 Huajie Zhao,1 Fan Yang,1 Liang Liu1
1Department of Pathogenic Biology, School of Basic Medical Sciences, Xinxiang Medical University, Xinxiang, People’s Republic of China; 2Faculty of Veterinary and Animal Sciences, Muhammad Nawaz Shareef University of Agriculture, Multan, Pakistan
*These authors contributed equally to this work
Correspondence: Liang Liu, Email [email protected]
Abstract: Bacteria communicate with their surroundings through diverse secretory systems, and the recently discovered Type VI Secretion System (T6SS) has gained significant attention. Klebsiella pneumoniae (K. pneumoniae), an opportunistic pathogen known for causing severe infections in both hospital and animal settings, possesses this intriguing T6SS. This system equips K. pneumoniae with a formidable armory of protein-based weaponry, enabling the delivery of toxins into neighboring cells, thus granting a substantial competitive advantage. Remarkably, the T6SS has also been associated with K. pneumoniae’s ability to form biofilms and acquire resistance against antibiotics. However, the precise effects of the T6SS on K. pneumoniae’s functions remain inadequately studied, despite research efforts to understand the intricacies of these mechanisms. This comprehensive review aims to provide an overview of the current knowledge regarding the biological functions and regulatory mechanisms of the T6SS in K. pneumoniae.
Keywords: Klebsiella pneumoniae, T6SS, competition, drug resistance, virulence
Introduction
Bacteria inhabit densely populated communities and consistently engage in competition with other bacterial species in order to acquire essential nutrients and occupy adequate space. The bacterial secretory system is a structure present on the surface of bacterial cells that allows bacteria to exchange information with their surrounding environment to obtain nutrients and transport effector proteins. As a result, the secretory system regulates the interaction between bacteria and their hosts.1,2 There are currently nine different secretion systems that have been identified in Gram-negative bacteria, ranging from type I (T1SS) to type IX (T9SS). The T7SS and T8SS predominantly play a role in the secretion of pili and fibers. The T9SS, on the other hand, is a recently discovered secretion system that has only been found in Bacteroides bacteria.3 First identified in V. cholerae by Pukatzki et al in 2006, the Type VI Secretion System (T6SS) is involved in the secretion of hemolysin co-regulatory protein (Hcp) and valine-glycine repeat protein G (VgrG).4 These secretions are associated with cytotoxic effects and require a cluster of intracellular multiplication protein F-related homologous genes.5,6 The initial discovery of T6SS effectors revealed their anti eukaryotic properties, leading to the disruption of the target host cell’s physiological processes.4 However, significant breakthroughs and subsequent advancements in T6SS research have unveiled a predominant antibacterial activity among these effectors. Recent investigations have further expanded our understanding by revealing that certain effectors possess antifungal properties, thus broadening the traditionally defined role of T6SS from solely antibacterial to encompassing antimicrobial capabilities.7,8 Beyond that, Lu et al designed antimicrobial peptides based on the bactericidal effect of T6SS effectors, which provided a new idea for the clinical treatment of multi-drug resistant bacteria.9 T6SS can also facilitate symbiotic or reciprocal relationships between bacteria and hosts, and play cooperative or competitive roles in bacterial interactions.10–12 Additionally, it has been demonstrated that the T6SS also serves as a general secretion system, facilitating the delivery of effector proteins into the extracellular environment to access shared resources, highlighting its versatility.13,14
K. pneumoniae, a Gram-negative opportunistic pathogenic bacterium belonging to the Enterobacteriaceae family, exhibits significant drug resistance. It ranks among the leading causes of nosocomial infections, encompassing conditions such as pneumonia, meningitis, liver abscesses, urinary tract infections (UTIs), wound infections, bacteremia, and sepsis.15,16 Carbapenem-resistant K. pneumoniae (CRKP) infections pose a significant public health threat, particularly in intensive care units (ICUs), where they can lead to higher mortality rates among critically ill and debilitated patients. The financial burden associated with hospitalization and treatment for these infections is a global concern.17,18 Recent studies have shown that T6SS may participate in the pathogenic process and play an important role in the virulence of K. pneumoniae.19–21 Consequently, with the increasing pathogenicity of K. pneumoniae, research on K. pneumoniae T6SS has attracted considerable attention. This article aims to provide a comprehensive review of the fundamental genes, biological functions, and regulatory mechanisms of K. pneumoniae T6SS to facilitate future research on this system.
The Typical T6SS
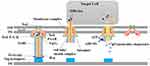
Current literature shows that T6SS is encoded within clusters containing at least 13 conserved core genes (tssA-M), which encode proteins that comprise the basic components of the secretion apparatus.22–24 Based on bioinformatics, these genes were classified into three main categories.25 The genes encode proteins that form the three-part architecture of T6SS, including bacteriophage-like injection apparatus, membrane complex, and molecular chaperone.26–28 The most distinctive feature of T6SS is its bacteriophage-injection device-like structure, which consists of a needle structure, a sheath complex, and a baseplate (Figure 1).29 The needle-like structure of the syringe includes Hcp and VgrG, which are translocator proteins secreted into the extracellular environment.30 In particular, Hcp secretion is commonly considered a marker of T6SS activity.31 In contrast, the VgrG homologous trimer is associated with the gp27/gp5 complex, a spike of the T4 phage.32 The Hcp protein forms the tail of the inner tube with the VgrG trimer.33 The length of sequences in the VgrG family proteins usually varies from approximately 750 to 1200 amino acids. The conserved domains are enclosed within the 750 amino acids. In addition, there is a variable C-terminal region in these proteins that often provides an effector function.34 The spike complex commonly consists of a trimer of VgrG proteins and a spike complex protein (PAAR) containing proline-alanine-alanine-arginine repeats.27,35,36 The needle structure (Hcp-VgrG-PAAR structure) is identified as the only part of T6SS that advances into the target cells. Hence, the T6SS effector is intimately linked with the needle structure.37,38 The cone-shaped PAAR is thought to enhance the sharpness of the T6SS spear, enabling effective penetration. However, the reasons for the necessity of PAAR in T6SS functions among certain bacteria, while being optional in others, are still not clearly understood.39 As an illustration, the effector could either be an appended structural domain fused to a needle component (eg, Hcp, VgrG, or PAAR) or a distinct protein that binds directly or indirectly to Hcp, VgrG, or PAAR.38,40 The sheath complex, constructed from the TssB and TssC subunits, has the capacity to form tubular structures bearing significant resemblance to gp18.41 The baseplate, composed of TssE, TssF, TssG, and TssK, bridges the transmembrane complex with the tail. It plays a pivotal role in triggering the polymerization of the tube/sheath complex and anchoring it to the membrane complex, thus initiating sheath contraction while keeping the entire assembled system solid during sheath contraction.42 It serves as a vital link that connects the tail to the membrane complex and initiates the formation of the needle. Interestingly, these proteins exhibit significant similarity to the baseplate proteins of the T4 bacteriophage, such as gp6, gp7, and gp25.43 Additionally, there is a protein called TssK, which shares certain structural characteristics with receptor-binding proteins found in siphophages. TssK interacts with TssG, and the stability of this interaction is ensured by the presence of TssF. Consequently, the TssKFG wedge complex is formed, representing an early building block of the T6SS baseplate that is analogous to the wedge complex of the T4 baseplate.44 Currently, evidence has demonstrated that the fundamental