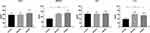Back to Journals » Journal of Inflammation Research » Volume 16
RNA Sequencing and Bioinformatics Analysis to Reveal Potential Biomarkers in Patients with Combined Allergic Rhinitis and Asthma Syndrome
Authors Mao ZD, Liu ZG, Qian Y, Shi YJ, Zhou LZ , Zhang Q , Qi CJ
Received 5 September 2023
Accepted for publication 7 December 2023
Published 19 December 2023 Volume 2023:16 Pages 6211—6225
DOI https://doi.org/10.2147/JIR.S438758
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Adam D Bachstetter
Zheng-Dao Mao,1,* Zhi-Guang Liu,1,* Yan Qian,1 Yu-Jia Shi,1 Lian-Zheng Zhou,1 Qian Zhang,1 Chun-Jian Qi2
1Department of Respiratory and Critical Care Medicine, Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University, Changzhou, People’s Republic of China; 2Central Laboratory, Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University, Changzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chun-Jian Qi, Laboratory of Oncology, Changzhou Second People’s Hospital, Changzhou Medical Center, Nanjing Medical University, Changzhou, 213003, People’s Republic of China, Tel +86-519-81087391, Email [email protected] Qian Zhang, Department of Respiratory and Critical Care Medicine, Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University, No. 29 Xinlong Road, Changzhou, 213003, People’s Republic of China, Tel +86-519-81087391, Email [email protected]
Introduction: Combined allergic rhinitis and asthma syndrome (CARAS) is a concurrent clinical or subclinical allergic symptom of diseases of the upper and lower respiratory tract. This study is the first to explore the expression profiles of mRNA, lncRNA, and circRNA in CARAS using RNA sequencing, which may provide insight into the mechanisms underlying CARAS.
Material and Methods: Whole blood samples from nine participants (three CARAS patients, three AR patients, and three normal control participants) were subjected to perform RNA sequencing, followed by identification of differentially expressed lncRNAs (DElncRNAs), circRNAs (DEcircRNAs) and mRNAs (DEmRNAs). Then, lncRNA/circRNA-mRNA regulatory pairs were constructed, followed by functional analysis, immune infiltration analysis, drug prediction, and expression validation with RT-qPCR and ELISA.
Results: The results showed that 61 DEmRNAs, 23 DElncRNAs and 3 DEcircRNAs may be related to the occurrence and development of CARAS. KRT8 may be implicated in the development of AR into CARAS. Three immunity-related mRNAs (IDO1, CYSLTR2, and TEC) and two hypoxia-related mRNAs (TKTL1 and VLDLR) were associated with the occurrence and development of CARAS. TEC may be considered a drug target for Dasatinib in treating CARAS. Several lncRNA/circRNA-mRNA regulatory pairs were identified in CARAS, including LINC00452/MIR4280HG/hsa_circ_0007272/hsa_circ_0070934-CLC, HEATR6-DT/LINC00639/LINC01783/hsa_circ_0008903-TEC, RP11-71L14.3-IDO1/SMPD3, RP11-178F10.2-IDO1/HRH4, and hsa_circ_0008903-CYSLTR2, which may indicate potential regulatory effects of lncRNAs/circRNAs in CARAS. Dysregulated levels of immune cell infiltration may be closely related to CARAS.
Conclusion: The regulating effect of lncRNA/circRNA-immunity/hypoxia-related mRNA regulatory pairs may be involved in the occurrence and development of CARAS.
Keywords: combined allergic rhinitis and asthma syndrome, lncRNA, circRNA, immunity, hypoxia
Introduction
Combined allergic rhinitis and asthma syndrome (CARAS), a new terminology introduced by the World Allergy Organization (WAO) in 2004, is an allergic reaction that occurs in the respiratory tract, including upper respiratory tract allergy (allergic rhinitis, AR) and lower respiratory tract allergy (asthma, AS).1,2 The incidence of AS in patients with AR is reported to be higher than that in normal populations.3 It has been proved that AR and AS are both manifestations of a single inflammatory process.4 AR is an infectious inflammatory disease of the nasal mucosa (mainly mediated by IgE).5 AS is a chronic inflammatory disease of the airway, and the occurrence and development of AS involves various cells and cellular components.6–8 The continuity of the nasal cavity and bronchus in anatomical structure and physiological function determines the relationship between AR and AS. In addition, rhinosinusitis, a disease similar to AR, often occurs in conjunction with AS and has been associated with severity of AS.9 In the vast majority of cases, the immune pathogenesis of the inflammatory process underlying AS and chronic rhinosinusitis is characterized by type 2 inflammation, which involves the process of cellular infiltration.10 At present, researchers working on CARAS are focusing on mechanisms underlying local pathogenesis, such as the synthesis mechanism of immunoglobulin E (IgE) in the local respiratory tract and the differentiation mechanism of selective T cells.5,11 These local mechanisms determine whether the inflammatory response to inhaling allergens is expressed in the upper or lower respiratory tract.
In addition, the pathogenesis of CARAS is associated with complex genetic background and environmental factors. Barrier systems such as airway epithelial cells receive stimulation, secrete mediators, transmit signals, activate innate immune system cells in the local tissue microenvironment, such as innate lymphoid cells (ILC), and many effector molecules are up-regulated or down-regulated.1,12 These changes in the immune microenvironment interact with the acquired immune system to start the disease process. The involvement of epigenome and mucosal microecological changes has been reported in CARAS initiation, development, and outcome.13,14 A deeper understanding of CARAS gene expression changes can help to shift from selecting treatment according to the patient’s disease phenotype to selecting patients suitable for treatment according to the disease mechanism. Hence, more reliable biomarkers are needed to select and evaluate treatment options.
