Back to Journals » Journal of Hepatocellular Carcinoma » Volume 11
PRMT1 Integrates Immune Microenvironment and Fatty Acid Metabolism Response in Progression of Hepatocellular Carcinoma
Authors Yan J, Li KX, Yu L, Yuan HY, Zhao ZM, Lin J, Wang CS
Received 26 October 2023
Accepted for publication 20 December 2023
Published 6 January 2024 Volume 2024:11 Pages 15—27
DOI https://doi.org/10.2147/JHC.S443130
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Mohamed Shaker
Jia Yan,1– 3,* Ke xin Li,2,* Lei Yu,2 Heng ye Yuan,2 Zhi min Zhao,2 Jing Lin,2,4 Chang Shan Wang2
1School of Basic Medicine, Inner Mongolia Medical University, Hohhot, Inner Mongolia, People’s Republic of China; 2College of Life Science, Inner Mongolia University, Hohhot, Inner Mongolia, People’s Republic of China; 3Medical Experimental Center of Basic Medical School, Inner Mongolia Medical University, Hohhot, Inner Mongolia, People’s Republic of China; 4Affiliated Hospital of Inner Mongolia Medical University, Hohhot, Inner Mongolia, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chang Shan Wang; Jing Lin, Email [email protected]; [email protected]
Background: Protein arginine methyltransferase (PRMT) family members have important roles in cancer processes. However, its functions in the regulation of cancer immunotherapy of hepatocellular carcinoma (HCC) are incompletely understood. This study aimed to investigate the roles of PRMT1 in HCC.
Methods: Single-cell RNA sequencing (scRNA-seq) and clinicopathological data were obtained and used to explore the diagnostic and prognostic value, cellular functions and roles in immune microenvironment regulation of PRMT1 in HCC. The functions of PRMT1 were explored using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO), as well as gene set enrichment analysis (GSEA). TIMER and CIBERSORT were used to analyze the relationships between PRMT1 expression and immune cell infiltration. The STRING database was used to construct a protein–protein interaction (PPI) network.
Results: PRMT1 was aberrantly expressed in HCC, which high expression was associated with tumor progression, worse overall survival (OS) and disease-free survival (DFS) of patients with HCC. PRMT1 was also associated with immune cell infiltration. Moreover, it was specifically expressed in immune cells, including exhausted CD8 T cells, B cells, and mono/macro cells in patients with immunotherapy. The expression of immune checkpoints was significantly increased in the high-PRMT1 expression groups of HCC patients. Regarding biological mechanisms, cell viability, migration and invasion, and the expression of genes related to fatty acid metabolism were suppressed in PRMT1 knockdown HCC cells. Moreover, genes co-expressed with PRMT1 were involved in the fatty acid metabolic process and enriched in fatty and drug-induced liver disease.
Conclusion: Taken together, these results indicate that PRMT1 might exert its oncogenic effects via immune microenvironment regulation and fatty acid metabolism in HCC. Our finding will provide a foundation for further studies and indicate a potential clinical therapeutic target for liver cancer.
Keywords: protein arginine methyltransferase, PRMT1, prognosis, tumor-infiltrating, fatty acid metabolism
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver malignancy worldwide and has increasing incidence and mortality.1 The pathophysiology of HCC involves in multistep processes. Owing to a lack of effective methods for early diagnosis, most HCC cases are diagnosed at an advanced stage.2,3 Currently, various therapies have been developed for the treatment of liver cancer, including combinations of radiotherapy, chemotherapy, hormonal therapy, and targeted therapy.1,3 In recent years, combinations of immunotherapy with targeted therapy have demonstrated practical clinical efficacy in HCC treatment.4 Post-translational modifications (PTMs), such as phosphorylation, acetylation, methylation, glycosylation, and ubiquitination have a major impact on protein function and have been shown to be associated with the development of various diseases including tumors, neurological diseases, and metabolic diseases.5
Protein methylation is a common PTM that has a pivotal role in regulation of cellular functions.6,7 In particular, accumulating evidence indicates that protein arginine methylation is an indispensable model of methylation related-PTM, which is regulated by a family of enzymes, named protein arginine methyltransferases 1–9 (PRMT1–9).7 PRMT1, PRMT2, PRMT3, PRMT4, CARM1 (coactivator associated arginine methyltransferase 1), PRMT6, and PRMT8 are involved in the formation of mono-methylarginine and asymmetric dimethylarginine; PRMT5 and PRMT9 engage in the mono-methylation and symmetric demethylation of arginine; and PRMT7 exclusively generates mono-methylarginine.7 PRMTs have crucial roles in transcription, splicing, DNA damage repair, RNA biology, and cellular metabolism.7,8 Moreover, dysregulation of PRMTs has been shown to be closely associated with immune response in several cancers.8 In recent years, PRMT inhibition has emerged as a viable therapeutic strategy, leading to the development of anticancer drugs intended to treat numerous malignancies.9,10 PRMT1 and PRMT5 inhibitors have entered the first phase of clinical trials and could pave the way for new targeted cancer interventions.10