components of T6SS include three proteins: TssL, TssM, and TssJ, which can collectively form a membrane complex.45 TssM and TssL are located in the inner membrane, while TssJ is located in the outer membrane. TssM is capable of interacting with TssL and TssJ to connect the inner and outer membranes, thus creating a ring-like channel structure that traverses the cell membrane, allowing the VgrG spike and Hcp tube to deliver a variety of effectors to the target cells.46,47 In bacterial secretion systems, ATPase or proton motive force is typically utilized to supply the energy required for the assembly of the secretion apparatus and the transfer of substrates.48 ClpV, a member of the AAA+ (ATPases associated with various cellular activities) protein family, which is part of the HSP100 (Heat Shock Proteins) family, plays a crucial role in this process.36,49 Upon the contraction of T6SS sheath-like structures, the exposed N-terminal structural domain of TssC is recognized by ClpV, leading to the disassembly of the structures into TssB and TssC monomers, a process that relies on ClpV-mediated ATP hydrolysis.36,50 Aside from the aforementioned genes, T6SS also incorporates some non-conserved genes, such as TssA and TagA. TssA is divided into three categories, each performing similar functions in T6SS despite their structural differences.51 TagA, a protein that interacts with TssA, halts the assembly of the sheath and maintains the sheath in an extended conformation.52 However, TagA, the “stopper”, is not conserved within the T6SS gene cluster,29 suggesting that different T6SS-positive strains may control tail-tube/sheath assembly and termination via distinct mechanisms.51–53 Figure 1 provides a schematic representation of the T6SS. Table 1 lists the core genes of T6SS as well as their encoded proteins and functions.
 |
Table 1 Genetic Organization and Coding Proteins of the Bacterial Type VI Secretion System |
T6SS of K. Pneumoniae
In recent years, continuous research has led to the discovery of 13 conserved genes encoded in T6SS loci in various Gram-negative bacteria, including K. pneumoniae.61 Lawlor et al have reported findings suggesting that the new virulence loci of K. pneumoniae appear to be homologous to the Yersinia pestis protein (YPO1467).62 Further evidence supporting this notion was presented by Sarris et al, who found that the protein YPO1467 is annotated as a T6SS core component and seems to be highly homologous (65.5%) with the putative K. pneumoniae protein. Concurrently, they detected 13 conserved genes of typical T6SS in K. pneumoniae. Sarris et al also proposed that a T6SS mechanism may assist the colonization and infection of the host by Klebsiella.63 In an effort to study T6SS more comprehensively, the SecReT6 (Type VI Secretion System Database) and related reports have proposed classifying T6SS clusters into types I, II, III and IV based on gene cluster structure and core component evolutionary relationships.64–67 Type I encompasses six subtypes (ie, I1, I2, I3, I4a, I4b, and I5), whereas type II is specific to the Francisella pathogenicity gene cluster; type III has been identified in Bacteroides. Type IV is newly discovered and represents a primordial system from which extracellular contractile injection systems, phages, and T6SSi−iii have evolved.68 According to SecReT6 data, the complete T6SS cluster in K. pneumoniae belongs to type I2.20,67 However, a comparative genomic analysis using Enterobacter cloacae—which contains a type I2 T6SS gene cluster—and K. pneumoniae revealed that some K. pneumoniae strains identified as containing a type I2 T6SS cluster lacked certain conserved T6SS genes. Notably, there are strains possessing only one unknown T6SS type, but the function of their T6SS clusters remains unclear.25 T6SS core genes are tightly clustered in one or more gene clusters. It has been reported that in some K. pneumoniae strains, T6SS genes are primarily grouped in two loci (eg, NTUH-K2044, Kpn2146, and HS11286); in others, they are found in three loci (eg, Kp52145, MGH78578, and Kp342).20,63,69 To facilitate more systematic T6SS research, Barbosa et al proposed using the TssA-M nomenclature to denote K. pneumoniae core T6SS components; they also reported that the majority of T6SS genes in the K. pneumoniae Kp52145 genome (33 genes, 87%) were clustered in three genomic loci, with only five (13%) being orphan genes. In their analysis of K. pneumoniae HS11286, 25 (86%) of the 29 T6SS genes were clustered in two main loci; only four (14%) were orphan genes.70 However, Barbosa et al’s data on K. pneumoniae T6SS genes were derived from computational analysis, indicating that further experimental studies are required to confirm these predictions.
In summary, the in-depth study of K. pneumoniae has led to the detection of T6SS core genes in its genome. More systematic genetic testing is urgently needed to enable a comprehensive investigation and confirm these computational speculations.
Biological Function of K. Pneumoniae’s T6SS
T6SS Mediates Competition for K. Pneumoniae Participation Among Species
T6SS plays a crucial role in the interspecies competition between K. pneumoniae and the host microbiota.19 Notably, T6SS exhibits robust antibacterial activity. Liu et al carried out competition assays to assess the antibacterial effectiveness of the T6SS in K. pneumoniae strain HS11286.20 The findings revealed that mutant strains deficient in the T6SS apparatus or effector exhibited a diminished killing activity compared to the wild type strain. This research also established K. pneumoniae’s significant advantage in T6SS-mediated interspecies competition with E. coli. Moreover, the study analyzed the T6SS gene cluster on chromosome HS11286, identifying the gene encoding for the effector protein Tle1KP. It was found that the Tli1KP immunoprotein, which is encoded within this gene cluster, could mitigate the toxicity of Tle1KP.20 T6SS in K. pneumoniae also has a role in mediating both intraspecies and interspecies bacterial competition. Research by Storey et al demonstrated that polymyxin B and antimicrobial peptides could stimulate and enhance the expression of T6SS in K. pneumoniae strain Kp52145, which consequently influenced bacterial competition.71 K. pneumoniae Kp52145 strain is particularly noteworthy as it contains highly virulent gene clones associated with human infections and encodes all virulence functions significantly linked to severe community-acquired diseases in humans.17,69 In the context of competition experiments with E. coli, it was observed that the killing activity of the Kp52145 ΔclpV mutant strain against E. coli, was markedly reduced compared to the clpV complementary strain. This suggests that the eradication of E. coli by Kp52145 is T6SS-dependent, and the clpV gene may have a crucial role in this process. Further, the Kp52145 strain was found capable of competing with fungi in a T6SS-dependent manner. This was in stark contrast to the ΔclpV mutant. The protein VgrG4 was identified as a critical player in this competition against fungi. The C-terminal extension of this protein, which contains the DUF2345 domain, was adequate to exhibit antibacterial and anti-eukaryotic effects.71 Yet, the homologous immune protein Sel1E was found to reduce the toxin function of VgrG4.61 Furthermore, the proteins VgrG1, VgrG2, and VgrG4 are essential in the T6SS-mediated killing mechanism of K. pneumoniae. However, the functionality of VgrG2 and VgrG4 is primarily contingent on the bacterial growth conditions.72 In a competition assay involving K. pneumoniae strain NTUH-K2044, Hsieh et al found that IcmF1, IcmF2, and Hcp augmented the bacterium’s antimicrobial activity. Conversely, strains with icmF1/icmF2 mutations demonstrated a decreased competitive ability in vivo.19 T6SS is capable of not only delivering effectors to inhibit the growth of other bacteria, but also of injecting effectors into to eukaryotic cells to promote bacterial survival by targeting the cytoskeleton. A recent investigation has demonstrated that the trans-kingdom antimicrobial T6SS effector VgrG4 from K. pneumoniae triggers the fragmentation of the mitochondrial network.73
In summary, K. pneumoniae effectively leverages T6SS to mediate both intraspecies and interspecies bacterial competition, thereby enhancing its survival. Nevertheless, the precise mechanisms underlying the competitive advantages conferred by T6SS remain elusive and warrant further investigation.
