Back to Journals » Infection and Drug Resistance » Volume 16
Metabolic Profiles of Clinical Isolates of Drug-Susceptible and Multidrug-Resistant Mycobacterium tuberculosis: A Metabolomics-Based Study
Authors Wang L, Ying R, Liu Y, Sun Q , Sha W
Received 4 February 2023
Accepted for publication 20 April 2023
Published 3 May 2023 Volume 2023:16 Pages 2667—2680
DOI https://doi.org/10.2147/IDR.S405987
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Li Wang,1,2 Ruoyan Ying,3 Yidian Liu,1,2 Qin Sun,1– 3 Wei Sha1– 3
1Clinic and Research Center of Tuberculosis, Shanghai Pulmonary Hospital, School of Medicine, Tongji University, Shanghai, People’s Republic of China; 2Department of Tuberculosis, Shanghai Pulmonary Hospital, School of Medicine, Tongji University, Shanghai, People’s Republic of China; 3Shanghai Key Laboratory of Tuberculosis, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, People’s Republic of China
Correspondence: Wei Sha; Qin Sun, Email [email protected]; [email protected]
Background: Mycobacterium tuberculosis (MTB) is a global and highly deleterious pathogen that creates an enormous pressure on global public health. Although several effective drugs have been used to treat tuberculosis, the emergence of multidrug-resistant Mycobacterium tuberculosis (MDR-MTB) has further increased the public health burden. The aim of this study was to describe in depth the metabolic changes in clinical isolates of drug-susceptible Mycobacterium tuberculosis (DS-MTB) and MDR-MTB and to provide clues to the mechanisms of drug resistance based on metabolic pathways.
Methods: Based on the minimum inhibition concentration (MIC) of multiple anti-tuberculosis drugs, two clinical isolates were selected, one DS-MTB isolate (isoniazid MIC=0.06 mg/L, rifampin MIC=0.25 mg/L) and one MDR-MTB isolate (isoniazid MIC=4 mg/L, rifampin MIC=8 mg/L). Through high-throughput metabolomics, the metabolic profiles of the DS-MTB isolate and the MDR-MTB isolate and their cultured supernatants were revealed.
Results: Compared with the DS-MTB isolate, 128 metabolites were significantly altered in the MDR-MTB isolate and 66 metabolites were significantly altered in the cultured supernatant. The differential metabolites were significantly enriched in pyrimidine metabolism, purine metabolism, nicotinate and nicotinamide metabolism, arginine acid metabolism, and phenylalanine metabolism. Furthermore, metabolomics analysis of the bacterial cultured supernatants showed a significant increase in 10 amino acids (L-citrulline, L-glutamic acid, L-aspartic acid, L-norleucine, L-phenylalanine, L-methionine, L-tyrosine, D-tryptophan, valylproline, and D-methionine) and a significant decrease in 2 amino acids (L-lysine and L-arginine) in MDR-MTB isolate.
Conclusion: The present study provided a metabolite alteration profile as well as a cultured supernatant metabolite alteration profile of MDR-MTB clinical isolate, providing clues to the potential metabolic pathways and mechanisms of multidrug resistance.
Keywords: Mycobacterium tuberculosis, multidrug-resistantMycobacterium tuberculosis, drug-susceptibleMycobacterium tuberculosis, metabolomics, metabolic pathway
Background
Tuberculosis (TB) is an ancient human disease caused by MTB, but its global epidemiological trajectory still presents a rather grim picture.1 TB is ranked as a major contributor to the global disease burden, causing more than one million deaths each year.2 There are about a quarter of the world’s population infected with MTB, equivalent to about 2 billion people.3 Globally in 2020, there were an estimated 1.3 million deaths among HIV-negative people, and an additional 214000 deaths among HIV-positive people.3
Concurrently, the treatment of TB has become more difficult and challenging with the emergence of drug-resistant variants.4 Resistance to the first-line antituberculosis drugs isoniazid and rifampicin is of greatest concern. And resistance to at least both drugs is defined as multidrug-resistant tuberculosis (MDR-TB). Globally in 2020, 71% (2.1/3.0 million) of patients diagnosed with pulmonary tuberculosis were tested for rifampicin resistance, identifying 132,222 cases of MDR-TB or rifampicin-resistant tuberculosis (RR-TB).3 And the treatment of MDR/RR-TB is not very promising, with a treatment success rate of only 59% for MDR/RR-TB in 2018.3
Metabolomics is key to the study of all life-critical metabolites in living organisms and can contribute to the screening of biomarkers, as well as to revealing regulatory pathways.5 Metabolomics focuses on global or system-level study of small molecules in specific biological systems and is therefore a promising research tool for the study of cell physiology, and biochemistry within pathogenic microorganisms. Metabolomic studies on MTB are still at an early stage.6 Currently, Xiao et al7 analyzed MTB stain H37Rv using non-targeted metabolomics and showed that H37Rv specifically induced tryptophan metabolism in human macrophages. Through widely targeted metabolite screening, Jiang et al8 found that glutamine is required for M1-like polarization of macrophages in response to MTB infection. Metabolomic analysis would help to clarify the metabolic signature of the MDR-MTB clinical isolate and generate a deeper understanding of the metabolic mechanism of drug-resistant mutations.