It has been reported that the dysregulation of PRMTs is associated with cancer development.11,12 PRMT1 is the most predominant enzyme which could upregulate EZH2 expression via methylation of Arg342 to increase the expression of EZH2 targeting genes, including HOXA10, DAB2IP, HOXA9, and HOXA7, resulting in cell migration and metastasis in breast cancer.13 Moreover, PRMT1 regulates methylation of transcription factor Twist1 to promote progression of non-small cell lung cancer.9 Several studies have documented aberrant expression of PRMTs in liver cancer.14–16 PRMT1 is a key protection factor against alcohol-associated liver cancer progression by regulating the IL-6-STAT3 axis.16
PRMTs also have important roles in tumor immune response, for example, regulation of activation of toll-like receptors (TLRs) and type I interferons (IFNs).17 Recently, potential functions of PRMT1-, CARM1-, PRMT5- and PRMT7 in regulating cancer immunity have attracted attention.17 PRMT5 is significantly associated with immune related-gene signatures,18 and PRMT7 inhibition combined with immune checkpoint inhibitors shows significant anti-tumor efficacy in melanomas.19 Moreover, a PRMT1 polymorphism, rs975484, may regulate levels of PD-L1 and PD-L2, as a predictor of immunotherapy efficiency in HCC.20 Despite this, the functions of PRMTs in the immune tumor microenvironment of HCC has not been reported. In this study, we comprehensively investigated the expression characteristics, diagnostic and prognostic values, tumor immune microenvironment and related signaling pathways, to obtain a PRMT1 better understanding of its role in liver cancer.
Materials and Methods
Gene Expression Analysis
PRMT1 expression in liver hepatocellular carcinoma (LIHC) was analyzed using data from the Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) and Genotype-Tissue Expression program (GTEx) (http://commonfund.nih.gov/GTEx/). The gene expression landscape of PRMT1 in LIHC was confirmed according to the Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn), and the Gene Set Cancer Analysis (GSCA) (http://bioinfo.life.hust.edu.cn) databases. The relationship of PRMT1 with pathological stage (stages I–IV) was determined using GEPIA2 (http://gepia.cancer-pku.cn/index.html), and its expression in tumors of different grades was assessed using the “tumor grade” module of UALCAN.
Kaplan–Meier Survival Analysis
The online Kaplan–Meier plotter (https://kmplot.com) analysis tool was used to evaluate the association between expression of PRMT1 and the prognosis of patients with liver cancer. The GEPIA database was also used to assess the relationships of PRMTs with overall survival (OS) and disease-free survival (DFS) in LIHC.
Immune Infiltration Analysis
The correlations of six types of tumor-infiltrating immune cells with PRMT1 expression in LIHC were determined using the tumor immune estimation resource (TIMER) (https://cistrome.shinyapps.io/timer/) database. Moreover, the correlations between immune cell infiltration and PRMT1 expression were assessed using the immune infiltrates module in GSCA (http://bioinfo.life.hust.edu.cn) database.
Analysis of PRMT1 Expression in scRNA-Seq Datasets
The scRNA-seq data set was obtained from GSE149614 in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The R package Seurat version 3.2.3 was used for single-cell data analysis. We used Seurat’s CCA algorithm to coordinate data and correct batch effects. Cells underwent dimensionality reduction with the uniform manifold approximation and projection method (UMAP).
Gene Set Analysis of Cancer Pathways
PRMT1 related pathways was assessed by GSCA based on RNA-seq data of TCGA_LIHC cancer.
Correlation and Functional Enrichment Analyses
Expression of PRMT1 related genes was analyzed in LIHC. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the cluster Profiler R package.
Construction of Protein–Protein Interaction Network and Screening of Hub Genes
Hallmark gene sets were obtained by gene set enrichment analysis GSEA (http://www.gsea-msigdb.org/gsea/index.jsp). STRING was used to predict a network for PRMT1-related proteins, and the results were visualized with Cytoscape (http://cytoscape.org; version 3.7.2) software.