Adhesion, Invasion, and Colonization of K. Pneumoniae Mediated by T6SS
The critical steps in its attack on host cells include adhesion, invasion, and colonization of K. pneumoniae to host cells. T6SS plays a crucial role in bacterial adhesion, invasion, and colonization, thus affecting the ability of K. pneumoniae to infect the host. In K. pneumoniae NTUH-K2044, the deletion of the T6SS gene significantly reduced bactericidal activity, expression of type-1 fimbriae, and epithelial cell adhesion and invasion.19 A study using a Galleria mellonella infection model illustrated that the virulence of Kp52145 mediated by T6SS was orchestrated by three types of VgrG proteins.61,71 The adhesion of K. pneumoniae to eukaryotic cells is linked to the production of type-1 fimbriae. In an experiment involving adhesion and invasion of K. pneumoniae in human colonic Caco-2 epithelial cells, it was discovered that after deleting the structural protein genes icmF1 and icmF2 of T6SS, the expression of type-1 fimbriae was down-regulated, which led to a significant reduction in adhesion and invasion to Caco-2 cells. Moreover, the bacterial load in infected mice’s intestines, spleens, and livers decreased markedly.71 The study conducted by Merciecca et al investigated the role of the Type VI secretion system (T6SS) in K. pneumoniae during long-term gastrointestinal colonization. Their findings revealed that T6SS isogenic mutants exhibited reduced efficiency in colonizing the upper gastrointestinal tract of mice compared to the parental strain over an extended period. Additionally, subsequent investigations demonstrated that T6SS had a negative impact on the richness and resilience of the gut microbiota.74 This suggests that the virulence, bacterial invasion, and intestinal colonization of T6SS icmF1 and icmF2 mutants would be diminished.
In general, T6SS-mediated expression of type-1 fimbriae, cell adhesion, invasion, and in vivo colonization are critical for the onset of K. pneumoniae infection.19 However, the current research on the colonization mechanism of K. pneumoniae T6SS is limited, and more detailed studies must be conducted.
T6SS Affects the Acquisition of Drug-Resistant Genes in K. Pneumoniae
The global misuse of antibiotics has resulted in K. pneumoniae’s multi-drug resistance, leading to significant healthcare challenges. Under the influence of antibiotics, the secretory function of T6SS can be induced, potentially providing a novel concept for the clinical development of targeted drugs for K. pneumoniae. Research on the ST11 carbapenem-resistant K. pneumoniae strain HS11286 demonstrated that clinical abuse of antibiotics might not inhibit its proliferation, but may induce T6SS activity, making it more invasive.20 PCR analysis of drug resistance genes in clinical isolates of K. pneumoniae by Liao et al showed that T6SS-positive strains had more drug resistance genes than T6SS-negative strains.75 Liu et al reported the clinical isolation of carbapenem-producing K. pneumoniae strain HS11286 from sputum samples. They discovered that the T6SS gene cluster on the chromosome of K. pneumoniae can encode the effector protein Hcp, which possesses antibacterial properties.20 Yet, when the bacteria were cultured in LB broth, there was no Hcp in the cells and supernatants. In contrast, Hcp could be detected in the cell components and supernatants of bacteria stimulated by β-lactam antibiotics, indicating that the T6SS secretory function occurred under the influence of antibiotics. Additionally, apramycin was used to determine that not only β-lactam antibiotics can induce T6SS secretion. In the competition experiment of the antibacterial activity of K. pneumoniae HS11286, it was found that the mortality of the non-antibiotic group was nearly two times lower than that of the meropenem or ceftazidime group, suggesting that the killing effect of T6SS activated by intercellular contact may be more potent under antibiotic stress.20 It has been demonstrated that V. cholerae can acquire new T6SS effector genes via horizontal transfer and use them to kill neighboring cells to promote horizontal gene transfer.76 Similarly, K. pneumoniae can acquire new T6SS effector genes via horizontal transfer and natural transformation. They can then be transcribed into proteins and utilized to kill target cells.77 Killing target cells via T6SS-mediated release DNA, which becomes accessible for uptake by naturally competent K. pneumoniae. Since antibiotic stimulation can activate the expression of T6SS, under antibiotic selective pressure, the coexistence of natural competence and T6SS-mediated bacterial killing may contribute to the highly efficient acquisition of drug-resistance genes in K. pneumoniae.77 However, some studies reported opposing results. Zhang et al reported that the drug resistance rate of T6SS-positive bacteria in clinical isolates of K. pneumoniae blood infection was lower than that of T6SS-negative bacteria, and a lower proportion of CRKP was also found in T6SS-positive strains.21 Similar findings were also reported by Liu et al in their study. They investigated a total of 169 K. pneumoniae strains obtained from patients with abscesses. Their results demonstrated that, with the exception of cefazolin and tegacycline, isolates positive for T6SS exhibited a reduced prevalence of antimicrobial resistance to other drugs.78
In conclusion, antibiotics induce the expression and secretion of T6SS, which may make attackers of T6SS-positive strains more aggressive in competitive growth. Simultaneously, the presence of T6SS is advantageous for strains to acquire drug resistance genes more efficiently, providing a competitive edge in biological competition. Nevertheless, some research indicates that the drug resistance rate of T6SS-positive strains is lower than that of T6SS-negative bacteria, and the internal relationship between T6SS and resistance genes in K. pneumoniae requires further discussion.