Non-coding RNAs, as exceptions of central dogma, have been reported over the past decades.15 They have been shown to play an important role in AR and AS. LncRNA FR215775 may encourage allergic inflammation by regulating the differentiation of Th2 cells in AR mice.16 LncRNA HCP5 participates in AR by promoting Tregs differentiation and proliferation.17 Hsa_circRNA_404013 had been reported as a potential diagnostic biomarker for patients with AR.18 LncRNA MALAT1 promotes the development of AS by promoting the proliferation and migration of airway smooth muscle cells.19 Liang et al suggested that circS100A11 may serve as a potential therapeutic and diagnostic target in children with AS.20 However, their exact effects in the development of CARAS remain unclear.
Therefore, in this study, we performed RNA sequencing on whole blood samples from CARAS patients, AR patients, and normal participants, followed by the identification of differentially expressed mRNAs (DEmRNAs), lncRNAs (DElncRNAs), and circRNAs (DEcircRNAs). Then, lncRNA/circRNA-mRNA regulatory pairs were constructed to further explore the roles of DElncRNAs/DEcircRNAs in the development of CARAS and their interactions with potential targeted mRNAs in CARAS. By doing this, we hope to discover potential molecular markers associated with the occurrence and development of AR into CARAS.
Materials and Methods
Participants
The patients with the allergic symptoms in upper and lower respiratory tract including rhinocnesmus, sneeze frequently, runny watery nose, rhinobyon, cough, and wheeze were enrolled as CARAS; the patients with the typical symptoms including paroxysmal sneeze, watery snot, rhinobyon, and rhinocnesmus were enrolled as AR; the healthy individuals with no above clinical symptoms were considered as normal controls. AR was diagnosed according to the Allergic Rhinitis Management Path (Chinese version) of Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines-2019.21 The mandatory inclusion criteria of CARAS patients were as follows: (1) based on clinical criteria for moderate-to-severe persistent AR in recent 2 years, patients were diagnosed with moderate persistent AS (such as runny nose, pruritus, sneezing, and congestion); (2) patients were sensitive to house dust mite (HDM), including Dermatophagoides pteronyssinus or D. farinae skin prick test positive (wheal diameter > 6 mm) and/or CAP-Pharmacia score > class 2; (3) forced expiratory volume in 1 s (FEV1) is within the normal range. The inclusion criteria for AR patients were as follows: (1) patients had more than two clinical symptoms, including sneezing, water-like runny nose, nasal congestion, and nasal itching. Furthermore, the symptoms should last or accumulate for more than 1 hr every day, which can be accompanied by eye symptoms (such as eye itching and conjunctival congestion); (2) patients had clinical signs of common nasal mucosa pale, edema, watery nasal secretion based on nasal endoscopy and sinus computed tomography (CT); (3) patients had positive skin prick test. Exclusion criteria included (1) patients with moderate persistent AS or anatomic abnormalities of the upper respiratory tract, (2) those undergoing chronic treatment with systemic steroids or with systemic immunological disorders, (3) patients with wheezing, shortness of breath, chest tightness, and cough due to other diseases. Healthy individuals with nasal diseases or a history of AS were excluded. The whole blood sample was collected from participants and used for RNA sequencing.
RNA Isolation, Sequencing, and Differential Expression Analysis
The whole blood sample was collected from participants with PAXgene® RNA Blood tubes and stored at −80°C prior to processing. Total RNA was isolated from the samples using PAXgene Blood RNA Kit. DNA fragments in total RNA samples were digested using DNase I. The reaction products were purified and recovered using magnetic beads. After removing the rRNA with the commercial kit, thermal interruption was performed in the polymerase chain reaction (PCR) instrument. After adding primers, a strand of cDNA was synthesized according to the corresponding procedure. Then, cDNA was doubly linked and digested by uracil-N-glycosylase (UNG) digestion reaction system through the synthesis of reverse transcription double-strand and terminal repair. After purification and recovery by magnetic beads, the quality of the RNA library was detected by Agilent 2100 Bioanalyzer (Agilent Technologies, USA) and ABI StepOnePlus real-time PCR system (Thermo Fisher Scientific, USA). Finally, the qualified library was denatured into a single chain by NaOH and diluted to optimal concentration on FlowCell, hybridized with the connector on FlowCell. Hybridized molecules were amplified by bridging PCR on cBot and sequenced using Illumina HiSeq X-Ten platform with library strategy of PE150. The adapter sequence reads were removed to ensure the accuracy of subsequent biological information analysis. Those bases with mass <20 and containing non AGCT at the 5’ end before shearing were trimmed. The reads with N greater than 10% was deleted. The small fragments with a length of less than 25 bp after the trimming of the adapter and mass were deleted. After quality control, statistical analysis of the data was performed. The reads were aligned with the GRCh38 genome by Hisat2 (v 2.1.0) software. The circRNAs were predicted by CIRI2. DElncRNAs, DEcircRNAs, and DEmRNAs between N, AR, and CARAS were identified under the screening criteria of |log2 Fold Change (FC)| >1 and p<0.05. The immune and hypoxia-related DEmRNAs were identified from ImmPort and Molecular Signatures Database (MSigDB) databases, respectively.
Functional Analysis of DEmRNAs
To investigate the function of DEmRNAs, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed with the David database. Gene Set Enrichment Analysis (GSEA) was also performed by “cluster profiler” package in R. Significantly enriched KEGG and GSEA terms were identified under the screening criterion of p<0.05. In addition, protein–protein interaction (PPI) and the transcription factor (TF) regulatory network were constructed. PPI network was constructed with the STRING database. TF subsets were obtained from the Cistrome (http://cistrome.org/) database. Corresponding TF expression values were obtained from expression matrix data. Pearson’s correlation coefficient was used to analyze the relationships between TFs and mRNAs. The TFs were identified under the threshold value of |r|>0.9 and p<0.05.