Cell Culture and Transfection
HCC cell lines (HepG2 and Huh7) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). These cells were cultured in Dulbecco’s modified eagle medium (DMEM) (Gibco, USA) containing 10% fetal bovine serum (FBS; Gibco, USA), 10 U/mL penicillin, and 10 mg/mL streptomycin (Sigma, USA). The cells were grown in a sterile incubator with a humidified atmosphere containing 5% CO2 at 37°C. A specific short hairpin RNA (shRNA) targeted to PRMT1 was synthesized by Gene Pharma (Shanghai, China). The empty vector was utilized as a negative control. Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, USA) was used for the transfection of all these vectors and reagents into cells.
Cell Viability Assays
Cell proliferation was evaluated by the MTS assay. Approximately 2×105 transfected cells per well were seeded into 96-well plates and cultured at 37°C. After incubation for 0, 24, 48, 72 and 96 h, MTS reagent (20 µL) was added, followed by further incubation for 4 h. The absorbance was determined at 490 nm. Each independent experiment was replicated at least three times.
Cell Invasion Assay
The transfected HCC cells (1×105) were suspended in 200 µL of serum-free medium and seeded into the upper chamber, and 600 µL of medium containing 10% FBS was added to the lower chamber. After incubation for 48 h at 37°C, the cells remaining on the upper surface were removed with cotton swabs. The membranes were fixed in methanol and stained with 0.5% crystal violet. Cells on the lower surface of the membrane were counted in randomly selected fields. Each independent experiment was replicated at least three times.
Cell Migration Assay
Cells were cultured in a six-well plate. After 24 hours transfection, when the cells had reached 90%~100% confluence, they were scratched with 100 μL pipette tips, and lines were drawn in parallel. Subsequently, the cells were washed twice times with phosphate-buffered saline (PBS) and cultured in serum-free DMEM at 37°C with 5% CO2. After 24 h, the wound was measured and photographed. The migration of cells was analyzed using ImageJ software. The wound healing rate was calculated as follows: 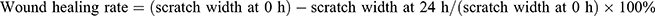 . Each independent experiment was repeated at least three times.
. Each independent experiment was repeated at least three times.
RNA Isolation and Quantitative Real-Time-PCR
Total RNA from cultured cells was extracted using TRIzol reagent (Invitrogen, CA) according to the manufacturer’s protocol. cDNA was obtained according to the protocol of Prime Script TM RT Master Mix Kit (Takara, Japan); then, real-time RT-PCR was performed using SYBR Premix Ex Taq™II (Takara, Japan) in a thermal cycler CFX6 system (Bio Rad, USA). Relative gene expression level was calculated using the 2−ΔΔCt method. Each independent experiment was replicated at least three times.
Statistical Analyses
Student’s t-test, multiple hypothesis correction (false discovery rate, FDR), and Spearman correlation analysis were performed to evaluate the relationships of PRMT1 expression with that of other genes. Kaplan–Meier curves were constructed to compare differences in survival times. P-values were calculated using Log rank tests, and values less than 0.05 were considered to indicate statistical significance. Immuno-correlation analysis in GSCA, P-values were adjusted using the false discovery rate (FDR)-method and less than 0.05 was considered statistically significant.
Results
The Clinicopathological Characteristics of PRMT1 in HCC
To explore the potential functions of PRMT1 in HCC, we first investigated its expression in patients with cancer. The GSCA results showed that PRMT1 was significantly unregulated in several cancers, including HCC (Figure 1A). We next analyzed the expression profiles of PRMT1 in liver cancer using a GEPIA dataset based on TCGA and GTEx data. As shown in Figure 1B, PRMT1 expression was increased in tumor tissues of HCC patients. Furthermore, we investigated the relationships between PRMT1 expression and clinicopathological characteristics, and found that the expression of PRMT1 was increased in stage III than that in stage II of LIHC, but significantly decreased in stage IV (P < 0.05) (Figure 1C). Moreover, the mRNA level of PRMT1 was significantly elevated in grade-3 HCC (Figure 1D). Taken together, these results indicate that levels of PRMT1 maybe be associated with pathological stage and histological grade in patients with HCC. Furthermore, clinical data and TCGA RNA-seq data were used to examine the prognostic value of PRMT1 in LIHC. We found that elevated PRMT1 level was significantly correlated with poor OS in patients with HCC (HR=1.7, P=0.0035) (Figure 1E). Moreover, PRMT1 expression was significantly associated with DFS in HCC (Figure 1F). Taken together, these results suggest that elevated PRMT1 expression could serve as a valuable biomarker for survival prediction and stage classification in HCC.
