Back to Journals » International Journal of Nanomedicine » Volume 16
Proteomics of Streptococcus mutans to Reveal the Antibiofilm Formation Mechanism of Ag/ZnO Nanocomposites with Light-Emitting Diode Radiation
Authors Jiang N , Zhao S, Wang S, Lu Z
Received 12 August 2021
Accepted for publication 30 October 2021
Published 19 November 2021 Volume 2021:16 Pages 7741—7757
DOI https://doi.org/10.2147/IJN.S333432
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Farooq A. Shiekh
Nan Jiang, Shuaiwei Zhao, Shilei Wang, Zhong Lu
Key Laboratory for Green Chemical Process of Ministry of Education, School of Environmental Ecology and Biological Engineering, Wuhan Institute of Technology, Wuhan, 430205, People’s Republic of China
Correspondence: Zhong Lu
Wuhan Institute of Technology, Wuhan, 430205, People’s Republic of China
Tel +86-18062036269
Fax +86-27-67905262
Email [email protected]
Introduction: As a biofilm-associated disease, dental caries benefits from nanoparticle (NP)-based therapies. Streptococcus mutans (S. mutans) is a primary aetiologic agent for dental caries development. We successfully applied a synergistic therapy of Ag/ZnO nanocomposites combined with light-emitting diode (LED) radiation to inhibit S. mutans biofilms. However, the antibiofilm mechanism has not been fully elucidated, and little is known about the biofilm formation ability of bacteria that survive NP-based therapies.
Methods: This study explored the antibiofilm formation mechanism of this synergistic therapy by an integrated approach based upon proteomics.
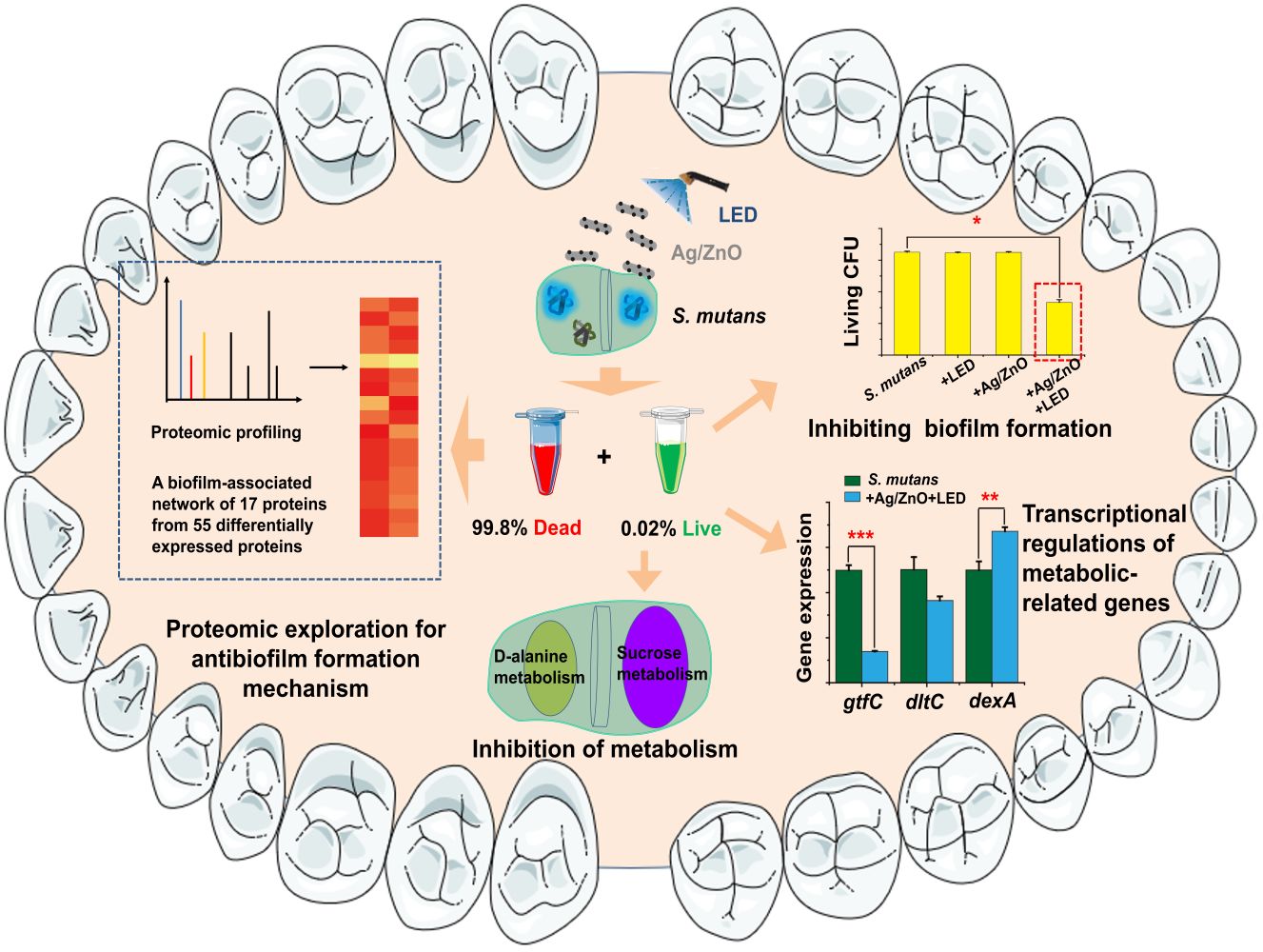
Results: Synergistic therapy killed 99.8% of bacteria, while the biofilm formation ability of 0.2% surviving bacteria was inhibited. The proteomic responses of S. mutans to synergistic therapy were comprehensively characterized to unveil the mechanism of bacterial death and biofilm formation inhibition of the surviving bacteria. In total, 55 differentially expressed proteins (12 upregulated and 43 downregulated) were recorded. The bioinformatic analysis demonstrated that cellular integrity damage and regulated expression of structure-associated proteins were the main reasons for bacterial death. In addition, the proteomic study indicated the potential inhibition of metabolism in surviving bacteria and provided a biofilm-related network consisting of 17 differentially expressed proteins, explaining the multiantibiofilm formation actions. Finally, we reported and verified the inhibitory effects of synergistic therapy on sucrose metabolism and D-alanine metabolism, which disturbed the biofilm formation of surviving bacteria.
Conclusion: Our findings demonstrated that synergistic therapy killed most bacteria and inhibited the surviving bacteria from forming biofilms. Furthermore, the antibiofilm formation mechanism was revealed by proteomics analysis of S. mutans after synergistic therapy and subsequent metabolic studies. Our success may provide a showcase to explore the antibiofilm formation mechanism of NP-based therapies using proteomic studies.
Keywords: antibiofilm, protein profiling, nanoparticles, S. mutans, biofilm-associated diseases
Graphical Abstract:
Introduction
Nanoparticle (NP)-based therapies are a highly promising treatment modality for diverse biofilm-associated diseases, especially oral diseases. Preventing oral biofilms is challenging for current broad-spectrum antimicrobial drugs due to the clearance of saliva and the poor penetration of exopolysaccharides (EPS) in biofilms. Conversely, chemically flexible and easily prepared NPs show exceptional promise to address such challenges.1–3 As a typical biofilm-mediated oral disease,4 dental caries is an excellent showcase for other biofilm-associated diseases that may benefit from NP-based therapies. Dental caries has Streptococcus mutans (S. mutans), a facultative anaerobic gram-positive bacterium, as one of its primary aetiologic agents.5,6 Nevertheless, the usage of antibiotics appears to be limited in oral clinics, while physical cleaning actions, such as brushing and flossing, still act as the main methods to remove oral biofilms.7,8 Herein, effective removal and antibiofilm formation approaches are urgently needed.