Identification of lncRNA/circRNA-mRNA Regulatory Pairs
Target mRNA prediction of lncRNA is divided into the following two parts: cis-regulation and trans-regulation. The Pearson correlation coefficient method was used to analyze the relationship between lncRNA and target mRNA under the threshold value of |r|≥0.8 and p<0.05 for cis-regulation and |r|≥0.9 and p<0.05 for trans-regulation. In addition, Pearson correlation coefficient method was applied to analyze the relationship between circRNA and mRNA under the screening criteria of |r|>0.8 and p<0.05. Cytoscape software was used to construct the lncRNA/circRNA-mRNA regulatory networks.
Immune Infiltration Analysis and Drug Prediction
CIBERSORT algorithm was performed to evaluate immune cell infiltration in N, AR, and CARAS. This algorithm can transform the normalized gene expression matrix into the composition of infiltrating immune cells. In addition, potential drugs related to DEmRNAs were predicted with the DGIdb database.
Validation in Real-Time qPCR (RT-qPCR) and Gene Expression Omnibus (GEO) Database
First, whole blood samples were collected from 12 patients with CARAS, 8 patients with AR, and 9 normal control participants. A total of 40 μL RNA was subjected for RT-qPCR. DNA fragments in total RNA samples were digested using DNase I to remove gDNA. A relative quantitative analysis of the data was performed by 2−ΔΔCT using the Gene-9660 quantitative PCR instrument. The primer sequences are listed in Table 1. GAPDH and ACTB were used as internal references. A two-sample T-test was used for statistical analysis. Second, GSE19187 (involving 11 normal controls, 14 AR, and 7 CARAS) and GSE141623 (involving 100 normal controls, 23 AR, and 33 CARAS) were downloaded from the GEO database for expression validation.
 |
Table 1 Primers Sequence in RT-qPCR |
Protein Expression Analysis by Enzyme-Linked Immunosorbent Assay (ELISA)
Serum samples from 23 participants (6 CARAS patients, 6 AR patients, and 11 healthy individuals) were collected to detect the protein expression by ELISA. The standard hole and a sample hole were set. A total of 50 μL of standard with different concentrations were added to the standard hole. The sample was added to the bottom of an enzyme-labeled plate hole and incubated at 37°C for 30 min. After discarding the liquid, each hole was filled with washing liquid and stood for 30 s. To each well was added 50 μL of enzyme-labeled reagent and incubated at 37°C for 30 min. After discarding the liquid, each hole was filled with washing liquid and stood for 30 s. To each hole was added 50 μL of color developing agent A and B and incubated at 37°C in the dark for 10 min. Thereafter, 50 μL of stopping solution was added to each well to stop the reaction. The absorbance (OD value) of each well at the wavelength of 450 nm was measured by SpectraMax M5 microplate reader.
Ethical Statement
The research protocol was in accordance with the tenets of the Declaration of Helsinki and was approved by the Ethics Committee of the Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University on February 25, 2019 (approval number [2019] KY032-01). We provided each participant with clear and detailed information about the research, including the purpose, methods, expected results, potential risks and benefits, and other available options. Each participant had been informed that they have the right to refuse to participate in the research or to withdraw their consent at any time without any negative consequences and their privacy and confidentiality will be protected during the research. Researchers adhered to the principles of non-deception, non-concealment, and non-misrepresentation during the informed consent process. Informed consent was obtained from each participant for clinical data use and publication.
Statistical Analysis
All data analyses were performed using R. To evaluate the correlations, Pearson’s correlation analysis was conducted. A two-sample T-test was utilized to determine significant differences. Kruskal–Wallis test was used to perform immune infiltration analysis. If the p-value is less than 0.05, it was considered statistically significant.
Results
Patient Characteristics
In this study, nine participants were enrolled, including three CARAS patients, three AR patients, and three normal controls (N). The demographic data of the participants are listed in Table 2.
 |
Table 2 The Demographic Data of Recruited Subjects |
DEmRNAs in AR vs N, CARAS vs AR, and CARAS vs N Groups
Compared with normal control group, 229 DEmRNAs, including 98 up-regulated mRNAs and 131 down-regulated mRNAs, were obtained in AR patients. Compared with AR group, 239 DEmRNAs were identified in CARAS patients, which included 165 up-regulated and 74 down-regulated mRNAs. Compared with normal control group, 232 DEmRNAs, including 110 up-regulated and 122 down-regulated mRNAs, were acquired in CARAS group. Venn diagram of DEmRNAs in CARAS, AR, and normal group is displayed in Figure 1A. A total of 40 common mRNAs were identified in AR vs N and CARAS vs AR groups. Among these, only keratin 8 (KRT8) was significantly down-regulated in CARAS vs AR and AR vs N groups. Furthermore, 61 DEmRNAs with the same expression trend were found in CARAS vs AR and CARAS vs N groups. A total of 25 mRNAs, such as ovochymase 1 (OVCH1), were down-regulated, and 36 mRNAs were up-regulated in CARAS, such as Charcot-Leyden crystal galectin (CLC), sphingomyelin phosphodiesterase 3 (SMPD3), and histamine receptor H4 (HRH4). The heat map of these 61 differentially expressed mRNAs is shown in Figure 1B. Among them, indoleamine 2, 3-dioxygenase 1 (IDO1, up-regulation), cysteinyl leukotriene receptor 2 (CYSLTR2, up-regulation), and tec protein tyrosine kinase (TEC, up-regulation) were immune-related mRNAs, and Transketolase-like 1 (TKTL1, up-regulation) and very low-density lipoprotein receptor (VLDLR, up-regulation) were hypoxia-related mRNAs.
DElncRNAs in AR vs N, CARAS vs AR, and CARAS vs N Groups
Compared with normal control group, 102 DElncRNAs (50 up-regulated and 52 down-regulated) were identified in AR patients. Compared with AR group, 106 DElncRNAs were obtained in CARAS patients, which included 63 up-regulated and 43 down-regulated lncRNAs. Compared with normal control group, 90 DElncRNAs (45 up-regulated and 45 down-regulated lncRNAs) were obtained in CARAS group. Venn diagram of DElncRNAs in CARAS, AR, and normal group is displayed in Figure 1C. The results showed that 21 common lncRNAs were identified in AR vs N and CARAS vs AR groups. Strangely, the expression trend of these 21 lncRNAs was opposite in AR vs N and CARAS vs AR groups. Further analysis showed that 23 DElncRNAs (15 up-regulated and 8 down-regulated) with the same expression trend were identified in CARAS vs AR and CARAS vs N groups, which may be associated with the occurrence and development of CARAS (Figure 1D).