Correlations Between PRMT1 Expression and Tumor Infiltration of Immune Cells in HCC
To investigate the correlations between PRMT1 expression and immune cells infiltration, we obtained the liver cancer scRNA-seq data from the GEO database and assessed the expression of PRMT1 in liver tissue of HCC patients in single cell level. PRMT1 was significantly expressed in most types of immune cell, including T cells, B cells, natural killer cells, dendritic cells, regulatory T cells (Tregs), mast cells, and monocyte (Figure 2A–D). Next, the relationships of PRMT1 expression with infiltration levels of immune cells were confirmed based on CIBERSORT. We found that PRMT1 was associated with the infiltration of most types of immune cell, including B cells, CD4+ T cells, CD8+ T cells, Tregs, NK, macrophages, neutrophils, and dendritic cells in HCC. The top four immune cells types associated with PRMT1 expression were Tregs, macrophages, T cell of follicular helper cells, and mast cells (Figure 2E).
Furthermore, we evaluated the association between PRMT1 expression and generated immune subtypes. High levels of PRMT1 were associated with macrophage regulation, wound healing, lymphocyte infiltration, and stromal fraction (Figure 2F). Next, we investigated the prognostic value of PRMT1 expression combined with infiltration of immune cells in HCC. The results showed that high level of PRMT1 and tumor-infiltrating macrophages indicated a significantly shorter OS in HCC (Figure 2G). Taken together, these results indicate that PRMT1 plays an important role in maintaining the HCC tumor immune microenvironment. We speculate that PRMT1 may mediate clinical response to immunotherapy in HCC.
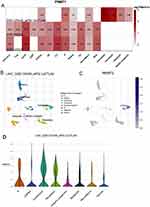
Immune checkpoint inhibitors are the best established and most widely used immunotherapeutic agents for tumors. To confirm the association of PRMT1 expression with immunotherapy response in HCC, we obtained scRNA-seq data from HCC patients treated with PD-L1 and CTLA4. PRMT1 expression was significantly increased in endothelial, fibroblasts, and malignant cells, whereas it was specifically expressed in immune cells, including exhausted CD8 T cells, B cells, mono/macro cells (Figure 3A–D). Taken together, these findings indicate that elevated PRMT1 expression could increase the potential for tumor immune escape and resistance to immunotherapy in HCC.
Relationship of PRMT1 Expression with Immune Checkpoints in HCC
To further evaluate the mechanisms of PRMT1 in immune microenvironment regulation, we investigated the relationship of PRMT1 expression with immune response-related marker genes, including chemokines, receptors, MHC, and immunomodulators in HCC. PRMT1 expression was positively associated with expression of most the immunomodulators, including immunoinhibitors and immunostimulators. Its expression was weakly associated with MHC-related molecules. However, PRMT1 expression was negatively correlated with chemokines and receptors, including CCL13, CCL14, CCL15, CXCL2, CXCL10, and CXCL11 (Figure 4A).
Consequently, we determined the correlations of these genes with all the immune checkpoints (Figure 4B). Moreover, the relationships of PRMT1 expression with key immune checkpoints including CD276, PDCD1, CTLA-4, and TIGIT were detected. All the genes showed significantly elevated levels of expression in the high-PRMT1 expression groups (Figure 4C). Moreover, the expression of tumor microenvironment (TME) immunosuppressive factors, including IL-10 and TGF-β, was also enhanced in the high-PRMT1 expression groups (Figure 4C). Therefore, high-PRMT1 is likely to contribute to the immune escape of tumor cells, resulting in poor prognosis of patients with HCC. PRMT1 may respond to immunotherapy via regulation of expression of the above immune checkpoint genes in HCC.
PRMT1 is Involved Cell Progression and Migration in HCC
To confirm the important role of PRMT1 in HCC progression, we performed PRMT1 functional verification in HCC via knockdown of PRMT1 in HepG2 and Huh7 cells using specific shRNAs. By MTT assay, we found that cell proliferation significantly decreased after down-regulation of PRMT1 expression (Figure 5A). Moreover, cell migration distances were significantly shorter (Figure 5B and C), and the cell invasion ratio was dramatically decreased in PRMT1-downregulated HCC cells (Figure 5D and E). Taken together, these results indicate that PRMT1 is involved in regulation of cell proliferation, migration, and invasion in HCC progression.