Zinc oxide (ZnO) has been widely used within the oral cavity with high safety and biocompatibility.9,10 ZnO at the nanoscale was preferred for better antimicrobial properties when incorporated into other metals, especially silver (Ag).11 Consequently, Ag/ZnO nanocomposites were formed by introducing Ag NPs into ZnO NPs.12 Given its intense antibacterial activity, a series of publications supported that Ag/ZnO nanocomposites can kill Pseudomonas aeruginosa (P. aeruginosa), Acinetobacter baumannii (A. baumannii), and Escherichia coli (E. coli)13,14 and inhibit biofilm formation.15 Our group has focused on the antimicrobial and antibiofilm formation effects of Ag/ZnO nanocomposites against S. mutans in recent years. We found that Ag/ZnO nanocomposites had enhanced antibacterial activity against S. mutans16 and downregulated biofilm-related gene expression effectively at sub-minimum inhibitory concentrations.17 We also developed a synergistic treatment by combining Ag/ZnO nanocomposites with light-emitting diode (LED) curing light radiation. In recent decades, LED radiation at 420−480 nm has been used to speed oral treatments, acting as a minimal heat generation instrument with relatively high emission intensity.18,19 This synergistic therapy can kill S. mutans bacteria and inhibit biofilm formation in a short period of time (5 min).20
Exploring the mechanism of antibiofilm formation is urgently needed for Ag/ZnO nanocomposites before they can be implemented in wide usage. Bacterial structural destruction and oxygen stress increases are widely accepted as antimicrobial and antibiofilm formation mechanisms for Ag/ZnO nanocomposites. In addition, disturbance of protein expression and DNA degradation were noted.20–22 Collectively, these current publications mainly focused on the antimicrobial properties of Ag/ZnO nanocomposites. However, the antibacterial mechanism at the molecular level has not been investigated thoroughly. Furthermore, the biofilm formation ability of the bacteria that survive NP-based treatment has not been considered.
Understanding the antibiofilm formation mechanism can be achieved by recording the comprehensive proteomic responses towards NP-based therapies. Due to high-performance mass spectrometry (MS), proteomics has developed rapidly to investigate proteins on a larger scale.23 Protein identification, quantification, and relevant bioinformatic analysis have provided comprehensive insights into exploring the toxicity and antibacterial actions of NPs.24–27 In this work, proteomic profiling and appropriate bioinformatic analysis of S. mutans were carried out to reveal the antibiofilm formation mechanism of Ag/ZnO combined with LED radiation. Furthermore, we investigated the biofilm formation ability and newly formed biofilm properties of the surviving bacteria after synergistic treatment. Based on proteomic results, the inhibition of the metabolism of sucrose and D-alanine in surviving bacteria was also unveiled. Thus, an integrated approach was built and applied to explore the antibiofilm formation mechanism of Ag/ZnO-based therapy, a representative NP-based therapy in biofilm-associated diseases (Figure 1).
Materials and Methods
Ag/ZnO Nanocomposite Preparation and Bacteria Culture
Zn(CH3COO)2·2H2O and silver nitrate (AgNO3) were purchased from Sinopharm Chemical Reagent Co., Ltd. (China) and Aladdin (China), respectively. Other chemicals were purchased from Sigma–Aldrich (USA) in analytical grades or otherwise stated. Then, ZnO nanorods were prepared via a solvothermal method using Zn(CH3COO)2·2H2O with sodium hydroxide. Ag/ZnO nanocomposites were synthesized via a deposition-precipitation method, strictly following the method described in our former publication.16 The characterization of this material is listed in the supporting information (SI) file (Figure S1). S. mutans bacteria were provided by the School of Stomatology, Wuhan University. The culture of this bacterial strain followed a standard method using brain heart infusion (BHI) medium (OXOID, UK) anaerobically at 37 °C.
LED Light Source and Radiation Treatment
LED radiation was provided by an LED curing light (Bluephase, Ivoclar Vivadent AG, Schaan/Liechtenstein, Austria) with a 420−480 nm wavelength. Then, we applied LED radiation following our previous protocol with slight modifications.20 Briefly, the sample was irradiated by an LED light source with a diameter of 1.0 cm and a power density of 170.0 mW/cm2. A distance of 0.8 cm was set between the sample surface and the light source. An individual tube covered by aluminium foil was used for each sample to avoid light scattering. The radiation cycle was kept at 30 s with a 10-s stop, and the total time was set at 5 min with 10 cycles. Each treated sample’s accumulated fluences were calculated as 51.0 J/cm2.
Treatment of Planktonic S. mutans by Ag/ZnO Combined with LED Radiation
S. mutans was cultured in BHI broth overnight and diluted to 107 colony-forming units (CFU)/mL. The bacterial suspension was incubated with 1.0 mg/mL Ag/ZnO nanocomposite for 5 min or without 5 min of LED radiation. At the same time, untreated samples acted as the blank control, and the total volume of each sample was set as 200 μL. Then, we estimated each sample’s living cell number using a standard dilution plating procedure.
Biofilm Formation Assay
We then estimated the antibiofilm formation effects of Ag/ZnO nanocomposites combined with LED radiation against S. mutans. These treated planktonic bacteria (107 CFU/mL) were collected and resuspended using the same volume (200 μL) of BHI broth supplemented with 1% (w/v) sucrose (BHI-1% sucrose). The samples of untreated bacteria (S. m. group), bacteria exposed to 5 min of LED radiation (+LED group), and bacteria incubated with 1.0 mg/mL Ag/ZnO nanocomposite for 5 min (+Ag/ZnO group) were diluted 1000-fold using BHI-1% sucrose. Dilutions of these three groups were required for the following experiments investigating newly formed biofilms. In addition, the other group of bacteria was treated with 1.0 mg/mL Ag/ZnO combined with 5 min of LED radiation (+Ag/ZnO+LED group). We then transferred these samples into wells of 96-well plates. After 24 h of anaerobic culture at 37 °C, the generated biofilms were subjected to phosphate-buffered saline (PBS) washing, methanol fixation (15 min), and 0.1% (w/v) crystal violet (CV) staining (5 min). DMSO was added, and the absorbance at 570 nm (OD570) was measured for each sample using a multiplate reader (RT-6000, Leidu Life Science Co., Ltd). Furthermore, we repeated the above procedures and detached the biofilms. A standard dilution plating procedure evaluated living cell numbers in these biofilms. All the results represent triplicate experiments.
Adherence Assay
The sucrose-independent and sucrose-dependent adherences of newly generated biofilms were measured following the protocol of Hasan et al.28 S. mutans samples of the S. m. group, +LED group, +Ag/ZnO group, and +Ag/ZnO+LED group were collected and resuspended in BHI or BHI-1% sucrose. Then, these bacteria were cultured at an angle of 30° in glass tubes for 24 h. Next, we discarded the planktonic cell suspension and washed the adhering cells using 0.5 mol/L NaOH. Finally, the adhering cells were resuspended in PBS for testing. The absorbance at 600 nm (OD600) was measured by a UV-Vis spectrophotometer (UV-2450, Shimadzu, Japan) for each sample, and the OD600 of the S. m. group normalized the obtained data. All the results represent triplicate experiments.