DEcircRNAs in AR vs N, CARAS vs AR, and CARAS vs N Groups
Compared with normal control group, 26 DEcircRNAs (9 up-regulated and 17 down-regulated) were identified in AR patients. Compared with AR group, 13 DEcircRNAs (7 up-regulated and 6 down-regulated) were obtained in CARAS group. Compared with normal control group, 16 DEcircRNAs were obtained in CARAS group, which included 9 up-regulated and 7 down-regulated circRNAs. Venn diagram of DEcircRNAs in CARAS, AR, and normal group is displayed in Figure 1E. It was worth mentioning that three circRNAs with the same expression trend (down-regulated hsa_circ_0007272 and hsa_circ_0070934 and up-regulated hsa_circ_0008903) were found in CARAS vs AR and CARAS vs N groups, meaning that these 3 circRNAs may be associated with the occurrence and development of CARAS (Figure 1F).
Functional Analysis of 61 DEmRNAs
To investigate the biological function of 61 DEmRNAs, functional analysis was performed. Based on the KEGG enrichment analysis, fibronectin 1 (FN1) was involved in the extracellular matrix (ECM) receptor interaction signal pathway. CLC and IDO1 were involved in regulating activated T cell proliferation (Figure 2A). GSEA showed that the gene set spliceosome was down-regulated in the CARAS group (Figure 2B).
PPI Network and TFs Correlation Analysis of 61 DEmRNAs
To explore protein interactions among the 61 DEmRNAs, a PPI network was constructed with the online database STRING (Figure 2C). CD24 and FN1 had the highest interaction score of 0.936. In addition, a total of 224 TFs were associated with 61 DEmRNAs (Figure 2D). There were eight TFs, including activating transcription factor 3 (ATF3), involved in the regulation of IDO1. One TF, a basic helix-loop-helix family member, e40 (BHLHE40), was involved in regulating CYSLTR2. Seven TFs were involved in regulating TKTL1, such as grainy head-like transcription factor 2 (GRHL2).
Identification of lncRNA/circRNA-mRNA Co-Expressed Interaction Pairs
To investigate the relationship between lncRNAs/circRNAs and mRNAs in the occurrence and development of CARAS, lncRNA/circRNA-mRNA regulatory pairs based on 23 lncRNAs, 3 circRNAs, and 61 mRNAs were analyzed. A total of 3 lncRNA-mRNA cis-regulatory pairs were screened, including OVCH1-AS1-OVCH1, RP11-71L14.3-SMPD3, and RP11-178F10.2-HRH4 (Figure 3A). In addition, 128 lncRNA-mRNA trans-regulatory pairs, such as LINC00452/MIR4280HG-CLC, HEATR6-DT/LINC00639/LINC01783-TEC, RP11-167J8.5-VLDLR, and RP11-178F10.2/RP11-71L14.3-IDO1, were identified (Figure 3B). Then, 37 circRNA-mRNA regulatory pairs were identified (Figure 3C), such as hsa_circ_0007272/hsa_circ_0070934-CLC, and hsa_circ_0008903-TEC/CYSLTR2.
Immune Infiltration Analysis and Drug Prediction
CIBERSORT was used to assess infiltration status of immune cells in AR vs N, CARAS vs AR, and CARAS vs N groups. The infiltration level of B-cell memory and plasma cells were significantly decreased and increased in AR and N group, respectively (Figure S1A). The infiltration level of NK cells resting and plasma cells was significantly increased and decreased in CARAS vs N group, and CARAS vs AR group, respectively (Figure S1B and C). It suggested that dysregulated levels of immune cell infiltration may be closely related to CARAS. Additionally, to provide a new perspective for CARAS treatment, potential drugs related to 61 DEmRNAs were screened based on the DGIdb database (Figure S2). Significantly, TEC was a drug target of DASATINIB, suggesting that TEC may be considered as a drug target for DASATINIB in the treatment of CARAS.
Validation in RT-qPCR and Online Dataset
The expression of four lncRNAs (HEATR6-DT, LINC00639, LINC01783, and RP11-178F10.2), two circRNAs (hsa_circ_0007272 and hsa_circ_0008903), and four mRNA (KRT8, CLC, OVCH1, and TEC) were detected by RT-qPCR (Figure 4). The result indicated that CLC, HEATR6-DT, LINC00639, LINC01783, and RP11-178F10.2 were up-regulated in CARAS vs N and CARAS vs AR groups, while KRT8 and hsa_circ_0007272 (with statistical significance in CARAS vs N group) were down-regulated in CARAS vs N and CARAS vs AR groups. In addition, OVCH1 and hsa_circ_0008903 were down-regulated in CARAS vs N group, while TEC was up-regulated in CARAS vs N group. These findings were in line with our results of RNA sequencing. In addition, GSE19187 and GSE141623 were used to validate the expression of four mRNAs (CYSLTR2, IDO1, OVCH1, and TEC) (Figure 5). The result showed that CYSLTR2 and IDO1 were up-regulated, and OVCH1 was down-regulated (with statistical significance in GSE141623) in CARAS vs N and CARAS vs AR groups in both two datasets. In GSE141623, TEC was up-regulated in both the CARAS vs N and CARAS vs AR groups, whereas in GSE19187, TEC was up-regulated only in the CARAS vs AR group. Although the results of this verification are still lacking in significance, the trend exists, which indicates that our results have a certain credibility.
 |
Figure 4 Validation in RT-qPCR. RT-qPCR, real time qPCR. Two-sample T-test was used for statistical analysis. **p < 0.01. |
 |
Figure 5 Validation in GSE19187 (A) and GSE141623 (B). ANOVA was used for statistical analysis. |
Protein Expression Analysis by ELISA
The protein expression of four mRNAs (IDO1, SMPD3, TEC, and CLC) was detected by ELISA (Figure 6). The protein expression of SMPD3 and CLC was significantly increased in the CARAS group compared with the N group. The protein expression tendency of DIO1 and TEC was up-regulated in the CARAS group compared with the N group. The protein expression result was basically consistent with the mRNA expression results.