T6SS Mediates the Virulence of K. Pneumoniae
The toxin delivery routes of T6SS can be categorized into the following two types: VgrG protein enters target cells directly as the effector protein of T6SS, and the Hcp channel transfers small molecules of virulence proteins, which are often not directly associated with T6SS-encoding gene clusters, to target cells.79 Liao et al analyzed the virulence genes of T6SS positive strains in clinical K. pneumoniae isolates, and the results revealed that compared to T6SS-negative strains, T6SS positive strains had a higher frequency of virulence genes (rmpA, fimH, entB, kfu, yetS).75 In a similar study, Liu et al studied the distribution of strain T6SS isolated from 169 liver abscess patients, and the study showed that the positive rate of T6SS was significantly higher in the highly virulent strains.78 The study conducted by Suchanda Dey et al also demonstrated a strong correlation between the T6SS and 76 virulence factors in the highly drug-resistant and epidemic clones of ST147.80 The T6SS gene frequency in clinical isolated K. pneumoniae was higher than that in blood and intestinal isolates and lower than that in pyogenic liver abscess (PLA) isolates.19,81 The virulence of hcp-positive strains may be more potent than that of T6SS-negative strains, and the invasive infection rate of hcp-positive strains is higher than that of T6SS-negative strains.75 Zhang et al investigated isolates obtained from patients with K. pneumoniae-induced blood infections and discovered that T6SS-positive strains were prevalent in patients with bloodstream infections.21 Concurrently, the study indicated that T6SS-positive strains had a high detection rate of virulence genes and were more likely to produce hypervirulent K. pneumoniae (hvKp). K. pneumoniae liver abscess (KPLA) with extrahepatic migratory infections is defined as invasive KPLA (IKPLA). Wang et al conducted a study on the relationship between T6SS and IKPLA. The results demonstrated a significant enrichment of T6SS-related genes in the IKPLA group. Additionally, the detection rate of T6SS-positive strains in the IKPLA group was higher compared to the KPLA group (97.1% versus 78.4%), and the expression level of hcp was notably increased in IKPLA isolates.82
In summary, T6SS-positive K. pneumoniae strains have a high frequency of virulence genes. However, the association between T6SS and the virulence genes of K. pneumoniae and the mechanism of this bacterium’s virulence effect remain unclear. Thus, further research is needed to explore the exact pathway and mechanism.
Regulation of T6SS in K. Pneumoniae
T6SS-positive bacteria can utilize T6SS for bacterial competition, cell invasion, and in vivo colonization. However, the function of T6SS requires the coordinated participation of numerous proteins for assembly and secretion. When bacteria encounter changes in external environmental conditions, they can precisely regulate the transcriptional, translational, and post-translational mechanisms of T6SS expression.83,84 The expression of T6SS is not fixed and can change under certain conditions. Recent studies have shown that the regulation of T6SSs can occur at three different levels, depending on the bacterial species: transcriptional, posttranscriptional, and post translational. Transcriptional regulation involves various factors such as quorum sensing (QS) systems, two-component systems (TCS), alternative sigma factors, and histone-like proteins. Post transcriptional regulation involves the participation of RNA-binding proteins and small regulatory RNAs that control the stability and translation of messenger RNAs (mRNAs). Lastly, post translational regulation mainly relies on the phosphorylation of T6SS structural proteins to regulate the assembly of the system. Nevertheless, the research on the regulation of T6SS in K. pneumoniae has primarily focused on the transcriptional level.28
Temperature Regulation
The negative regulator histone-like nucleoid-structuring protein (H-NS) of T6SS gene transcription can directly bind to the AT-rich region of DNA, acting as a silencer.85 Its activity depends on temperature and osmotic pressure. Barbosa et al hypothesized that the H-NS binding site is widespread and conserved in the T6SS promoter of K. pneumoniae, potentially inhibiting the expression of significant virulence factors such as type 3 fimbriae and biofilms.70,86,87 However, these inferences still need experimental verification. Additionally, H-NS binding sites have been detected upstream of the T6SS gene in three K. pneumoniae species, leading to speculation that H-NS binds to and silences the tssD gene promoter of the T6SS component in strain NTUH-K2044.70 Furthermore, deletion of h-ns increases the transcription of the hcp gene.19 A macrophage infection assay showed that K. pneumoniae H-NS and T6SS expression decreased in the early stage of infection.88
Osmotic Pressure Regulation
PH, osmolarity, and NaCl can regulate the expression of T6SS encoded by hypervirulent K. pneumoniae. The PhoQ periplasmic domain senses acidic pH, divalent cations, and antimicrobial peptides.71 The acidic residues in the periplasmic domain of PhoQ have been confirmed to activate T6SS, increasing the T6SS activity of Kp52145 in co-culture experiments with a competitive strain having active T6SS.71 OmpR, the response regulator of the two-component system (EnvZ-OmpR) with the sensor kinase, has been previously reported to bind to the promoter region of a Yersinia pseudotuberculosis T6SS, participating in bacterial survival under high osmolarity conditions, resistance to deoxycholate, and pH homeostasis.89,90 K. pneumoniae has been shown to use OmpR to respond to osmotic stress by regulating the cdi-GMP signaling pathway, expressing type 3 bacterial fimbriae, and forming biofilms.91
Oxidative Stress Regulation
K. pneumoniae T6SS may also be activated under oxidative stress conditions, aiding the bacteria in processing reactive oxygen species (ROS) and thereby reducing the damage these species can cause to bacterial cells.70,92 OxyR is a conserved oxidative stress response transcriptional regulator, and bacteria detoxify ROS by producing ROS-detoxifying enzymes, repairing DNA, sequestering metal ions, and employing mechanisms involving catalases.70 K. pneumoniae OxyR has been shown to regulate biofilm formation, fimbrial genes, antibiotic resistance, and adhesion to epithelial cells.93,94 Barbosa and Lery et al predicted that most K. pneumoniae T6SS promoter regions have conserved OxyR binding sites.70
Overall, K. pneumoniae T6SS may be activated under oxidative stress conditions, contributing to the bacterial processing of reactive oxygen species. The study also found that OxyR might regulate each gene encoding tubes, sheaths, and ATPase components. It is suggested that at least two signals are required to express K. pneumoniae T6SS. However, due to experimental limitations, many predictions remain unconfirmed.