EPS Estimation
The planktonic S. mutans samples of the S. m. group, +LED group, +Ag/ZnO group, and +Ag/ZnO+LED group were prepared as described above to form biofilms. We then detached and collected these biofilms for EPS estimation. The biofilm was dispersed and washed 3 times with water and then centrifuged at 14,000 × g. All supernatants were collected for water-soluble EPS analysis. Then, the precipitate was resuspended in 1 mol/L NaOH. This suspension was incubated at 37 °C for 2 h, and supernatants were collected after centrifugation at 14,000 × g for insoluble EPS analysis. The extraction steps were performed in triplicate, and all supernatants were combined for each sample. The polysaccharides of both water-soluble and insoluble EPS extractions were precipitated by adding 3-fold volumes of ice-cold 95% ethanol and overnight incubation at 4 °C. We then collected the precipitants after centrifugation at 10,000 × g, washed them 3 times using ice-cold 75% ethanol, and resuspended them in water (water-soluble EPS) or 1 mol/L NaOH solution (water-insoluble EPS). Finally, a phenol-sulfuric acid colorimetric assay was carried out for each sample.29 Again, all the results represent triplicate experiments.
Total Proteome Analysis
We collected approximately 50 mg (wet weight) of planktonic S. mutans with a 5-min incubation in 1.0 mg/mL Ag/ZnO combined with LED radiation. The same amount of untreated bacteria acted as the control. First, according to the manufacturer’s instructions, peptide mixtures (100 μg each sample) were extracted and subjected to tandem mass tag (TMT) labelling (Thermo Fisher Scientific, USA). We then loaded these peptide mixtures onto a reverse-phase nanoViper C18 trap column (Thermo Scientific Acclaim PepMap100, 100 μm×2 cm) connected to a C18 reversed-phase analytical column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin) in buffer A (0.1% formic acid). Next, these peptides were separated with a linear gradient of buffer B (84% acetonitrile and 0.1% formic acid) at a 300 nL/min flow rate controlled by IntelliFlow technology.
Tandem mass spectrometry (MS/MS) analysis was carried out using a Q Exactive mass spectrometer (Thermo Fisher Scientific, USA) coupled with Easy nLC (Thermo Fisher Scientific, USA). We selected the positive ion mode of the mass spectrometer and acquired MS data using a data-dependent top 10 method, which could choose the most abundant precursor ions from the survey scan (300–1800 m/z) for higher energy collisional dissociation (HCD) fragmentation. The automatic gain control (AGC) target was set to 3e6, and the maximum injection time was set to 10 ms. With a dynamic exclusion duration of 40.0 s, we acquired survey scans at a resolution of 70,000 at m/z 200. The HCD spectral resolution was kept at 17,500 at m/z 200, and the isolation width was 2 m/z. Finally, the normalized collision energy was 30 eV, and the underfill ratio was defined as 0.1%. These procedures were performed with peptide recognition mode enabled. All the results represent triplicate experiments.
The obtained MS data were treated using the MASCOT engine (version 2.2, Matrix Science, UK) embedded in Proteome Discoverer 1.4. The parameters were set as follows: search type as MS/MS ion search, enzyme as trypsin, mass values as monoisotopic, max missed cleavages as 2, fixed modifications as Carbamidomethyl (C), TMT 6plex (N-term), and TMT 6plex (K), variable modifications as Oxidation (M), and TMT 6plex (Y), peptide mass tolerance as ± 20 ppm, fragment mass tolerance as 0.1 Da, protein mass as unrestricted, database pattern as a decoy, and peptide FDR as ≤0.01. We calculated the protein ratios as the median of unique peptides of the protein and normalized all peptide ratios by the median protein ratio (Figure S2). Proteins with fold change>1.2 and p < 0.05 were denoted as differentially expressed proteins. Further bioinformatics analyses were carried out for these differentially expressed proteins, including annotation and functional enrichment analysis of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and clustering. The following software was used during the investigation: Blast2GO10 (version 3.3.5), Cluster3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm), Java Treeview software (http://jtreeview.sourceforge.net), STRING (http://string-db.org/), Cytoscape (version 3.5.1), and Hemi (version 1.0).
Transcriptional Analysis of Metabolic-Related Genes by RT-qPCR
We measured the expression levels of gtfC, dexA, and dltc via quantitative real-time PCR (RT-qPCR). A total of 108 CFU/mL planktonic S. mutans was subjected to a 5-min incubation of 1.0 mg/mL Ag/ZnO combined with LED radiation, while the untreated bacteria acted as the control. We first extracted and purified the total RNA from each sample using an RNAprep Pure Cell/Bacteria Kit (TianGen, China) and reverse-transcribed the mRNA into cDNA using the GoScript™ Reverse Transcription System (Promega, USA). The total volume of each PCR reagent was set as 50 μL, including 25 μL GoTaq® qPCR Master Mix (Promega, USA), 100 ng cDNA samples, and 0.6 μL forward and reverse primers (5 μM). We carried out all RT-qPCR experiments in a LightCycler® 480 system (Roche Life Science, Switzerland). 16S rRNA was used as the normalization control, and the 2−ΔΔCT method was used for data treatment.30 Table 1 shows the primer sequences for 16S rRNA and target genes. All the assays were performed in triplicate.
 |
Table 1 Primer Sequences of 16S rRNA and Target Genes |
Bacterial Sucrose Consumption After Synergistic Therapy
We prepared planktonic S. mutans samples of the S. m. group, +LED group, +Ag/ZnO group, and +Ag/ZnO+LED group as described above. These bacteria were cultured using BHI-1% sucrose for 0–24 h, and we serially removed the culture medium at 6, 12, 18, and 24 h. A sucrose assay kit was used to determine the sucrose content in the culture medium (Nanjing Jiancheng Bioengineering Institute, China). Given that the initial sucrose content was 1%, we calculated the sucrose consumption for each sample. All the results represent triplicate experiments.
Preparation of Teichoic Acid Extract and Supplementation in S. mutans Culture
We extracted poly(ribitol) phosphate teichoic acids (TA) from S. mutans using a classic approach with modifications.31 First, 8 g bacterial cells (wet weight) were harvested and resuspended in 800 mL PBS with a magnetic stirrer overnight at 4 °C. The mixture was centrifuged to obtain the sediment. Next, we washed the residue using 200 mL cold PBS and extracted the samples using cold 10% trichloroacetic acid (TCA) 3 times (100 mL each run). These homogenates were combined and centrifuged to obtain the sedimented bacterial debris. Next, we collected the residue of the previous extraction for further extraction. This debris was mixed with 100 mL 10% cold TCA and homogenized for 5 min at 4 °C. Then, we kept the homogenate at 4 °C overnight and homogenized this again at the highest speed for 2 min, continuing with filtering by a Gelman 0.2-μm filter. Next, 1 vol cold ethanol was added to the column before centrifugation. Ten millilitres of acetone was added to the sediment, and the sample was centrifuged again. The supernatant was discarded, and 10 mL diethyl ether was added to the sample. The sample was mixed and centrifuged again at 4 °C. Then, 4 mL water was added to dissolve the sediment and mixed at 4 °C overnight. Finally, we collected the supernatant after centrifugation and precipitated poly(ribitol) phosphate TA by adding 2 vol acetone. The mixture was kept at 4 °C overnight and subjected to centrifugation. The resin-like material was obtained by drying the final sediment, and then a 2 mg/mL poly(ribitol) phosphate stock solution was prepared for further use.