PRMT1 Participates in Metabolism-Related Pathways to Regulate Progression of HCC
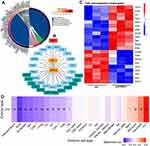
We further profiled the functions of PRMT1 in LIHC. GSEA results indicated that PRMT1 was involved in immune response and fatty acid metabolic processes in HCC (Figure 6A), and KEGG results showed that PRMT1 was enriched in metabolic-related pathways including amino acid metabolism, fatty acid degradation, and drug metabolism (Figure 6B). Furthermore, 297 up-regulated and 207 down-regulated PRMT1 related genes were found in LIHC (Figure S1A). The top 50 of these genes are shown as heatmaps in Figure S1B. KEGG enrichment analysis showed that some pathways involved in RNA splicing, and DNA-templated transcription and fatty acid metabolic processes were significantly associated with PRMT1 expression (Figure 6C). Furthermore, DisGeNET database analysis showed that PRMT1-related genes were also associated with drug-induced liver disease, fatty liver disease, and liver injury (Figure 6D).
Furthermore, we constructed the regulatory network of PRMT1 in metabolic pathway. As shown in Figure 7A, several PRMT1-associated genes were involved in metabolic pathways, especially fatty acid-related pathways. Based on these findings, we constructed a network of the interactions between PRMT1 and fatty acid metabolic-related genes. PRMT1 directly interacted with SIRT1 and PPARCC1A, which were the hub genes of the metabolic-related gene set in HCC (Figure 7B). Moreover, we observed the expression of fatty-acid-related genes after down-regulation of PRMT1 in HCC cells (Figure 7C). In addition, we evaluated the associations of gene sets related to PRMT1 and fatty acid metabolism with immune infiltration. These gene sets were significantly associated with infiltration of several immune cells, including B cells, exhausted cells, NK, CD8 cells and Th17 cells (Figure 7D). Taken together, these results indicate that PRMT1 was involved in metabolism-related pathways to regulate progression of HCC.
Discussion
Due to the complex heterogeneity within and among individual patients with HCC, more biomarkers for predicting prognosis and treatment strategies need to be explored. PRMT1 is critical in methylation of arginine residues, approximately 90% of total arginine methylation is catalyzed by PRMT1,8,9 suggesting that PRMT1 perhaps a biomarker for predicting HCC prognosis and treatment. In current study, we found that PRMT1 was significantly overexpressed and associated with tumor progression and clinical features in HCC, suggesting that it could act as potential diagnostic biomarkers in HCC. In addition, previous studies have shown that PRMT genes were dysregulated expression in several cancers and play crucial roles in transcriptions regulation, DNA damage, and immune response.21,22 Meanwhile, PRMT1 maybe act as a potential therapeutic target in cancers.23,24 Taken together, this study highlights the fact that the prognostic value of PRMT1 and its potential as a novel therapeutic target in HCC.
The immune cells within the tumor microenvironment are important in tumorigenesis.25,26 Increasing evidences have shown that PRMTs are involved in the immune response. PRMT1 is crucial for B cell activation.27 Moreover, it has been reported that PRMT1 is highly expressed in T helper cells and increase IL-10 and IL-6 cytokines expression to represses tumor growth in HCC.16 Additionally, PRMT5 inhibition induces lymphocytes infiltration and the expression of several MHC II molecule, such as H2-Ab1, H2-Aa, and Cd74.16,28 Therefore, manipulating PRMTs could be a feasible strategy to modulate tumor microenvironment for immunotherapy. In this study, scRNA-seq data showed that PRMT1 was expressed in most of immune cells and associated with immune cells infiltration, including B cells, Tregs, macrophages and mast cells. High level of PRMT1 was correlated with macrophages regulation, wound healing, lymphocyte infiltration, and stromal fraction. Moreover, it indicated significantly shorter OS in HCC. Taken together, we speculated that PRMT1 plays an important role in maintaining the HCC tumor immune microenvironment.
Recently, immunotherapy based on regulation of the tumor immune microenvironment has emerged a novel treatment choice for patients with HCC.29,30 The immune checkpoints inhibitors have been reported to enhance antitumor immune responses,31,32 and inhibitors such as nivolumab and pembrolizumab targeting PD-1 and PD-L1 have been recommended as second-line treatments for advanced HCC so far.33,34 Therefore, developing effective combination therapies for HCC immunotherapy may be a focus in the future.35 As a novel small-molecule inhibitor of type I PRMTs has been reported that could suppress PD-L1 expression in pancreatic cancer cells,36,37 and the polymorphism rs975484 of PRMT1 has been confirmed to regulate the expression of immune checkpoint genes PD-L1 and PD-L2 in HCC.20 In this study, we found that PRMT1 was specifically expressed in immune cells, including CD8tex, B cells, mono/macro cells after PDL1 and CTLA4 treatment in LIHC. Moreover, levels of key immune checkpoints including CD276, PDCD1, CTLA-4, and TIGIT were all significantly elevated in the high-PRMT1 expression groups. Therefore, PRMT1 could be combined with immune checkpoints to effectively inhibit cancer progression in immunotherapy, providing a novel target to mediate clinical response to immunotherapy in HCC.