Other Regulatory Factors
Barbosa and Lery et al also predicted that RcsAB, GcvA, and Fis are regulators of K. pneumoniae T6SS.70 RcsAB is a unique regulatory system that binds an rcsAB box and regulates the K. pneumoniae galF gene, affecting capsule expression and virulence.95,96 GcvA, a transcriptional regulator of the glycine cleavage system, is involved in amino acid metabolism and is required for F. tularensis fitness and full virulence.97 However, GcvA has not been studied in K. pneumoniae. Fis is a transcriptional regulator that responds to changes in the nutritional environment of intestinal bacteria.98 Zhang et al identified BolA, an important virulence regulator in K. pneumoniae, and found that the expression of T6SS related proteins was also down-regulated after BolA deletion. Furthermore, Li reported that the regulation of T6SS involves the negative regulators RcsB and MgrB, as well as the positive regulator PhoPQ. The upregulation of PhoPQ controlled the augmented activity of T6SS in mutants lacking mgrB and rcsB.99 However, it is not clear whether this regulator directly regulates the expression of T6SS related genes.100 Some antibiotics and antimicrobial peptides can also activate and enhance the expression and activity of different K. pneumoniae strains of T6SS.20,71 It has been found that sub-inhibitory concentrations of β-lactam antibiotics (Meropenem 4mg/L or Ceftazidime 32mg/L) induced the expression of T6SS in K. pneumoniae HS11286, while T6SS was inactive in LB medium or M9 medium without antibiotics. In addition, similar results were obtained using apramycin, indicating that not only β-lactam antibiotics can induce T6SS secretion.20
However, the expression of K. pneumoniae T6SS appears to be associated with various environmental regulators. Whether these regulatory mechanisms are conserved in T6SS-positive strains of K. pneumoniae remains to be determined. At the same time, antibiotic misuse makes combating T6SS-positive K. pneumoniae more challenging. Consequently, there is an urgent need to explore the functions and mechanisms of K. pneumoniae T6SS to provide new targets for studying targeted drugs for clinical treatment.
Conclusion
In recent years, the emergence of multi-drug resistance and hvKp has posed a significant challenge to clinical management and treatment. The rapid spread and increasing prevalence of drug-resistant and virulence genes have created an urgent need for novel therapeutic options. As research on K. pneumoniae T6SS has progressed, it has been discovered that this secretion system may be associated with K. pneumoniae’s virulence effects and the propagation of its drug-resistance genes. However, compared with other pathogens, T6SS has not been studied deeply in K. pneumoniae. In this review, we summarize the main role and research progress of T6SS in virulence and resistance and regulation of K. pneumoniae. Current studies have primarily focused on analyzing T6SS gene clusters and their functions in whole-genome sequencing, numerous questions remain unanswered. For instance, what are the molecular mechanisms by which K. pneumoniae T6SS recognizes target cells? As K. pneumoniae T6SS secretes multiple effectors, are they all secreted simultaneously, or are there selective regulatory mechanisms? Are there any new effectors to be discovered? Can K. pneumoniae T6SS benefit neighboring bacteria without being competitive? What is the molecular mechanism by which T6SS mediates K. pneumoniae’s competitive action? Moreover, T6SS mediates multiple biological effects of K. pneumoniae, especially the virulence effects of the bacterium, which may be closely linked to this system. This system’s critical role in bacterial pathogenesis and bacterial-host interactions offers promising prospects. For example, T6SS may become a new target for the treatment of K. pneumoniae infection, leading to the development of new targeted drugs for effective treatment in the face of emerging multi-drug resistant K. pneumoniae due to antibiotic misuse.
Funding
This research was supported by the Doctoral Scientific Research Foundation of Xinxiang Medical University (grant XYBSKYZZ202137), Science and Technology Research Project of Henan Province (grant 182102310553, 222102520036), the Project of Basic Medical College of Xinxiang Medical University (grant JCYXYKY202117).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yu KW, Xue P, Fu Y, Yang L. T6SS mediated stress responses for bacterial environmental survival and host adaptation. Int J Mol Sci. 2021;22(2). doi:10.3390/ijms22020478
2. Le NH, Pinedo V, Lopez J, Cava F, Feldman MF. Killing of Gram-negative and Gram-positive bacteria by a bifunctional cell wall-targeting T6SS effector. Proc Natl Acad Sci USA. 2021;118(40). doi:10.1073/pnas.2106555118
3. Yang M, Zhou X, Bao Y, et al. Comprehensive genomic analysis reveals extensive diversity of type I and type IV secretion systems in Klebsiella pneumoniae. Curr Microbiol. 2023;80(8):270. doi:10.1007/s00284-023-03362-5
4. Pukatzki S, Ma AT, Sturtevant D, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103(5):1528–1533. doi:10.1073/pnas.0510322103
5. Broms JE, Meyer L, Lavander M, Larsson P, Sjostedt A. DotU and VgrG, core components of type VI secretion systems, are essential for Francisella LVS pathogenicity. PLoS One. 2012;7(4):e34639. doi:10.1371/journal.pone.0034639
6. Blondel CJ, Amaya FA, Bustamante P, Santiviago CA, Pezoa D. Identification and distribution of new candidate T6SS effectors encoded in salmonella pathogenicity Island 6. Front Microbiol. 2023;14:1252344. doi:10.3389/fmicb.2023.1252344
7. Trunk K, Peltier J, Liu YC, et al. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol. 2018;3(8):920–931. doi:10.1038/s41564-018-0191-x
8. Fridman CM, Keppel K, Gerlic M, Bosis E, Salomon D. A comparative genomics methodology reveals a widespread family of membrane-disrupting T6SS effectors. Nat Commun. 2020;11(1):1085. doi:10.1038/s41467-020-14951-4
9. Lu W, Lu H, Wang C, Wang G, Dong W, Tan C. Effectors of the type VI secretion system have the potential to be modified into antimicrobial peptides. Microbiol Spectr. 2023;11(4):e0030823. doi:10.1128/spectrum.00308-23
10. Decoin V, Barbey C, Bergeau D, et al. A type VI secretion system is involved in Pseudomonas fluorescens bacterial competition. PLoS One. 2014;9(2):e89411. doi:10.1371/journal.pone.0089411
11. Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe. 2010;8(1):2–6. doi:10.1016/j.chom.2010.06.012
12. Tashiro Y, Yawata Y, Toyofuku M, Uchiyama H, Nomura N. Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 2013;28(1):13–24. doi:10.1264/jsme2.ME12167
13. Yang X, Liu H, Zhang Y, Shen X. Roles of type VI secretion system in transport of metal ions. Front Microbiol. 2021;12:756136. doi:10.3389/fmicb.2021.756136
14. Kanarek K, Fridman CM, Bosis E, Salomon D. The RIX domain defines a class of polymorphic T6SS effectors and secreted adaptors. Nat Commun. 2023;14(1):4983. doi:10.1038/s41467-023-40659-2
15. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
16. Ahmadi M, Ranjbar R, Behzadi P, Mohammadian T. Virulence factors, antibiotic resistance patterns, and molecular types of clinical isolates of Klebsiella Pneumoniae. Expert Rev Anti Infect Ther. 2022;20(3):463–472. doi:10.1080/14787210.2022.1990040
17. Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA. 2015;112(27):E3574–3581. doi:10.1073/pnas.1501049112
18. Karampatakis T, Tsergouli K, Behzadi P. Carbapenem-resistant Klebsiella pneumoniae: virulence factors, molecular epidemiology and latest updates in treatment options. Antibiotics. 2023;12:2.