The planktonic S. mutans samples of the S. m. group and +Ag/ZnO+LED group were prepared as described above. Subsequently, these samples were incubated with 0–100 μg/mL TA for 24 h of culture. The generated biofilms were evaluated by CV staining. We also prepared planktonic S. mutans samples of the S. m. group, +LED group, +Ag/ZnO group, and +Ag/ZnO+LED group and incubated them with 100 μg/mL TA for 24-h culture and used the dilution plating procedure to evaluate the live cell numbers in the generated biofilms. All the assays were performed in triplicate.
Biofilm Imaging
Planktonic S. mutans samples of the S. m. group, +LED group, +Ag/ZnO group, and +Ag/ZnO+LED group were prepared in a 96-well plate with or without 100 μg/mL TA to form biofilms. We first used an ordinary camera to capture an image of the generated biofilms in a 96-well plate. Then, the generated biofilms were subjected to a LIVE/DEAD® BacLight™ bacterial viability assay (Thermo Fisher Scientific, USA). Green fluorescence (SYTO 9) and red fluorescence (propidium iodide, PI) were excited by the BP460–480 and BA495-540 sections, respectively. All images were captured by fluorescence microscopy (IX73, Olympus, Japan) with a 40X objective (NA 0.6, WD 2.7–4.0 mm).
The biofilm generated after +Ag/ZnO+LED treatment was further imaged by field-emission scanning electron microscopy (FE-SEM). We prepared planktonic S. mutans samples of the S. m. group, +LED group, +Ag/ZnO group, and +Ag/ZnO+LED group as described above. These bacteria were cultured on coverslips to form biofilms and then fixed with 2.5% glutaraldehyde solution at 4 °C overnight. All samples were dehydrated with a graded ethanol series (50, 75, 85, 95, and 100%), sputter-coated with platinum, and imaged by FE-SEM (GeminiSEM 300, Zeiss, Germany) with extra-high tension at 3.0 kV.
Results
Bacterial Killing Efficiency of the Synergistic Therapy
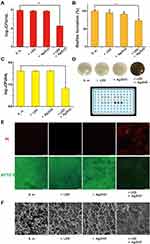
Our previous study demonstrated that synergistic therapy has excellent antibiofilm effects against S. mutans with high bacteria-killing efficiency.20 To understand its antibiofilm formation mechanism, the first aim of this study was to quantify the percentages of dead and surviving bacteria. As shown in Figure 2A, treatment with Ag/ZnO (1 mg/mL) combined with 5-min LED radiation caused 99.8% bacterial death and 0.2% survival, as calculated by a 3.35 log reduction in living bacteria CFU. However, only Ag/ZnO nanocomposites or LED radiation failed to kill most bacteria alone.
Biofilm Formation Ability of the Bacteria That Survived Synergistic Therapy
Subsequently, we focused on evaluating the biofilm formation ability of these 0.2% surviving bacteria after treatment with Ag/ZnO combined with LED radiation. Compared with the S. m. group, biofilm formation decreased significantly (p = 0.021 in a t-test) in the +Ag/ZnO+LED group, from 100% to 72.7%, as evaluated by CV staining (Figure 2B). This decreasing trend was further confirmed by counting the living bacterial CFU in these newly generated biofilms. The CFU in the +Ag/ZnO+LED group decreased by 0.79 log, indicating a 42.5% reduction in the bacterial number in the new biofilm compared with that of the S. m. group (Figure 2C). The reduction in biofilm formation was notable even in normal observations. All three groups except for the +Ag/ZnO+LED group could form thick and yellow biofilm layers. In contrast, the black background of the 96-well plate could be viewed through the newly developed thin biofilm of the +Ag/ZnO+LED group (Figure 2D). We further stained the biofilm generated by surviving bacteria with a live/dead bacterial viability kit containing SYTO 9 (green fluorescence) and PI (red fluorescence). Figure 2E shows that the green fluorescence was strong in the three groups except for the +Ag/ZnO+LED group. However, very dim green fluorescence was observed in the +Ag/ZnO+LED group, suggesting fewer living bacteria in the biofilm generated by surviving bacteria after treatment with Ag/ZnO combined with LED radiation. FE-SEM imaging further confirmed the destruction of biofilm formation by synergistic therapy (Figure 2F). In the S. m. group, +LED group, and +Ag/ZnO group, we observed intact biofilms by FE-SEM, consisting of bacteria with plump rod shapes. In contrast, no intact bacteria were found in the +Ag/ZnO+LED group.
Properties of Newly Formed Biofilms by Surviving Bacteria After Synergistic Therapy
We then estimated the properties of newly generated biofilms by recording adherence and EPS contents. As shown in Figure 3A, Ag/ZnO nanocomposites and LED radiation slightly reduced sucrose-independent and sucrose-dependent adherence (<20%) compared with the S. m. group. However, the adherence in the +Ag/ZnO+LED group was reduced most significantly (a 48.5% decrease in sucrose-independent samples and a 69.7% decrease in sucrose-dependent samples). Similar to adherence, both water-soluble and water-insoluble EPS contents in the biofilm formed by surviving bacteria were notably reduced with synergistic therapy. Compared with the S. m. group, the water-soluble EPS decreased 41.5% (p = 0.021 in a t-test), and the water-insoluble EPS decreased 65.5% (p = 0.020 in a t-test) in the +Ag/ZnO+LED group. Meanwhile, only Ag/ZnO nanocomposites or LED radiation had almost no effects on water-soluble EPS and only slightly reduced water-insoluble EPS (Figure 3B).
Proteomic Profiling of Bacteria After Synergistic Therapy
We executed a quantitative proteomic analysis based on TMT labelling for bacteria before and after synergistic therapy. In total, 1552 proteins were identified and quantified. The incubation of Ag/ZnO nanocomposites (1 mg/mL) combined with LED radiation (5 min) significantly regulated 55 proteins in S. mutans bacteria (fold change>1.2, p < 0.05 in a t-test) (Figure 4A). Among these 55 differentially expressed proteins, 12 (21.8%) were upregulated, and 43 (78.2%) were downregulated, as listed in Figure 4B and Table S1.
Functional Categorization of Differentially Expressed Proteins
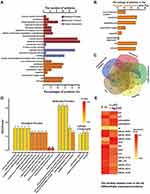
An array of bioinformatic analyses was carried out based upon the proteomic data to explain the death of S. mutans bacteria and the biofilm formation inhibition found in surviving bacteria. Figure 5A lists the GO analysis results (only GO terms in level 2 are presented). Briefly, the 55 differentially expressed proteins were involved in 10 biological process terms, 6 molecular function terms, and 8 cellular component terms. Among them, the “metabolic” term (GO:0008152) contained 28 proteins (50.9%), the “catalytic activity” term (GO:0003824) contained 24 proteins (43.6%), the “membrane” term (GO:0016020) and the “cell” term (GO:0005623) contained 15 proteins (43.6%) (Table S2). The GO terms in the cellular component section are compared in Figure 5B and C. Twenty-seven differentially expressed proteins were classified according to their locations in the cellular architectures, including 6 in macromolecular complexes (22.2%), 15 in membranes (55.5%), 6 in extracellular regions (22.2%) and 3 (11.1%) in organelles. Among the overlapping proteins, our Venn analysis reported that 50S ribosomal protein L7/L12 (rplL), elongation factor Ts (tsf), and 50S ribosomal protein L33 (rpmG) appeared in both macromolecular complexes and organelles. Meanwhile, glucan-binding protein D with lipase activity BglB-like (gbpD), A0A0A0RAT8 (a kind of protein antigen (PA)), levansucrase (SMU82_04583), and a putative Zn-dependent protease (SMU29_04639) were located in both the membrane and extracellular regions. We further carried out enrichment analysis for GO terms and listed the top 20 terms in Figure 5D and Table S3, with rich factors ranging from 0.051 to 1.00 (p < 0.05 in a t-test). These represented biological processes that were mainly associated with DNA replication, the immune system, the defence response, and metabolic processes. Among them, cellular macromolecule metabolic process (GO:0044260) and macromolecule metabolic process (GO:0043170) contained the most differentially expressed proteins (41.8% of 55 proteins). In the molecular function category, the presented terms were mainly involved in prosthetic group binding, prosthetic group of acyl carrier proteins (ACP) phosphopantetheine attachment site binding, phosphopantetheine binding, and modified amino acid binding. Every term contained 2 differentially expressed proteins, while two kinds of acyl carrier proteins, acpP-1 and acpP-2, were involved in these functional terms. In the enrichment analysis of cellular components, the terms extracellular region, cell wall, and external encapsulating structure appeared to be most significant.