Recently, non-alcoholic steatohepatitis (NASH) has made metabolic disorders as major risk factors for the development of HCC.38 Previous studies have confirmed that PRMT1 can also protect against NASH induced liver injury,39,40 enhancing hepatic lipogenesis by increasing lipogenic gene expression via the TXNIP/PRMT1/PGC-1α pathway.39 Moreover, PRMT1 was required to induce PGC-1α activation via recruitment of HNF-4α and hence attenuated HFD-induced hepatic steatosis.40 In this study, we found that genes-related to fatty acid metabolism were differentially expressed in shPRMT1 transfected HCC cells. These results were consistent with the results of bioinformatics analysis showing that PRMT1-associated genes were involved in metabolic pathways, especially in fatty acid-related pathways. Moreover, PRMT1 could directly interacts with SIRT1 and PPARCC1A proteins, which have important roles in metabolic-related pathways in HCC. Combine with our results that the function of PRMT1 was confirmed to involve in the cell proliferation, migration and invasion ability in HCC, PRMT1 may accelerate the transition through fatty acid related metabolic regulation in the progression of non-alcoholic fatty liver disease (NAFLD) to HCC. However, the molecular mechanism by which PRMT1 is involved protein methylation in NAFLD-induced HCC remains to be explored.
Multiple studies have shown that fatty acid metabolism has multifaceted roles in the proliferation and function of tumor-infiltrating immune cells in the TME.41 Moreover, key genes related to fatty acid metabolism pathways have been reported to be significantly related to immune-cell infiltration and to act as regulators of immune checkpoint inhibitor treatment.38 In this study, we found that PRMT1 was involved in fatty acid metabolic pathways and regulates expression of genes related to fatty acid metabolism. Taken together, these findings indicate that PRMT1 is likely to integrate fatty acid metabolic pathways to affect the proliferation and function of immune cells in HCC progression.
Conclusion
The results of this study suggest that PRMT1 could be a potential biomarker for diagnosis and prediction of prognosis of HCC patients. It is also associated with immune cell infiltration in the tumor microenvironment and could thus serves as a potential target for immunotherapy in HCC. PRMT1 was confirmed to have functions involved in the regulation of cell proliferation, migration, and invasion, and expression of genes related to fatty acid metabolism in HCC. Our finding will lay a foundation for further studies and provide a valuable information for the development of targeted therapies for liver cancer.
Data Sharing Statement
The data used to support the findings of this study are included within the article and Supplementary Materials (PRMT1 related genes-based RNA-seq in LIHC from TCGA database). The datasets analyzed during this study were available in public open access repositories. The list of open access repositories is as follows:
The data of gene expression from TCGA database, which can be found at https://www.cancer.gov/tcga.
The single cell sequencing data set was obtained from GEO database https://www.ncbi.nlm.nih.gov/geo/. Accessions number: GSE149614, GSE125449, GSE110228, GSE1240228.
The data of immune cell infiltration estimation can be found at http://timer.cistrome.org/; http://bioinfo.life.hust.edu.cn.
The immune-related gene list can be found at http://cis.hku.hk/TISIDB/.
All data is already released and could obtain using above web links.
Ethical Approval and Consent to Participate
The study was conducted in accordance with the Declaration of Helsinki. Our study is based on open-source data in public database TCGA and GEO, which were reviewed and approved by the Ethics Committee of Inner Mongolia Medical University. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for research and publish relevant articles.
Acknowledgments
The authors manifest great appreciation to all those involved in this research.
Funding
This work was supported by Natural Science Foundation of Inner Mongolia (2023MS08033), National Natural Science Foundation of China (CN) (grant Nos. 81660024), the Joint Project of Inner Mongolia Medical University (grant Nos. YKD2021LH001), Key Project of Natural Science for Colleges and Universities in Inner Mongolia (grant Nos. NJZZ22686) and Inner Mongolia Medical University Science and Technology Innovation Team (grant Nos. YKD2022TD031).
Disclosure
The authors declare that they have no conflict of interest.