19. Hsieh PF, Lu YR, Lin TL, Lai LY, Wang JT. Klebsiella pneumoniae type VI secretion system contributes to bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization. J Infect Dis. 2019;219(4):637–647. doi:10.1093/infdis/jiy534
20. Liu L, Ye M, Li X, et al. Identification and characterization of an antibacterial type VI secretion system in the carbapenem-resistant strain Klebsiella pneumoniae HS11286. Front Cell Infect Microbiol. 2017;7:442. doi:10.3389/fcimb.2017.00442
21. Zhang Y, Xu Y, Huang Y. Virulence genotype and correlation of clinical severeness with presence of the type VI secretion system in Klebsiella pneumoniae isolates causing bloodstream infections. Infect Drug Resist. 2022;15:1487–1497. doi:10.2147/IDR.S353858
22. Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104. doi:10.1186/1471-2164-10-104
23. Zheng J, Leung KY. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol. 2007;66(5):1192–1206. doi:10.1111/j.1365-2958.2007.05993.x
24. Singh RP, Kumari K. Bacterial type VI secretion system (T6SS): an evolved molecular weapon with diverse functionality. Biotechnol Lett. 2023;45(3):309–331. doi:10.1007/s10529-023-03354-2
25. Li W, Liu X, Tsui W, et al. Identification and comparative genomic analysis of type VI secretion systems and effectors in Klebsiella pneumoniae. Front Microbiol. 2022;13:853744. doi:10.3389/fmicb.2022.853744
26. Cascales E, Cambillau C. Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci. 2012;367(1592):1102–1111. doi:10.1098/rstb.2011.0209
27. Navarro-Garcia F, Ruiz-Perez F, Cataldi A, Larzabal M. Type VI secretion system in pathogenic Escherichia coli: structure, role in virulence, and acquisition. Front Microbiol. 2019;10:1965. doi:10.3389/fmicb.2019.01965
28. Hespanhol JT, Nobrega-Silva L, Bayer-Santos E. Regulation of type VI secretion systems at the transcriptional, posttranscriptional and posttranslational level. Microbiology. 2023;169(8). doi:10.1099/mic.0.001376
29. Nguyen VS, Douzi B, Durand E, Roussel A, Cascales E, Cambillau C. Towards a complete structural deciphering of Type VI secretion system. Curr Opin Struct Biol. 2018;49:77–84. doi:10.1016/j.sbi.2018.01.007
30. Cianfanelli FR, Alcoforado Diniz J, Guo M, De Cesare V, Trost M, Coulthurst SJ. VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 2016;12(6):e1005735. doi:10.1371/journal.ppat.1005735
31. Weber BS, Kinsella RL, Harding CM, Feldman MF. The secrets of Acinetobacter secretion. Trends Microbiol. 2017;25(7):532–545. doi:10.1016/j.tim.2017.01.005
32. Leiman PG, Basler M, Ramagopal UA, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106(11):4154–4159. doi:10.1073/pnas.0813360106
33. Douzi B, Logger L, Spinelli S, Blangy S, Cambillau C, Cascales E. Structure-function analysis of the C-terminal domain of the type VI secretion TssB tail sheath subunit. J Mol Biol. 2018;430(3):297–309. doi:10.1016/j.jmb.2017.11.015
34. Do Nascimento Soares T, Silva Valadares V, Cardoso Amorim G, et al. The C-terminal extension of VgrG4 from Klebsiella pneumoniae remodels host cell microfilaments. Proteins. 2022;90(9):1655–1668. doi:10.1002/prot.26344
35. Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500(7462):350–353. doi:10.1038/nature12453
36. Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483(7388):182–186. doi:10.1038/nature10846
37. Howard SA, Furniss RCD, Bonini D, et al. The breadth and molecular basis of hcp-driven type VI Secretion system effector delivery. mBio. 2021;12(3):e0026221. doi:10.1128/mBio.00262-21
38. Durand E, Cambillau C, Cascales E, Journet L. VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 2014;22(9):498–507. doi:10.1016/j.tim.2014.06.004
39. Liang X, Zheng HY, Zhao YJ, et al. VgrG Spike Dictates PAAR Requirement for the assembly of the type VI secretion system. J Bacteriol. 2023;205(2):e0035622. doi:10.1128/jb.00356-22
40. Alcoforado Diniz J, Liu YC, Coulthurst SJ. Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cell Microbiol. 2015;17(12):1742–1751. doi:10.1111/cmi.12532
41. Chen L, Zou Y, She P, Wu Y. Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol Res. 2015;172:19–25. doi:10.1016/j.micres.2015.01.004
42. Cherrak Y, Rapisarda C, Pellarin R, et al. Biogenesis and structure of a type VI secretion baseplate. Nat Microbiol. 2018;3(12):1404–1416. doi:10.1038/s41564-018-0260-1
43. Nguyen VS, Logger L, Spinelli S, et al. Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex. Nat Microbiol. 2017;2:17103. doi:10.1038/nmicrobiol.2017.103
44. Cherrak Y, Filella-Merce I, Schmidt V, et al. Inhibiting type VI secretion system activity with a biomimetic peptide designed to target the baseplate wedge complex. mBio. 2021;12(4):e0134821. doi:10.1128/mBio.01348-21
45. Rapisarda C, Cherrak Y, Kooger R, et al. In situ and high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex. EMBO J. 2019;38(10). doi:10.15252/embj.2018100886
46. Yin M, Yan Z, Li X. Architecture of type VI secretion system membrane core complex. Cell Res. 2019;29(3):251–253. doi:10.1038/s41422-018-0130-7
47. Cherrak Y, Flaugnatti N, Durand E, Journet L, Cascales E. Structure and activity of the type VI secretion system. Microbiol Spectr. 2019;7(4). doi:10.1128/microbiolspec.PSIB-0031-2019
48. Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28(4):315–325. doi:10.1038/emboj.2008.269
49. Schlieker C, Zentgraf H, Dersch P, Mogk A. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol Chem. 2005;386(11):1115–1127. doi:10.1515/BC.2005.128
50. Kapitein N, Bonemann G, Pietrosiuk A, et al. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol. 2013;87(5):1013–1028. doi:10.1111/mmi.12147
51. Dix SR, Owen HJ, Sun R, et al. Structural insights into the function of type VI secretion system TssA subunits. Nat Commun. 2018;9(1):4765. doi:10.1038/s41467-018-07247-1
52. Santin YG, Doan T, Lebrun R, Espinosa L, Journet L, Cascales E. In vivo TssA proximity labelling during type VI secretion biogenesis reveals TagA as a protein that stops and holds the sheath. Nat Microbiol. 2018;3(11):1304–1313. doi:10.1038/s41564-018-0234-3
53. Zoued A, Durand E, Santin YG, et al. TssA: the cap protein of the Type VI secretion system tail. Bioessays. 2017;39(10). doi:10.1002/bies.201600262
54. Planamente S, Salih O, Manoli E, Albesa-Jove D, Freemont PS, Filloux A. TssA forms a gp6-like ring attached to the type VI secretion sheath. EMBO J. 2016;35(15):1613–1627. doi:10.15252/embj.201694024
55. Stietz MS, Liang X, Li H, Zhang X, Dong TG. TssA-TssM-TagA interaction modulates type VI secretion system sheath-tube assembly in Vibrio cholerae. Nat Commun. 2020;11(1):5065. doi:10.1038/s41467-020-18807-9
56. Pira H, Risdian C, Musken M, Schupp PJ, Wink J. Photobacterium arenosum WH24, isolated from the gill of pacific oyster Crassostrea gigas from the North Sea of Germany: co-cultivation and prediction of virulence. Curr Microbiol. 2022;79(8):219. doi:10.1007/s00284-022-02909-2
57. Brunet YR, Zoued A, Boyer F, Douzi B, Cascales E. The Type VI Secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet. 2015;11(10):e1005545. doi:10.1371/journal.pgen.1005545
58. Liebl D, Robert-Genthon M, Job V, Cogoni V, Attree I. Baseplate component TssK and spatio-temporal assembly of T6SS in pseudomonas aeruginosa. Front Microbiol. 2019;10:1615. doi:10.3389/fmicb.2019.01615
59. Zoued A, Cassaro CJ, Durand E, et al. Structure-function analysis of the TssL cytoplasmic domain reveals a new interaction between the type VI secretion baseplate and membrane complexes. J Mol Biol. 2016;428(22):4413–4423. doi:10.1016/j.jmb.2016.08.030
60. Zheng HY, Yang L, Dong T. More than just a spearhead: diverse functions of PAAR for assembly and delivery of toxins of the contractile injection systems. mSystems. 2021;6(6):e0138621. doi:10.1128/msystems.01386-21
61. Y G, J L, W H. Advances in Klebsiella pneumoniae and Acinetobacter baumannii type VI secretion systems. Chin J Microbiol Immunol. 2021;41(08):640–644.