Pathway Analysis
In addition to GO analysis, we mapped 6 differentially expressed proteins into 5 KEGG pathways (Table 2). Among them, D-alanine-poly(phosphoribitol) ligase (dltC) was in the D-alanine metabolism pathway, while SMU82_04583 and dextranase (dexA) were in the starch and sucrose metabolism pathway. These two pathways were reported to be highly related to biofilm formation.32–34 Given this importance, we listed the involved GO terms dltC and dexA. dexA was located in the terms metabolic process, catalytic activity, extracellular region, cell, cell part, extracellular region, external encapsulating structure, and extracellular region. Similarly, dltC was located in the terms single-organism process, biological regulation, regulation of biological process, cellular component organization or biogenesis, developmental process, cellular process, metabolic process, D-alanyl carrier activity, binding, cellular macromolecule metabolic process, and macromolecule metabolic process.
 |
Table 2 The KEGG Pathways Related to Differentially Expressed Proteins |
Biofilm-Related Protein Network
Finally, a heat map of biofilm-related proteins was constructed to visualize the antibiofilm formation mechanism, as shown in Figure 5E. Remarkably, approximately one-third of differentially expressed proteins (17 proteins) have been reported to be associated with biofilms. In this biofilm-related protein network, 5 proteins (29.4%) were upregulated, and 12 (70.6%) were downregulated. With fold changes ranging from 0.30 to 1.84, their differential expression can disturb bacterial adhesion,35 dental plaque development,36 and biofilm architecture (Table S1).37
Transcriptional Levels of Metabolic-Related Genes
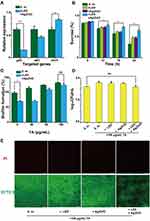
Data from biofilm formation and proteomics suggested that Ag/ZnO nanocomposites with LED radiation killed most S. mutans bacteria and inhibited the biofilm formation of the surviving bacteria. Furthermore, our bioinformatic analysis based upon proteomics indicated that synergistic therapy regulated the expression of essential proteins involved in the D-alanine metabolism pathway and the sucrose metabolism pathway. Such regulation may inhibit related metabolic pathways and contribute significantly to antibiofilm formation actions on surviving bacteria. Herein, we selected 2 metabolism-related proteins, dltC, and dexA, based on their importance in biofilm formation, D-alanine metabolism, and sucrose metabolism.32–34 The transcriptional levels of their corresponding genes were estimated using RT-qPCR, which further validated their up- or downregulated expression.38,39 Meanwhile, the same RT-qPCR assay was performed to assess the transcriptional level of glucosyltransferase-SI (gtfC). Our former study demonstrated the close relationship among biofilm formation, Ag/ZnO-based therapy and the expression regulation of genes such as gtfC.17,20 gtfC acted as a representative for these genes. Similar to the proteomic response, gtfC was downregulated 72.2%, dltC was downregulated 27.2%, and dexA was upregulated 34.5% with synergistic therapy (Figure 6A).
Inhibition of Sucrose Metabolism
Subsequently, we directly demonstrated the inhibitory effects of synergistic therapy on sucrose metabolism. Beginning with 1% sucrose in culture medium, we tested the sucrose consumption for the S. m. group, +LED group, +Ag/ZnO group, and +Ag/ZnO+LED group at 6, 12, 18, and 24 h after treatments. The differences between the S. m. group and the +Ag/ZnO+LED group occurred most notably (p = 0.036 in a t-test) at 12 h. At 24 h, the sucrose consumption of bacteria was reduced by 48.3% (p = 0.0065 in a t-test) with synergistic therapy. Greater than 2-fold more sucrose remained in the +Ag/ZnO+LED group than in the S. m. group (Figure 6B).
Inhibition of D-Alanine Metabolism by Synergistic Therapy and Recovery of Biofilm Formation by TA Supplementation
The downregulated expression of dltC can decrease the amount of poly(ribitol) phosphate TA biosynthesis, the critical product of the D-alanine metabolism pathway.40 To estimate the effects of this issue on biofilm formation, we added poly(ribitol) phosphate TA extracts to the bacterial cultures. With the gradual supplementation of TA extracts, the significant reduction in biofilm formation almost disappeared in the +Ag/ZnO+LED group (Figure 6C). The biofilm formation of surviving bacteria was further estimated by colony counting. Compared with the S. m. group, no significant difference was observed in the +Ag/ZnO+LED group with 100 μg/mL TA extracts (Figure 6D). The recovering effects of TA extracts on biofilm formation also agreed with the biofilm images. For bacteria treated by synergistic therapy, thicker and more compacted biofilms were obtained by a supplementation with 100 μg/mL TA extracts, as demonstrated by the comparatively strong green fluorescence and dim red fluorescence shown in Figure 6E.
Discussion
In a review of the literature, Ag/ZnO nanocomposites have been shown to act as excellent antimicrobial reagents against diverse bacteria, including S. mutans, even with low dosage use.16,17 Combined with 5-min LED radiation, Ag/ZnO nanocomposites can effectively inhibit the biofilm formation of S. mutans.20 However, these publications mainly focused on the antimicrobial properties of Ag/ZnO-based therapies, while we need to consider deep exploration of the antibiofilm formation mechanism. Furthermore, the biofilm formation ability of surviving bacteria after synergistic therapy was more critical for evaluating the potential applications of Ag/ZnO-based therapies. Ag/ZnO-based therapies induced comprehensive proteomic responses in bacteria, but only several biofilm-related genes were selected for RT-qPCR quantification, limited to current reports and our knowledge.17,20,22 Since proteomics has developed rapidly for studies of the toxicity and antimicrobial actions of NPs,24–27 an integrated approach centred on proteomics can benefit us in exploring the antibiofilm formation mechanism against S. mutans.