References
1. Kelley KR, Greten FT, Phimister EG. Hepatocellular carcinoma-origins and outcomes. N Engl J Med. 2021;385(3):280–282. doi:10.1056/NEJMcibr2106594
2. Xie DY, Ren ZG, Zhou J, Fan J, Gao Q. 2019 Chinese clinical guidelines for the management of hepatocellular carcinoma: updates and insights. Hepatobiliary Surg Nutr. 2020;9(4):452–463. doi:10.21037/hbsn-20-480
3. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
4. Zheng Y, Wang S, Cai J, Ke A, Fan J. The progress of immune checkpoint therapy in primary liver cancer. Biochim Biophys Acta Rev Cancer. 2021;1876(2):188638. doi:10.1016/j.bbcan.2021.188638
5. Wang J, Wang F, Wang N, Zhang MY, Wang HY, Huang GL. Diagnostic and prognostic value of protein post-translational modifications in hepatocellular carcinoma. J Clin Transl Hepatol. 2023;11(5):1192–1200. doi:10.14218/JCTH.2022.00006S
6. Peng C, Wong CC. The story of protein arginine methylation: characterization, regulation, and function. Expert Rev Proteomics. 2017;14(2):157–170. doi:10.1080/14789450.2017.1275573
7. Kaniskan HÜ, Konze KD, Jin J. Selective inhibitors of protein methyltransferases. J Med Chem. 2015;58(4):1596–1629. doi:10.1021/jm501234a
8. Jarrold J, Davies CC. PRMTs and arginine methylation: cancer’s best-kept secret? Trends Mol Med. 2019;25(11):993–1009. doi:10.1016/j.molmed.2019.05.007
9. Wu Q, Schapira M, Arrowsmith CH, Barsyte-Lovejoy D. Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat Rev Drug Discov. 2021;20(7):509–530. doi:10.1038/s41573-021-00159-8
10. Li X, Wang C, Jiang H, Luo C. A patent review of arginine methyltransferase inhibitors (2010–2018). Expert Opin Ther Pat. 2019;29(2):97–114. doi:10.1080/13543776.2019.1567711
11. Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65(1):8–24. doi:10.1016/j.molcel.2016.11.003
12. Hwang JW, Cho Y, Bae GU, Kim SN, Kim YK. Protein arginine methyltransferases: promising targets for cancer therapy. Exp Mol Med. 2021;53(5):788–808. doi:10.1038/s12276-021-00613-y
13. Li Z, Wang D, Lu J, et al. Methylation of EZH2 by PRMT1 regulates its stability and promotes breast cancer metastasis. Cell Death Differ. 2020;27(12):3226–3242. doi:10.1038/s41418-020-00615-9
14. Li B, Liu L, Li X, Wu L. miR‐503 suppresses metastasis of hepatocellular carcinoma cell by targeting PRMT1. Biochem Biophys Res Commun. 2015;464(4):982–987. doi:10.1016/j.bbrc.2015.06.169
15. Zhao J, O’Neil M, Schonfeld M, Komatz A, Weinman SA, Tikhanovich I. Hepatocellular protein arginine methyltransferase 1 suppresses alcohol-induced hepatocellular carcinoma formation by inhibition of inducible nitric oxide synthase. Hepatol Commun. 2020;4(6):790–808. doi:10.1002/hep4.1488
16. Zhao J, O’Neil M, Vittal A, Weinman SA, Tikhanovich I. PRMT1-dependent macrophage IL-6 production is required for alcohol-induced HCC progression. Gene Expr. 2019;19(2):137–150. doi:10.3727/105221618X15372014086197
17. Parry RV, Ward SG. Protein arginine methylation: a new handle on T lymphocytes? Trends Immunol. 2010;31(4):164–169. doi:10.1016/j.it.2010.01.006
18. Kim H, Kim H, Feng Y, et al. PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci Transl Med. 2020;12(551):eaaz5683. doi:10.1126/scitranslmed.aaz5683
19. Srour N, Villarreal OD, Hardikar S, et al. PRMT7 ablation stimulates anti-tumor immunity and sensitizes melanoma to immune checkpoint blockade. Cell Rep. 2022;38(13):110582. doi:10.1016/j.celrep.2022.110582
20. Schonfeld M, Zhao J, Komatz A, Weinman SA, Tikhanovich I. The polymorphism rs975484 in the protein arginine methyltransferase 1 gene modulates expression of immune checkpoint genes in hepatocellular carcinoma. J Biol Chem. 2020;295(20):7126–7137. doi:10.1074/jbc.RA120.013401
21. Charlène T, Louisane E, Coralie P, Muriel LR. Structure, activity, and function of PRMT1. Life. 2021;11(11):1147. doi:10.3390/life11111147