Discussion
In recent years, epidemiological studies have demonstrated that the incidence of AR and AS is increasing yearly.22 In this study, 61 mRNAs, 23 lncRNAs, and 3 circRNAs with the same expression trend were found in CARAS vs AR and CARAS vs N groups, which may be related to the occurrence and development of CARAS. Then, three immunity-related mRNAs and two hypoxia-related mRNAs with the same expression trend were identified. Several lncRNA/circRNA-mRNA regulatory pairs were identified in CARAS, which may indicate potential regulatory effects of lncRNAs/circRNAs in CARAS. In addition, immune infiltration analysis and drug prediction were performed to further provide novel insight into the underlying mechanisms in CARAS.
Inflammation of nasal and bronchial mucosa plays a critical role in the pathogenesis of AR and AS. Although there are different inflammatory indicators in AR and AS, the chronic allergic inflammation in the upper and lower respiratory tract is similar. In addition, the participating types of inflammatory cells, cytokines, and various inflammatory mediators are similar.23,24 TGF-β is an important inflammatory cytokine involved in the AR and AS process.24,25 KRT8 is one of the keratin genes.26 Strunz et al recently studied the RNA expression characters in 28 cell types and reported that the KRT8+ state appeared in several independent models of lung injury and persisted in human lung fibrosis.27 Therefore, it creates a distinct cell–cell communication network with mesenchymes and macrophages during repair. This result indicated the involvement of KRT8 in the repair function of the lungs. Differential expression analysis of mRNAs showed that KRT8 was down-regulated stepwise in AR and CARAS patients, suggesting a potential role of KRT8 in the development of AR into CARAS.
CLC, a valuable biomarker of eosinophil/basophil, is involved in AS and AR.28 Genetic variation in CLC had been associated with AR.29 CLC was dramatically up-regulated in AR group compared with healthy control.30 Expression of CLC is increased in patients with eosinophilic AS.31 Up-regulated CLC was detected in the peripheral blood of patients with aspirin-induced AS.32 LINC00452 and hsa_circ_0070934 are involved in ovarian and hepatocellular carcinoma.33,34 LINC00639, expressed in bronchial epithelial cells, is involved in the development of chronic obstructive pulmonary disease.35 LINC01783, a rarely studied novel lncRNA, has been found to be highly up-regulated in serum samples.36 Up to now, there are no reports about MIR4280HG, HEATR6-DT, and hsa_circ_0007272 in any disease. In the present study, CLC and TEC were up-regulated in CARAS. Moreover, CLC was a trans-acted mRNA of LINC00452 and MIR4280HG and under the regulation of hsa_circ_0007272 and hsa_circ_0070934. TEC was a trans-acted mRNA of HEATR6-DT, LINC00639, and LINC01783 and under the regulation of hsa_circ_0008903. Our results indicate that the regulatory pairs of LINC00452/MIR4280HG/hsa_circ_0007272/hsa_circ_0070934-CLC and HEATR6-DT/LINC00639/LINC01783/hsa_circ_0008903-TEC may be involved in the development of CARAS.
IDO1, involved in inflammation, is significantly associated with chronic rhinosinusitis.37 Increased expression of IDO1 occurs in patients with allergic AS.38 In addition, IDO1 can suppress T cell activation.39 In our analysis, when compared with AR patients, the expression of IDO1 was up-regulated in CARAS patients. Moreover, IDO1 is involved in the signaling pathway of regulation of activated T cells proliferation. T lymphocytes play leading roles in regulating airway inflammatory response. The toll-like receptor (TLR) is an innate immune pathogen pattern recognition receptor. Activation of the TLR signal transduction pathway will aggravate airway inflammation and airway hyperresponsiveness and cause the release of inflammatory factors.40 Overstimulation of TLR leads to chronic inflammation such as AS and negative regulation of TLR signal ensures that the damage to the body is reduced during immune activation.41 The expression of SMPD3 reduces the TLR-induced inflammatory response of macrophages,42 and SMPD3 is up-regulated in allergic AS.38 Furthermore, HRH4 is up-regulated in AS, dermatitis, and rhinitis multimorbidity in children and adolescents.43 RP11-178F10.2 is a potential serum biomarker and predictor of patient survival with alcoholic cirrhosis.44 In this study, IDO1 and SMPD3 were, respectively, the trans-acted and cis-acted mRNA of RP11-71L14.3. IDO1 was also the trans-acted mRNA of RP11-178F10.2. Consistent with the study of Lemonnier, lncRNA RP11-178F10.2 up-regulated HRH4 in our study.43 Currently, to the best of our knowledge, no study has been reported on the function of lncRNA RF11-71L14.3. Thus, specific regulatory mechanisms need to be clarified in future research.