62. Lawlor MS, Hsu J, Rick PD, Miller VL. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol. 2005;58(4):1054–1073. doi:10.1111/j.1365-2958.2005.04918.x
63. Sarris PF, Zoumadakis C, Panopoulos NJ, Scoulica EV. Distribution of the putative type VI secretion system core genes in Klebsiella spp. Infect Genet Evol. 2011;11(1):157–166. doi:10.1016/j.meegid.2010.09.006
64. Barret M, Egan F, Fargier E, Morrissey JP, O’Gara F. Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology. 2011;157(Pt 6):1726–1739. doi:10.1099/mic.0.048645-0
65. Barret M, Egan F, O’Gara F. Distribution and diversity of bacterial secretion systems across metagenomic datasets. Environ Microbiol Rep. 2013;5(1):117–126. doi:10.1111/j.1758-2229.2012.00394.x
66. Russell AB, Wexler AG, Harding BN, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16(2):227–236. doi:10.1016/j.chom.2014.07.007
67. Zhang J, Guan J, Wang M, et al. SecReT6 update: a comprehensive resource of bacterial Type VI Secretion Systems. Sci China Life Sci. 2023;66(3):626–634. doi:10.1007/s11427-022-2172-x
68. Bock D, Medeiros JM, Tsao HF, et al. In situ architecture, function, and evolution of a contractile injection system. Science. 2017;357(6352):713–717. doi:10.1126/science.aan7904
69. Lery LM, Frangeul L, Tomas A, et al. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 2014;12:41. doi:10.1186/1741-7007-12-41
70. Barbosa VAA, Lery LMS. Insights into Klebsiella pneumoniae type VI secretion system transcriptional regulation. BMC Genomics. 2019;20(1):506. doi:10.1186/s12864-019-5885-9
71. Storey D, McNally A, Astrand M, et al. Klebsiella pneumoniae type VI secretion system-mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PLoS Pathog. 2020;16(3):e1007969. doi:10.1371/journal.ppat.1007969
72. Fitzsimons TC, Lewis JM, Wright A, et al. Identification of novel Acinetobacter baumannii type VI Secretion system antibacterial effector and immunity pairs. Infect Immun. 2018;86(8). doi:10.1128/IAI.00297-18
73. Sa-Pessoa J, Lopez-Montesino S, Przybyszewska K, et al. A trans-kingdom T6SS effector induces the fragmentation of the mitochondrial network and activates innate immune receptor NLRX1 to promote infection. Nat Commun. 2023;14(1):871. doi:10.1038/s41467-023-36629-3
74. Merciecca T, Bornes S, Nakusi L, et al. Role of Klebsiella pneumoniae Type VI secretion system (T6SS) in long-term gastrointestinal colonization. Sci Rep. 2022;12(1):16968. doi:10.1038/s41598-022-21396-w
75. Liao W, Huang HH, Huang QS, et al. Distribution of type VI secretion system (T6SS) in clinical Klebsiella pneumoniae strains from a Chinese hospital and its potential relationship with virulence and drug resistance. Microb Pathog. 2022;162:105085. doi:10.1016/j.micpath.2021.105085
76. Thomas J, Watve SS, Ratcliff WC, Hammer BK. Horizontal gene transfer of functional type VI killing genes by natural transformation. mBio. 2017;8(4). doi:10.1128/mBio.00654-17
77. Chen F, Zhang W, Schwarz S, et al. Genetic characterization of an MDR/virulence genomic element carrying two T6SS gene clusters in a clinical Klebsiella pneumoniae isolate of swine origin. J Antimicrob Chemother. 2019;74(6):1539–1544. doi:10.1093/jac/dkz093
78. Liu P, Yang A, Tang B, et al. Molecular epidemiology and clinical characteristics of the type VI secretion system in Klebsiella pneumoniae causing abscesses. Front Microbiol. 2023;14:1181701. doi:10.3389/fmicb.2023.1181701
79. Kapitein N, Mogk A. Deadly syringes: type VI secretion system activities in pathogenicity and interbacterial competition. Curr Opin Microbiol. 2013;16(1):52–58. doi:10.1016/j.mib.2012.11.009
80. Dey S, Gaur M, Sykes EME, et al. Unravelling the evolutionary dynamics of high-risk Klebsiella pneumoniae ST147 clones: insights from comparative pangenome analysis. Genes. 2023;14(5):1037. doi:10.3390/genes14051037
81. Zhou M, Lan Y, Wang S, et al. Epidemiology and molecular characteristics of the type VI secretion system in Klebsiella pneumoniae isolated from bloodstream infections. J Clin Lab Anal. 2020;34(11):e23459. doi:10.1002/jcla.23459
82. Wang H, Guo Y, Liu Z, Chang Z. The type VI secretion system contributes to the invasiveness of liver abscess caused by Klebsiella pneumoniae. J Infect Dis. 2023;228:1127–1136. doi:10.1093/infdis/jiad166
83. Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and regulation of the type VI secretion system. Annu Rev Microbiol. 2012;66:453–472. doi:10.1146/annurev-micro-121809-151619
84. Miyata ST, Bachmann V, Pukatzki S. Type VI secretion system regulation as a consequence of evolutionary pressure. J Med Microbiol. 2013;62(Pt 5):663–676. doi:10.1099/jmm.0.053983-0
85. Ali SS, Xia B, Liu J, Navarre WW. Silencing of foreign DNA in bacteria. Curr Opin Microbiol. 2012;15(2):175–181. doi:10.1016/j.mib.2011.12.014
86. Ares MA, Fernandez-Vazquez JL, Rosales-Reyes R, et al. H-NS nucleoid protein controls virulence features of Klebsiella pneumoniae by regulating the expression of type 3 Pili and the capsule polysaccharide. Front Cell Infect Microbiol. 2016;6:13. doi:10.3389/fcimb.2016.00013