In this study, 99.8% of planktonic bacteria were killed with synergistic therapy, while 0.2% of bacteria survived the treatment. Similar to our former study,20 we attributed the antibiofilm formation actions of Ag/ZnO-based therapies to bacterial killing. However, the surviving 0.2% of bacteria may proliferate to generate new biofilms, and little has been known about these newly formed biofilms until now. In this work, we first confirmed that the biofilm formation of surviving bacteria was significantly reduced (Figure 2). Furthermore, the newly formed biofilms exhibited properties of lower adherence (both sucrose-independent and sucrose-dependent biofilms) and EPS contents (both water-soluble and insoluble biofilms) (Figure 3). Lower adherence confirmed the direct inhibition of biofilm formation. The biofilm formation of S. mutans begins with adhesion.29 However, daily tooth cleaning can easily remove newly generated biofilms with low adhesion. For EPS content, higher EPS could inhibit drug access by increasing biofilm adhesion and stability.41,42 When subjected to synergistic therapy, both insoluble (primarily α1,3-linked glucans) and soluble (mostly α1,6-linked glucans) EPS were notably reduced in the newly formed biofilms. Therefore, synergistic therapy led to low-quality biofilms even though some bacteria survived. Moreover, fluorescent images showed that biofilms developed by the surviving bacteria were thin and low-density. The FE-SEM images further validated the destruction of biofilm formation. In summary, synergistic therapy caused most S. mutans to die and inhibited the biofilm formation capability of the surviving biofilms, which could only generate thin and low-density biofilms with lower adhesion and EPS.
Ag/ZnO-based therapies regulate the expression of biofilm-related genes and damage bacterial protein synthesis.17,20,22 This inspired us to explore the antibiofilm formation mechanism from the proteomic response towards synergistic therapy. Among 55 differentially expressed proteins, the number of downregulated proteins was approximately 3.6-fold that of upregulated proteins (Figure 4). In the relevant bioinformatic study, structural destruction and expression regulation of critical structure-related proteins were indicated as the main reasons for bacterial death. Several differentially expressed proteins were found to be essential in DNA replication, immune system, defence response, binding, and metabolic processes. Finally, a biofilm-related network of differentially expressed proteins was mined, thus explaining the multiantibiofilm formation actions of Ag/ZnO-based therapy.
According to the GO analysis, the terms in the cellular component section indicated that structural destruction and critical structure-related protein expression regulation should be the main causes of bacterial death following treatment. Similar to Ag NP toxicity to P. aeruginosa,24 remarkably, 15 membrane proteins were observed among the 55 differentially expressed proteins, which were highly related to transfructosylation, carbohydrate uptake, metallic transport, and biofilm formation. Significantly, downregulated SMU82_04583, sortase (srtA), and sorbitol-specific enzyme IIA (SMU82_07124) disturb the transfructosylation of levan,43 surface protein modification,44 and carbohydrate utilization.45 Several metallic enzymes, such as SMU29_04639, and metallic transporters, such as putative Mg2+/citrate transporters (SMU29_08849), were also regulated, which may assist with Ag/ZnO transport and bacteria killing. A0A0A0R886 and A0A0A0RAT8 were found to be upregulated, as they are responsible for the sucrose-independent initial adhesion process.35,46 In addition to those membrane proteins, the differentially expressed proteins in macromolecular complexes and organelles were reported to be related to DNA replication (DNA polymerase III (SMU29_09019)), transcription (MepR (mepR)47), and translation (rplL, rpmG, and tsf48). Using GO enrichment analysis, the top three terms in the cellular component terms were extracellular region, cell wall, and external encapsulating structure. These terms all partly overlapped with membrane proteins, further indicating the importance of the identified membrane proteins. Imaging by scanning electron microscopy (SEM) revealed that the synergistic therapy resulted in the scattering of membrane components of S. mutans bacteria from their original ordered structure, and the rod-like morphology of bacteria was completely disorganized.20 Therefore, the proteomic changes detected in the cellular components lead us to speculate that Ag/ZnO interacted with specific cell components, destroyed bacterial integrity, and affected the expression of related proteins, especially membrane proteins.
Furthermore, GO and KEGG analyses indicated that several differentially expressed proteins were associated with bacterial DNA replication, immune system, defence response, binding of essential groups, and metabolic processes. Such findings supported the reason for bacterial death, described the bacterial responses towards Ag/ZnO-based therapy, and indicated the potential inhibition of metabolism in the surviving bacteria. Typically, CRISPR-associated endonuclease Cas9 (Cas9) was mostly downregulated (fold change = 0.07), which ultimately inhibited DNA replication.49 This partly explained the bacterial death and proliferation inhibition shown in Figure 2A and C. The biological process category of the GO enrichment analysis suggested that several terms, such as response to virus and immune system process, may also be significant. These terms might be regarded as the various responses of bacteria to extracellular stimuli, such as temperature stimuli.50,51 In the molecular function category, acpP-1 and acpP-2 were involved in all main terms, affecting the binding of prosthetic groups, ACP phosphopantetheine attachment sites, phosphopantetheines, and modified amino acids. The downregulation of these two proteins (fold change = 0.72) might inhibit bacterial fatty acid and polyketide biosynthesis.52 We noted two differentially expressed proteins in the analysis of metabolic processes, ie, dexA and dltC, for their importance and close relationship with biofilm formation. In GO analysis, they were involved in multiple terms related to cellular components and metabolic processes. In KEGG analysis, dexA and dltC were separately involved in sucrose metabolism and D-alanine metabolism pathways (Table 2). Furthermore, inhibition of sucrose metabolism and D-alanine metabolism can disturb biofilm formation.32–34 Thus, the expression regulation of dexA and dltC could disturb sucrose metabolism and D-alanine metabolism, which resulted in the antibiofilm formation actions detected in surviving bacteria.
We generated a biofilm-related protein network consisting of 17 differentially expressed proteins from the proteomic analysis. Glycosyltransferases (GTFs)53 and glucan-binding proteins (GBPs)5 appeared to be typical due to their importance in sucrose-dependent adhesion. Similarly, EPS synthesis mainly uses GTFs and fructosyltransferases (FTFs).54 Herein, the reductions in adhesion and EPS shown in Figure 3A and B were explained by the up- or downregulation of glucan-binding protein A (SMU82_00485), gbpD, glucan-binding protein C (SMU82_05402), glucosyltransferase-SI (fragment) (SMU82_09792), and gtfC. In addition, several other biofilm-related proteins were mentioned above, including two PAs (A0A0A0R886 and A0A0A0RAT8),35,46 tsf,36 and srtA.44 The notable proteomic changes shown in the heat map in Figure 5E demonstrated the multiple antibiofilm formation actions of the synergistic therapy against S. mutans.
Finally, the regulation of the expression of metabolism-related dexA and dltC was validated to be responsible for the biofilm formation inhibition of the surviving bacteria. As several biofilm-associated genes, such as gtfC, have been subjected to RT-qPCR in our former Ag/ZnO-related studies,17,20 we selected gtfC as a representative and quantified the transcription levels of dexA, dltC and gtfC. The trends of the expression levels of these three genes were consistent with the fold changes in protein expression. The up- or downregulated transcription levels further validated the differential expression of these three proteins.
Then, we reported the inhibition of sucrose metabolism and D-alanine metabolism in surviving bacteria after synergistic therapy. As a type of glucanase, dexA catalyses the degradation of dextran to low-molecular-weight fractions. The increase in dexA expression can disturb dextran-dependent aggregation in biofilm formation.33,34 The sucrose consumption decrease directly confirmed the inhibition of sucrose metabolism. With a 24-h culture, synergistic therapy reduced sucrose consumption by approximately half (Figure 6B). Such inhibition of sucrose metabolism, induced by an increase in dexA expression, could ultimately result in lower EPS contents and adherences, in agreement with related reports.33 For dltC, its downregulation reduces poly(ribitol) phosphate TA biosynthesis, which disturbs the D-alanine metabolism pathway.40,55 The essential role of TA in biofilm formation has been reported, and TA-deficient cells have a reduced capability for biofilm formation.56 The application of tunicamycin successfully inhibited Staphylococcus aureus (S. aureus) bacteria from forming biofilms by disturbing TA synthesis.57 For surviving bacteria after synergistic therapy, we supplemented poly(ribitol) phosphate TA extracts and observed the recovery of biofilm formation (Figure 6). The recovery indicated that the synergistic therapy had a similar antibiofilm formation effect as that reported for tunicamycin. The combination of Ag/ZnO nanocomposites with LED radiation inhibited the D-alanine metabolism pathway by reducing poly(ribitol) phosphate TA biosynthesis, and such inhibition reduced the biofilm formation capability of surviving bacteria. These findings unveiled and validated the antibiofilm formation actions of synergistic therapy on surviving bacteria via metabolic pathways for the first time.