CYSLTR2, related to allergic diseases and the immune response, is up-regulated in patients with allergic AS.38,45 In this study, CYSLTR2 was regulated by hsa_circ_0008903 in CARAS. At present, no study has reported the function and role of hsa_circ_0008903 in AR and AS. This could provide information on some clinical findings and mechanisms of CARAS, and this also needs to be verified in future studies. Based on KEGG analysis, we found that 61 DEmRNA was involved in the ECM receptor interaction signal pathway. The thickening of the asthmatic subepithelial layer is due to exaggerated deposition of ECM proteins.46–50 Epithelial hyperplasia and degradation of the matrix along with the accumulation of plasma proteins occurred in AR and bronchial AS.51,52
Based on immune infiltration analysis, we found that the infiltration level of NK cells resting and plasma cells was significantly increased and decreased in CARAS vs N and CARAS vs AR, respectively. Studies have reported that the immune regulatory function of NK cells plays an important role in childhood-onset AS and chronic rhinosinusitis.53–55 Although the importance of eosinophils and basophils had been indicated in AS, no difference between eosinophils and basophils among groups was observed in this present study.56,57 The balance of Th1/Th2 is associated with AR. Th2 can induce activation of B cells and IgE class switching, which leads to B-cell differentiation into plasma cells that produce allergen-specific IgE.58 It is indicated that dysregulated infiltration levels of NK cells resting and plasma cells may be closely associated with the development of CARAS. In addition, we found that TEC was a drug target for Dasatinib. A previous study has demonstrated that Dasatinib is beneficial in AS improvement.59 Thus, TEC may be considered as a potential drug target for Dasatinib in the treatment of CARAS.
This study has some limitations. The sample sizes for RNA sequencing and RT-qPCR were small and thus introduced greater sample heterogeneity. The validation results in RT-qPCR and GEO database are lacking in significance. A larger number of samples are needed for further studies. Further functional experiments at cellular and animal levels are needed to investigate the potential mechanism of identified mRNAs, lncRNAs, and circRNAs in CARAS. Therefore, the possible mechanisms of candidate mRNAs, lncRNAs, and circRNAs in CARAS would be the focus of our future studies.
Conclusion
Our study identified three immunity-related mRNAs (IDO1, CYSLTR2, and TEC) and two hypoxia-related mRNAs (TKTL1 and VLDLR), which may be associated with the occurrence and development of CARAS. Several lncRNA/circRNA-mRNA regulatory pairs, such as RP11-71L14.3-IDO1/SMPD3, RP11-178F10.2-IDO1, LINC00452/MIR4280HG/hsa_circ_0007272/hsa_circ_0070934-CLC, HEATR6-DT/LINC00639/LINC01783/hsa_circ_0008903-TEC, and hsa_circ_0008903-CYSLTR2, were identified in this study, which may be implicated in the development of AR into CARAS.
Data Sharing Statement
All data generated or analyzed during this study are available from the corresponding author on reasonable request.
Ethics Approval
The Ethics Committee of the Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University approved this study (approval number [2019] KY032-01). All experiments were performed in accordance with relevant guidelines and regulations. Informed consent was obtained from each patient for clinical data use and publication.
Consent for Publication
The subjects gave written informed consent for the publication of any associated data.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was funded by the Changzhou Sci & Tech Program (CJ20190099 to ZDM, CE20205023 to QZ), the China Postdoctoral Science Foundation (2020M670011ZX to ZDM), the Jiangsu province social development project (BE2020651 to QZ), and the Jiangsu province “333 talents” project (BRA2020015 to QZ).
Disclosure
The authors report no conflict of interest.
References
1. Paiva Ferreira LKD, Paiva Ferreira LAM, Monteiro TM, Bezerra GC, Bernardo LR, Piuvezam MR. Combined allergic rhinitis and asthma syndrome (CARAS). Int Immunopharmacol. 2019;74:105718.
2. Xia S, Zhu Z, Guan WJ, et al. Correlation between upper and lower airway inflammations in patients with combined allergic rhinitis and asthma syndrome: a comparison of patients initially presenting with allergic rhinitis and those initially presenting with asthma. Exp Ther Med. 2018;15(2):1761–1767. doi:10.3892/etm.2017.5536
3. Bergeron C, Hamid Q. Relationship between asthma and rhinitis: epidemiologic, pathophysiologic, and therapeutic aspects. Allergy Asthma Clin Immunol. 2005;1(2):81–87. doi:10.1186/1710-1492-1-2-81
4. Rahman MA, Chakraborty R, Ferdousi KR, Alam A, Chowdhury MK, Paul BK. New therapeutic approach to treat allergic rhinitis and bronchial asthma, considering these two as one united airway disease. Mymensingh Med J. 2017;26(1):216–221.
5. Bousquet J, Anto JM, Bachert C, et al. Allergic rhinitis. Nat Rev Dis Primers. 2020;6(1):95. doi:10.1038/s41572-020-00227-0
6. Alobaidi AH, Alsamarai AM, Alsamarai MA. Inflammation in asthma pathogenesis: role of T cells, macrophages, epithelial cells and type 2 inflammation. Antiinflamm Antiallergy Agents Med Chem. 2021;20(4):317–332. doi:10.2174/1871523020666210920100707
7. Erle DJ, Sheppard D. The cell biology of asthma. J Cell Biol. 2014;205(5):621–631. doi:10.1083/jcb.201401050
8. Rupani H, Fong WCG, Kyyaly MA, Kurukulaaratchy RJ. Recent insights into the management of inflammation in asthma. J Inflamm Res. 2021;14:4371–4397. doi:10.2147/JIR.S295038
9. Marseglia GL, Caimmi S, Marseglia A, et al. Rhinosinusitis and asthma. Int J Immunopathol Pharmacol. 2010;23(1 Suppl):29–31. doi:10.1177/03946320100230S108
10. Matucci A, Bormioli S, Nencini F, et al. Asthma and chronic rhinosinusitis: how similar are they in pathogenesis and treatment responses? Int J Mol Sci. 2021;22(7):3340. doi:10.3390/ijms22073340
11. Wheatley LM, Togias A, Solomon CG. Clinical practice. Allergic rhinitis. N Engl J Med. 2015;372(5):456–463. doi:10.1056/NEJMcp1412282
12. Pawankar R, Mori S, Ozu C, Kimura S. Overview on the pathomechanisms of allergic rhinitis. Asia Pac Allergy. 2011;1(3):157–167. doi:10.5415/apallergy.2011.1.3.157
13. Alashkar Alhamwe B, Alhamdan F, Ruhl A, Potaczek DP, Renz H. The role of epigenetics in allergy and asthma development. Curr Opin Allergy Clin Immunol. 2020;20(1):48–55. doi:10.1097/ACI.0000000000000598
14. Kabesch M, Tost J. Recent findings in the genetics and epigenetics of asthma and allergy. Semin Immunopathol. 2020;42(1):43–60. doi:10.1007/s00281-019-00777-w