87. Ares MA, Fernandez-Vazquez JL, Pacheco S, et al. Additional regulatory activities of MrkH for the transcriptional expression of the Klebsiella pneumoniae mrk genes: antagonist of H-NS and repressor. PLoS One. 2017;12(3):e0173285. doi:10.1371/journal.pone.0173285
88. Bent ZW, Poorey K, LaBauve AE, Hamblin R, Williams KP, Meagher RJ. A rapid spin column-based method to enrich pathogen transcripts from eukaryotic host cells prior to sequencing. PLoS One. 2016;11(12):e0168788. doi:10.1371/journal.pone.0168788
89. Gueguen E, Durand E, Zhang XY, d’Amalric Q, Journet L, Cascales E. Expression of a Yersinia pseudotuberculosis type VI secretion system is responsive to envelope stresses through the OmpR transcriptional activator. PLoS One. 2013;8(6):e66615. doi:10.1371/journal.pone.0066615
90. Zhang W, Wang Y, Song Y, et al. A type VI secretion system regulated by OmpR in Yersinia pseudotuberculosis functions to maintain intracellular pH homeostasis. Environ Microbiol. 2013;15(2):557–569. doi:10.1111/1462-2920.12005
91. Lin TH, Chen Y, Kuo JT, et al. Phosphorylated OmpR is required for type 3 fimbriae expression in Klebsiella pneumoniae under hypertonic conditions. Front Microbiol. 2018;9:2405. doi:10.3389/fmicb.2018.02405
92. Wong Fok Lung T, Charytonowicz D, Beaumont KG, et al. Klebsiella pneumoniae induces host metabolic stress that promotes tolerance to pulmonary infection. Cell Metab. 2022;34(5):761–774 e769. doi:10.1016/j.cmet.2022.03.009
93. Hennequin C, Forestier C. oxyR, a LysR-type regulator involved in Klebsiella pneumoniae mucosal and abiotic colonization. Infect Immun. 2009;77(12):5449–5457. doi:10.1128/IAI.00837-09
94. Srinivasan VB, Mondal A, Venkataramaiah M, Chauhan NK, Rajamohan G. Role of oxyRKP, a novel LysR-family transcriptional regulator, in antimicrobial resistance and virulence in Klebsiella pneumoniae. Microbiology. 2013;159(Pt 7):1301–1314. doi:10.1099/mic.0.065052-0
95. Peng D, Li X, Liu P, et al. Transcriptional regulation of galF by RcsAB affects capsular polysaccharide formation in Klebsiella pneumoniae NTUH-K2044. Microbiol Res. 2018;216:70–78. doi:10.1016/j.micres.2018.08.010
96. Su K, Zhou X, Luo M, et al. Genome-wide identification of genes regulated by RcsA, RcsB, and RcsAB phosphorelay regulators in Klebsiella pneumoniae NTUH-K2044. Microb Pathog. 2018;123:36–41. doi:10.1016/j.micpath.2018.06.036
97. Brown MJ, Russo BC, O’Dee DM, Schmitt DM, Nau GJ. The contribution of the glycine cleavage system to the pathogenesis of Francisella tularensis. Microbes Infect. 2014;16(4):300–309. doi:10.1016/j.micinf.2013.12.003
98. Mallik P, Pratt TS, Beach MB, Bradley MD, Undamatla J, Osuna R. Growth phase-dependent regulation and stringent control of fis are conserved processes in enteric bacteria and involve a single promoter (fis P) in Escherichia coli. J Bacteriol. 2004;186(1):122–135. doi:10.1128/JB.186.1.122-135.2004
99. Li L, Ma J, Cheng P, et al. Roles of two-component regulatory systems in Klebsiella pneumoniae: regulation of virulence, antibiotic resistance, and stress responses. Microbiol Res. 2023;272:127374. doi:10.1016/j.micres.2023.127374
100. Zhang F, Yan X, Bai J, et al. Identification of the BolA protein reveals a novel virulence factor in K. pneumoniae that contributes to survival in host. Microbiol Spectr. 2022;10(5):e0037822. doi:10.1128/spectrum.00378-22
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Epidemiology, Drug Resistance, and Risk Factors for Mortality Among Hematopoietic Stem Cell Transplantation Recipients with Hospital-Acquired Klebsiella pneumoniae Infections: A Single-Center Retrospective Study from China
Liu YF, Liu Y, Chen X, Jia Y
Infection and Drug Resistance 2022, 15:5011-5021
Published Date: 30 August 2022
Genetic and Phenotypic Characteristics of Carbapenem-Resistant Klebsiella pneumoniae Isolates from a Tertiary Hospital in Beijing
Ni Q, Yao X, Li J, Ma J, Wang K, Liu X, Li P, Yang L, Li P, Li S
Infection and Drug Resistance 2022, 15:7503-7508
Published Date: 19 December 2022
Long Term Characteristics of Clinical Distribution and Resistance Trends of Carbapenem-Resistant and Extended-Spectrum β-Lactamase Klebsiella pneumoniae Infections: 2014–2022
Wang N, Zhan M, Wang T, Liu J, Li C, Li B, Han X, Li H, Liu S, Cao J, Zhong X, Lei C, Zhang W, Zhang Z
Infection and Drug Resistance 2023, 16:1279-1295
Published Date: 4 March 2023
Molecular Profiling of a Multi-Strain Hypervirulent Klebsiella pneumoniae Infection Within a Single Patient
Cao H, Liang S, Zhang C, Liu B, Fei Y
Infection and Drug Resistance 2023, 16:1367-1380
Published Date: 11 March 2023
Clinical Characteristics, Drug Resistance, and Risk Factors for Death of Klebsiella pneumoniae Infection in Patients with Acute Pancreatitis: A Single-Center Retrospective Study from China
Jia Y, Liu Y, Huang Y, Wang J, Wang H, Tan S, Shi Y, Wang Q, Peng J
Infection and Drug Resistance 2023, 16:5039-5053
Published Date: 7 August 2023