22. Xu J, Richard S. Cellular pathways influenced by protein arginine methylation: implications for cancer. Mol Cell. 2021;81(21):4357–4368. doi:10.1016/j.molcel.2021.09.011
23. Zhao Y, Lu Q, Li C, et al. PRMT1 regulates the tumour-initiating properties of esophageal squamous cell carcinoma through histone H4 arginine methylation coupled with transcriptional activation. Cell Death Dis. 2019;10(5):359. doi:10.1038/s41419-019-1595-0
24. Liang Z, Liu L, Wen C, et al. Clinicopathological and prognostic significance of PRMT5 in cancers: a system review and meta-analysis. Cancer Control. 2021;28:107327482110505. doi:10.1177/10732748211050583
25. Lei X, Lei Y, Li JK, et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi:10.1016/j.canlet.2019.11.009
26. Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology. 2018;68(3):1025–1041. doi:10.1002/hep.29904
27. Infantino S, Light A, O’Donnell K, et al. Tarlinton D Arginine methylation catalyzed by PRMT1 is required for B cell activation and differentiation. Nat Commun. 2017;8(1):891. doi:10.1038/s41467-017-01009-1
28. Luo Y, Gao Y, Liu W, et al. MYC-protein arginine methyltransferase 5 axis defines the tumorigenesis and immune response in hepatocellular carcinoma. Hepatology. 2021;74(4):1932–1951. doi:10.1002/hep.31864
29. Waidmann O. Recent developments with immunotherapy for hepatocellular carcinoma. Expert Opin Biol Ther. 2018;18(8):905–910. doi:10.1080/14712598.2018.1499722
30. Lee JH, Lee JH, Lim YS, et al. Sustained efficacy of adjuvant immunotherapy with cytokine-induced killer cells for hepatocellular carcinoma: an extended 5-year follow-up. Cancer Immunol Immunother. 2019;68(1):23–32. doi:10.1007/s00262-018-2247-4
31. Liu HT, Jiang MJ, Deng ZJ, et al. Immune checkpoint inhibitors in hepatocellular carcinoma: current progresses and challenges. Front Oncol. 2021;11:737497. doi:10.3389/fonc.2021.737497
32. Calderaro J, Rousseau B, Amaddeo G, et al. Programmed death ligand 1 expression in hepatocellular carcinoma: relationship with clinical and pathological features. Hepatology. 2016;64(6):2038–2046. doi:10.1002/hep.28710
33. Heimbach JK, Kulik LM, Finn RS, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–380. doi:10.1002/hep.29086
34. Zhou J, Sun H, Wang Z, et al. Guidelines for the diagnosis and treatment of hepatocellular carcinoma (2019 Edition). Liver Cancer. 2020;9(6):682–720. doi:10.1159/000509424
35. Sangro B, Gomez-Martin C, de la Mata M, et al. A clinical trial of CTLA-4 blockade with tremelimumab in patients with hepatocellular carcinoma and chronic hepatitis C. J Hepatol. 2013;59(1):81–88. doi:10.1016/j.jhep.2013.02.022
36. Lastwika KJ, Wilson W 3rd, Li QK, et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76(2):227–238. doi:10.1158/0008-5472.CAN-14-3362
37. Zheng NN, Zhou M, Sun F, et al. Combining protein arginine methyltransferase inhibitor and anti-programmed death-ligand-1 inhibits pancreatic cancer progression. World J Gastroenterol. 2020;26(26):3737–3749. doi:10.3748/wjg.v26.i26.3737
38. Du D, Liu C, Qin M, et al. Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharm Sin B. 2022;12(2):558–580. doi:10.1016/j.apsb.2021.09.019
39. Park MJ, Kim DI, Lim SK, et al. Thioredoxin-interacting protein mediates hepatic lipogenesis and inflammation via PRMT1 and PGC-1alpha regulation in vitro and in vivo. J Hepatol. 2014;61(5):1151–1157. doi:10.1016/j.jhep.2014.06.032
40. Xu L, Huang Z, Lo TH, et al. Hepatic PRMT1 ameliorates diet-induced hepatic steatosis via induction of PGC1α. Theranostics. 2022;12(6):2502–2518. doi:10.7150/thno.63824
41. Luo Y, Wang H, Liu B, Wei J. Fatty acid metabolism and cancer immunotherapy. Curr Oncol Rep. 2022;24(5):659–670. doi:10.1007/s11912-022-01223-1
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.