Conclusion
In conclusion, the antibiofilm formation mechanism of Ag/ZnO combined with LED irradiation against S. mutans involved killing most bacteria and reducing the biofilm formation ability and decreasing the regrown biofilm quality of surviving bacteria. Our proteomic analysis validated that synergistic therapy notably regulated the expression of proteins related to bacterial structure, metabolic processes, and biofilm formation. The subsequent metabolic study demonstrated that the inhibition of sucrose and D-alanine metabolism prevented the surviving bacteria from forming high-quality biofilms. These findings elucidated the antibiofilm formation mechanism at the molecular level. Furthermore, our success can provide a feasible showcase for exploring the antibiofilm formation mechanism of NP-based therapy against microorganisms in addition to S. mutans.
Acknowledgments
We thank the Research Grants of the Wuhan Institute of Technology (K202063), Science and Technology Research Project of the Hubei Provincial Department of Education (Q20201511).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Benoit DS, Sims KR
2. Han C, Romero N, Fischer S, et al. Recent developments in the use of nanoparticles for treatment of biofilms. Nanotechnol Rev. 2017;6(5):383–404. doi:10.1515/ntrev-2016-0054
3. Yousef MS, Abdelhamid HN, Hidalgo M, et al. Antimicrobial activity of silver-carbon nanoparticles on the bacterial flora of bull semen. Theriogenology. 2021;161:219–227. doi:10.1016/j.theriogenology.2020.12.006
4. Pitts NB, Zero DT, Marsh PD, et al. Dental caries. Nat Rev Dis Primers. 2017;3(1):17030. doi:10.1038/nrdp.2017.30
5. Matsumoto-Nakano M. Role of Streptococcus mutans surface proteins for biofilm formation. Jpn Dent Sci Rev. 2018;54(1):22–29. doi:10.1016/j.jdsr.2017.08.002
6. Covarrubias C, Trepiana D, Corral C. Synthesis of hybrid copper-chitosan nano-particles with antibacterial activity against cariogenic Streptococcus mutans. Dent Mater J. 2018;37(3):379–384. doi:10.4012/dmj.2017-195
7. Morita Y, Tomida J, Kawamura Y. Responses of Pseudomonas aeruginosa to antimicrobials. Front Microbiol. 2014;4:422. doi:10.3389/fmicb.2013.00422
8. Mazhari F, Boskabady M, Moeintaghavi A, et al. The effect of toothbrushing and flossing sequence on interdental plaque reduction and fluoride retention: a randomized controlled clinical trial. J Periodontol. 2018;89(7):824–832. doi:10.1002/JPER.17-0149
9. Javidi M, Zarei M, Naghavi N, et al. Zinc oxide nanoparticles as sealer in endodontics and its sealing ability. Contemp Clin Dent. 2014;5(1):20–24. doi:10.4103/0976-237X.128656
10. Li X, Luo C, Fu Q, et al. A transparent, wearable fluorescent mouthguard for high-sensitive visualization and accurate localization of hidden dental lesion sites. Adv Mater. 2020;32(21):2000060. doi:10.1002/adma.202000060
11. Mousavi SA, Ghotaslou R, Akbarzadeh A, et al. Evaluation of antibacterial and antifungal properties of a tissue conditioner used in complete dentures after incorporation of ZnO-Ag nano-particles. J Dent Res Dent Clin Dent Prospects. 2019;13(1):11–18. doi:10.15171/joddd.2019.002
12. Liu H, Zhong L, Govindaraju S, et al. ZnO rod decorated with Ag nanoparticles for enhanced photocatalytic degradation of methylene blue. J Phys Chem Solids. 2019;129:46–53. doi:10.1016/j.jpcs.2018.12.040
13. Huang J, Huang G, An C, et al. Exploring the use of ceramic disk filter coated with Ag/ZnO nanocomposites as an innovative approach for removing Escherichia coli from household drinking water. Chemosphere. 2020;245:125545. doi:10.1016/j.chemosphere.2019.125545
14. Khatami M, Varma RS, Zafarnia N, et al. Applications of green synthesized Ag, ZnO and Ag/ZnO nanoparticles for making clinical antimicrobial wound-healing bandages. Sustain Chem Pharm. 2018;10:9–15. doi:10.1016/j.scp.2018.08.001
15. Naskar A, Khan H, Sarkar R, et al. Anti-biofilm activity and food packaging application of room temperature solution process based polyethylene glycol capped Ag-ZnO-graphene nanocomposite. Mat Sci Eng C Mater. 2018;91:743–753. doi:10.1016/j.msec.2018.06.009
16. Wang S, Wu J, Yang H, et al. Antibacterial activity and mechanism of Ag/ZnO nanocomposite against anaerobic oral pathogen Streptococcus mutans. J Mater Sci Mater M. 2017;28(1):23. doi:10.1007/s10856-016-5837-8
17. Huang Q, Wang S, Sun Y, et al. Effects of Ag/ZnO nanocomposite at sub-minimum inhibitory concentrations on virulence factors of Streptococcus mutans. Arch Oral Biol. 2020;111:104640. doi:10.1016/j.archoralbio.2019.104640
18. Alasiri RA, Algarni HA, Alasiri RA. Ocular hazards of curing light units used in dental practice-a systematic review. Saudi Dent J. 2019;31(2):173–180. doi:10.1016/j.sdentj.2019.02.031
19. Price RB. Light curing in dentistry. Dent Clin North Am. 2017;61(4):751–778. doi:10.1016/j.cden.2017.06.008
20. Wang S, Huang Q, Liu X, et al. Rapid antibiofilm effect of Ag/ZnO nanocomposites assisted by dental LED curing light against facultative anaerobic oral pathogen Streptococcus mutans. ACS Biomater Sci Eng. 2019;5(4):2030–2040. doi:10.1021/acsbiomaterials.9b00118
21. Khan I, Paul P, Behera SK, et al. To decipher the antibacterial mechanism and promotion of wound healing activity by hydrogels embedded with biogenic Ag@ZnO core-shell nanocomposites. Chem Eng J. 2021;417:128025. doi:10.1016/j.cej.2020.128025
22. Wei X, Li Q, Wu C, et al. Preparation, characterization and antibacterial mechanism of the chitosan coatings modified by Ag/ZnO microspheres. J Sci Food Agric. 2020;100(15):5527–5538. doi:10.1002/jsfa.10605
23. Vidova V, Spacil Z. A review on mass spectrometry-based quantitative proteomics: targeted and data independent acquisition. Anal Chim Acta. 2017;964:7–23. doi:10.1016/j.aca.2017.01.059
24. Yan X, He B, Liu L, et al. Antibacterial mechanism of silver nanoparticles in Pseudomonas aeruginosa: proteomics approach. Metallomics. 2018;10(4):557–564. doi:10.1039/C7MT00328E
25. Abdelhamid HN, Wu HF. Proteomics analysis of the mode of antibacterial action of nanoparticles and their interactions with proteins. Trends Analyt Chem. 2015;65:30–46. doi:10.1016/j.trac.2014.09.010