15. Hombach S, Kretz M. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17.
16. Ma Y, Shi L, Zhao K, Zheng C, Mora-Montes H. lncRNA FR215775 regulates Th2 differentiation in murine allergic rhinitis. J Immunol Res. 2022;2022:7783481. doi:10.1155/2022/7783481
17. Yang C, Shangguan C, Cai C, Xu J, Qian X. LncRNA HCP5 participates in the tregs functions in allergic rhinitis and drives airway mucosal inflammatory response in the nasal epithelial cells. Inflammation. 2022;45(3):1281–1297. doi:10.1007/s10753-022-01620-5
18. Jin P, Sun K, Zhang Q, et al. Peripheral circular RNA profiling from patients with allergic rhinitis identified hsa_circRNA_404013 as a potential diagnostic biomarker. Int Arch Allergy Immunol. 2022;183(10):1078–1088. doi:10.1159/000525791
19. Huang J, Wang FH, Wang L, Li Y, Lu J, Chen J. LncRNA MALAT1 promotes proliferation and migration of airway smooth muscle cells in asthma by downregulating microRNA-216a. Saudi J Biol Sci. 2021;28(8):4124–4131. doi:10.1016/j.sjbs.2021.03.076
20. Liang Q, Fu J, Wang X, et al. circS100A11 enhances M2a macrophage activation and lung inflammation in children with asthma. Allergy. 2022;78(6):1459–1472. doi:10.1111/all.15515
21. Zhang L, Wang X, Wang C, et al. Allergic rhinitis management path (Chinese version) of Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines-2019. Chin Arch Otolaryngol Head Neck Surg. 2019;26(12):690–699.
22. Dierick BJH, van der Molen T, Flokstra-de Blok BMJ, et al. Burden and socioeconomics of asthma, allergic rhinitis, atopic dermatitis and food allergy. Expert Rev Pharmacoecon Outcomes Res. 2020;20(5):437–453. doi:10.1080/14737167.2020.1819793
23. Eiringhaus K, Renz H, Matricardi P, Skevaki C. Component-resolved diagnosis in allergic rhinitis and asthma. J Appl Lab Med. 2019;3(5):883–898. doi:10.1373/jalm.2018.026526
24. Kappen JH, Durham SR, Veen HI, Shamji MH. Applications and mechanisms of immunotherapy in allergic rhinitis and asthma. Ther Adv Respir Dis. 2017;11(1):73–86. doi:10.1177/1753465816669662
25. Paiva Ferreira LKD, Paiva Ferreira LAM, Bezerra Barros GC, et al. MHTP, a synthetic alkaloid, attenuates combined allergic rhinitis and asthma syndrome through downregulation of the p38/ERK1/2 MAPK signaling pathway in mice. Int Immunopharmacol. 2021;96:107590. doi:10.1016/j.intimp.2021.107590
26. Bragulla HH, Homberger DG. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J Anat. 2009;214(4):516–559. doi:10.1111/j.1469-7580.2009.01066.x
27. Strunz M, Simon LM, Ansari M, et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat Commun. 2020;11(1):3559. doi:10.1038/s41467-020-17358-3
28. Acharya KR, Ackerman SJ. Eosinophil granule proteins: form and function. J Biol Chem. 2014;289(25):17406–17415. doi:10.1074/jbc.R113.546218
29. Bryborn M, Halldén C, Säll T, Cardell LO. CLC- a novel susceptibility gene for allergic rhinitis? Allergy. 2010;65(2):220–228. doi:10.1111/j.1398-9995.2009.02141.x
30. Lei Y, Guo P, An J, Guo C, Lu F, Liu M. Identification of pathogenic genes and upstream regulators in allergic rhinitis. Int J Pediatr Otorhinolaryngol. 2018;115:97–103. doi:10.1016/j.ijporl.2018.09.005
31. Baines KJ, Simpson JL, Wood LG, et al. Sputum gene expression signature of 6 biomarkers discriminates asthma inflammatory phenotypes. J Allergy Clin Immunol. 2014;133(4):997–1007. doi:10.1016/j.jaci.2013.12.1091
32. Devouassoux G, Pachot A, Laforest L, et al. Galectin-10 mRNA is overexpressed in peripheral blood of aspirin-induced asthma. Allergy. 2008;63(1):125–131. doi:10.1111/j.1398-9995.2007.01558.x
33. Yang J, Wang WG, Zhang KQ. LINC00452 promotes ovarian carcinogenesis through increasing ROCK1 by sponging miR-501-3p and suppressing ubiquitin-mediated degradation. Aging. 2020;12(21):21129–21146. doi:10.18632/aging.103758
34. Morovat P, Morovat S, Ashrafi AM, Teimourian S. Identification of potentially functional circular RNAs hsa_circ_0070934 and hsa_circ_0004315 as prognostic factors of hepatocellular carcinoma by integrated bioinformatics analysis. Sci Rep. 2022;12(1):4933. doi:10.1038/s41598-022-08867-w
35. Mostafaei S, Kazemnejad A, Azimzadeh Jamalkandi S, Amirhashchi S. Identification of novel genes in human airway epithelial cells associated with Chronic Obstructive Pulmonary Disease (COPD) using machine-based learning algorithms. Sci Rep. 2018;8(1):15775. doi:10.1038/s41598-018-33986-8
36. Umu SU, Langseth H, Bucher-Johannessen C. A comprehensive profile of circulating RNAs in human serum. RNA Biol. 2018;15(2):242–250. doi:10.1080/15476286.2017.1403003
37. Min JY, Tan BK. Risk factors for chronic rhinosinusitis. Curr Opin Allergy Clin Immunol. 2015;15(1):1–13. doi:10.1097/ACI.0000000000000128