26. Zhang Y, Pan X, Liao S, et al. Quantitative proteomics reveals the mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa biofilms. J Proteome Res. 2020;19(8):3109–3122. doi:10.1021/acs.jproteome.0c00114
27. Gedda G, Abdelhamid HN, Khan MS, et al. ZnO nanoparticle-modified polymethyl methacrylate-assisted dispersive liquid–liquid microextraction coupled with MALDI-MS for rapid pathogenic bacteria analysis. RSC Adv. 2014;4(86):45973–45983. doi:10.1039/C4RA03391D
28. Hasan S, Danishuddin M, Adil M, et al. Efficacy of E. officinalis on the cariogenic properties of Streptococcus mutans: a novel and alternative approach to suppress quorum-sensing mechanism. PLoS One. 2012;7(7):e40319. doi:10.1371/journal.pone.0040319
29. Liu Y, Xu Y, Song Q, et al. Anti-biofilm activities from Bergenia crassifolia leaves against Streptococcus mutans. Front Microbiol. 2017;8:1738. doi:10.3389/fmicb.2017.01738
30. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
31. Nowotny A. Extraction of teichoic acid. In: Basic Exercises in Immunochemistry. Berlin: Heidelberg; 1979:78–80. doi:10.1007/978-3-642-67356-6_24
32. Nilsson M, Rybtke M, Givskov M, et al. The dlt genes play a role in antimicrobial tolerance of Streptococcus mutans biofilms. Int J Antimicrob Agents. 2016;48(3):298–304. doi:10.1016/j.ijantimicag.2016.06.019
33. Yang Y, Mao M, Lei L, et al. Regulation of water-soluble glucan synthesis by the Streptococcus mutans dexA gene effects biofilm aggregation and cariogenic pathogenicity. Mol Oral Microbiol. 2019;34(2):51–63. doi:10.1111/omi.12253
34. Ebaya MMA, El-Mowafy M, Adel El-Sokkary MM, et al. Purification, characterization, and biocatalytic and antibiofilm activity of a novel dextranase from Talaromyces sp. Int J Microbiol. 2020;2020(9):9198048. doi:10.1155/2020/9198048
35. Lapirattanakul J, Nomura R, Matsumoto-Nakano M, et al. Variation of expression defects in cell surface 190-kDa protein antigen of Streptococcus mutans. Int J Med Microbiol. 2015;305(3):383–391. doi:10.1016/j.ijmm.2015.03.001
36. Yi L, Wang Y, Ma Z, et al. Biofilm formation of Streptococcus equi ssp. zooepidemicus and comparative proteomic analysis of biofilm and planktonic cells. Curr Microbiol. 2014;69(3):227–233. doi:10.1007/s00284-014-0574-z
37. Yadav P, Verma S, Bauer R, et al. Deciphering streptococcal biofilms. Microorganisms. 2020;8(11):1835. doi:10.3390/microorganisms8111835
38. Wang Y, Ma C, Zhang J, et al. Investigation of cobalt bioaccumulation in Rhodopseudomonas palustris by proteomics analysis. Anal Methods. 2019;11(33):4280–4288. doi:10.1039/C9AY00406H
39. Fernández de Mera IG, Chaligiannis I, Hernández-Jarguín A, et al. Combination of RT-PCR and proteomics for the identification of Crimean-Congo hemorrhagic fever virus in ticks. Heliyon. 2017;3(7):e00353. doi:10.1016/j.heliyon.2017.e00353
40. Mazda Y, Kawada-Matsuo M, Kanbara K, et al. Association of CiaRH with resistance of Streptococcus mutans to antimicrobial peptides in biofilms. Mol Oral Microbiol. 2012;27(2):124–135. doi:10.1111/j.2041-1014.2012.00637.x
41. Costa Oliveira BE, Cury JA, Ricomini Filho AP. Biofilm extracellular polysaccharides degradation during starvation and enamel demineralization. PLoS One. 2017;12(7):e0181168. doi:10.1371/journal.pone.0181168
42. Heredia-Ponce Z, Gutiérrez-Barranquero JA, Purtschert-Montenegro G, et al. Role of extracellular matrix components in the formation of biofilms and their contribution to the biocontrol activity of Pseudomonas chlororaphis PCL1606. Environ Microbiol. 2020;23(4):2086–2101. doi:10.1111/1462-2920.15355
43. Xu W, Ni D, Zhang W, et al. Recent advances in levansucrase and inulosucrase: evolution, characteristics, and application. Crit Rev Food Sci. 2019;59(22):3630–3647. doi:10.1080/10408398.2018.1506421
44. Hu P, Huang P, Chen MW. Curcumin reduces Streptococcus mutans biofilm formation by inhibiting sortase A activity. Arch Oral Biol. 2013;58(10):1343–1348. doi:10.1016/j.archoralbio.2013.05.004
45. Higgins MA, Hamilton AM, Boraston AB. Structural characterization of the PTS IIA and IIB proteins associated with pneumococcal fucose utilization. Proteins. 2017;85(5):963–968. doi:10.1002/prot.25264
46. Rivera-Quiroga RE, Cardona N, Padilla L, et al. In silico selection and in vitro evaluation of new molecules that inhibit the adhesion of Streptococcus mutants through antigen I/II. Int J Mol Sci. 2021;22(1):377. doi:10.3390/ijms22010377
47. Deochand DK, Grove A. MarR family transcription factors: dynamic variations on a common scaffold. Crit Rev Biochem Mol. 2017;52(6):595–613. doi:10.1080/10409238.2017.1344612
48. Gromadski KB, Wieden HJ, Rodnina MV. Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry. 2002;41(1):162–169. doi:10.1021/bi015712w
49. Allen F, Crepaldi L, Alsinet C, et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol. 2019;37(1):64–72. doi:10.1038/nbt.4317
50. Emery O, Schmidt K, Engel P. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol Ecol. 2017;26(9):2576–2590. doi:10.1111/mec.14058
51. Yang JJ, Ma YF, Yin Y, et al. Antioxidative defense response of Ulva prolifera under high or low-temperature stimulus. Algal Res. 2019;44:101703. doi:10.1016/j.algal.2019.101703
52. Ma JC, Wu YQ, Cao D, et al. Only acyl carrier protein 1 (AcpP1) functions in Pseudomonas aeruginosa fatty acid synthesis. Front Microbiol. 2017;8:2186. doi:10.3389/fmicb.2017.02186
53. Scharnow AM, Solinski AE, Wuest WM. Targeting S. mutans biofilms: a perspective on preventing dental caries. Med Chem Comm. 2019;10(7):1057–1067. doi:10.1039/C9MD00015A
54. Karygianni L, Ren Z, Koo H, et al. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 2020;28(8):668–681. doi:10.1016/j.tim.2020.03.016
55. Wang Z, Shen Y, Haapasalo M. Antibiofilm peptides against oral biofilms. J Oral Microbiol. 2017;9(1):1327308. doi:10.1080/20002297.2017.1327308
56. Brown S, Santa Maria JP, Walker S. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol. 2013;67(1):313–336. doi:10.1146/annurev-micro-092412-155620
57. Zhu X, Liu D, Singh AK, et al. Tunicamycin mediated inhibition of wall teichoic acid affects Staphylococcus aureus and Listeria monocytogenes cell morphology, biofilm formation and virulence. Front Microbiol. 2018;9:1352. doi:10.3389/fmicb.2018.01352
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.