38. Elena-Perez S, Heredero-Jung DH, Garcia-Sanchez A, et al. Molecular analysis of IL-5 receptor subunit alpha as a possible pharmacogenetic biomarker in asthma. Front Med. 2020;7:624576. doi:10.3389/fmed.2020.624576
39. Ju JM, Nam G, Lee YK, et al. IDO1 scavenges reactive oxygen species in myeloid-derived suppressor cells to prevent graft-versus-host disease. Proc Natl Acad Sci U S A. 2021;118(10):e2011170118. doi:10.1073/pnas.2011170118
40. McLetchie S, Volpp BD, Dinauer MC, Blum JS. Hyper-responsive toll-like receptor 7 and 9 activation in NADPH oxidase-deficient B lymphoblasts. Immunology. 2015;146(4):595–606. doi:10.1111/imm.12530
41. Wang J, Hu Y, Deng WW, Sun B. Negative regulation of toll-like receptor signaling pathway. Microbes Infect. 2009;11(3):321–327. doi:10.1016/j.micinf.2008.12.011
42. Liu F, Li X, Yue H, et al. TLR-induced SMPD3 defects enhance inflammatory response of B cell and macrophage in the pathogenesis of SLE. Scand J Immunol. 2017;86(5):377–388. doi:10.1111/sji.12611
43. Lemonnier N, Melén E, Jiang Y, et al. A novel whole blood gene expression signature for asthma, dermatitis, and rhinitis multimorbidity in children and adolescents. Allergy. 2020;75(12):3248–3260. doi:10.1111/all.14314
44. Yang Z, Ross RA, Zhao S, Tu W, Liangpunsakul S, Wang L. LncRNA AK054921 and AK128652 are potential serum biomarkers and predictors of patient survival with alcoholic cirrhosis. Hepatol Commun. 2017;1(6):513–523. doi:10.1002/hep4.1061
45. Thompson MD, Capra V, Clunes MT, et al. Cysteinyl leukotrienes pathway genes, atopic asthma and drug response: from population isolates to large genome-wide association studies. Front Pharmacol. 2016;7:299. doi:10.3389/fphar.2016.00299
46. Yamauchi K, Inoue H. Airway remodeling in asthma and irreversible airflow limitation-ECM deposition in airway and possible therapy for remodeling. Allergol Int. 2007;56(4):321–329. doi:10.2332/allergolint.R-07-151
47. Royce SG, Tan L, Koek AA, Tang MLK. Effect of extracellular matrix composition on airway epithelial cell and fibroblast structure: implications for airway remodeling in asthma. Ann Allergy Asthma Immunol. 2009;102(3):238–246. doi:10.1016/S1081-1206(10)60087-7
48. Todorova L, Bjermer L, Westergren-Thorsson G, Miller-Larsson A. TGFβ-induced matrix production by bronchial fibroblasts in asthma: budesonide and formoterol effects. Respir Med. 2011;105(9):1296–1307. doi:10.1016/j.rmed.2011.03.020
49. Shifren A, Witt C, Christie C, Castro M. Mechanisms of remodeling in asthmatic airways. J Allergy. 2012;2012:316049. doi:10.1155/2012/316049
50. Reeves SR, Kolstad T, Lien TY, et al. Asthmatic airway epithelial cells differentially regulate fibroblast expression of extracellular matrix components. J Allergy Clin Immunol. 2014;134(3):663–670 e661. doi:10.1016/j.jaci.2014.04.007
51. Eliseeva TI, Tush EV, Krasilnikova SV, Kuznetsova SV, Ignatov SK. Metabolism of the extracellular matrix in bronchial asthma (Review). Sovrem Tekhnologii Med. 2018;10(4):220. doi:10.17691/stm2018.10.4.25
52. Bousquet J, Jacot W, Vignola AM, Bachert C, Van Cauwenberge P. Allergic rhinitis: a disease remodeling the upper airways? J Allergy Clin Immunol. 2004;113(1):43–49. doi:10.1016/j.jaci.2003.09.047
53. Karimi K, Forsythe P. Natural killer cells in asthma. Front Immunol. 2013;4:159. doi:10.3389/fimmu.2013.00159
54. Kim JH, Jang YJ. Role of natural killer cells in airway inflammation. Allergy Asthma Immunol Res. 2018;10(5):448–456. doi:10.4168/aair.2018.10.5.448
55. Kim JH, Choi GE, Lee BJ, et al. Natural killer cells regulate eosinophilic inflammation in chronic rhinosinusitis. Sci Rep. 2016;6:27615. doi:10.1038/srep27615
56. Nadif R, Zerimech F, Bouzigon E, Matran R. The role of eosinophils and basophils in allergic diseases considering genetic findings. Curr Opin Allergy Clin Immunol. 2013;13(5):507–513. doi:10.1097/ACI.0b013e328364e9c0
57. Poddighe D, Mathias CB, Brambilla I, Marseglia GL, Oettgen HC. Importance of basophils in eosinophilic asthma: the murine counterpart. J Biol Regul Homeost Agents. 2018;32(2):335–339.
58. Luo C, Peng S, Li M, Ao X, Liu Z. The efficacy and safety of probiotics for allergic rhinitis: a systematic review and meta-analysis. Front Immunol. 2022;13:848279. doi:10.3389/fimmu.2022.848279
59. da Silva AL, Magalhaes RF, Branco VC, et al. The tyrosine kinase inhibitor dasatinib reduces lung inflammation and remodelling in experimental allergic asthma. Br J Pharmacol. 2016;173(7):1236–1247. doi:10.1111/bph.13430
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.