In this study, metabolomics was carried out on clinical isolates of drug-susceptible Mycobacterium tuberculosis and multidrug-resistant MTB, as well as on their cultured supernatant, to discover the underlying drug-resistance mechanisms.
Methods
Isolation and Culture of MTB
Based on the MIC of multiple anti-tuberculosis drugs, two clinical isolates of MTB were sampled for subsequent experiment: one DS-MTB clinical isolate and one MDR-MTB clinical isolate (Supplemental Figure 1). As shown in Table 1, the DS-MTB clinical isolate used in the study was susceptible to anti-tuberculosis drugs, such as isoniazid (MIC=0.06 mg/L) and rifampin (MIC=0.25 mg/L). The MDR-MTB clinical isolate was resistant to multiple anti-tuberculosis drugs, such as isoniazid (MIC=4 mg/L) and rifampin (MIC=8 mg/L). The isolates were obtained using a BD BACTEC™ MGIT™ 960 system (Becton Dickinson, Franklin Lakes, NJ, USA). The MIC results were interpreted based on CLSI breakpoints.9,10 All isolates were stored in 7H9 broth (Becton Dickinson, Franklin Lakes, NJ, USA) containing 15% glycerol in a freezer at –80°C until used.
 |
Table 1 MIC of Multi-Drugs Against DS-MTB and MDR-MTB Isolate (Mg/L) |
Samples for metabolomics analysis were obtained from four independent cultures of DS-MTB and MDR-MTB isolate, respectively. Both isolates were cultured in a biosafety LEVEL 3 laboratory at 37 °C in Middlebrook 7H9 broth (Difco/Becton Dickinson, Franklin Lakes, NJ) to reach the mid-log phase of growth. After centrifugation, the bacterial suspension was divided into two parts, precipitation and supernatant. The supernatant was filtered through a 0.22 μm filter membrane to filter out the suspended bacteria (supernatant inactivate) so that the samples could be safely taken out of the LEVEL 3 laboratory.
The present study was approved by the Ethics Committee of Shanghai Pulmonary Hospital (Shanghai, China, approval no. K16-300). Informed consent was obtained from all study participants. The study was performed in accordance with the Declaration of Helsinki Principles and informed consent was obtained from all study participants.
Metabolite Extractions
To extract the metabolites, 1200 μL of cold extraction solvent methanol/acetonitrile (1:1, v/v) was added to 300 μL of the sample and vortexed for 30s. Subsequently, the sample was incubated at-20°C for 1 h and centrifuged at 14000g for 20min.Then the supernatant was collected and dried in a vacuum centrifuge at 4°C. For LC-MS analysis, the sample was re-dissolved in 100 μL acetonitrile/water (1:1, v/v) solvent and transferred to LC vials.
High Performance Liquid Chromatography and Mass Spectrometric Analysis
Samples were first separated by an ultra-high performance liquid chromatography system Vanquish (Thermo Scientific, Waltham, Massachusetts, USA) using a HILIC column (Waters, ACQUITY UPLC BEH Amide 1.7 μm, 2.1×100 mm column). Subsequently, the samples were analyzed using a Q Exactive HF-X mass spectrometer (Thermo Scientific, Waltham, Massachusetts, USA). Next, the raw data collected by the mass spectrometry analysis were searched in the database by the Thermo Compound Discoverer software (Version 3.2). Finally, the metabolite identification results of the samples are obtained. The total ion chromatograms were attached in Supplemental file 2.
Qualitative and Quantitative Analysis of Metabolites
The RAW data (RAW files) collected by mass spectrometry were retrieved by Compound Discoverer software (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA) and the identification information of the samples was obtained.
Differential Metabolites Analysis
Based on univariate analysis, all metabolites detected in positive and negative ion mode were analyzed for differences. The difference screening criteria were p-value<0.05, fold-change >1.5 or fold-change <0.67. All metabolites are visualized in the form of a volcano map (Supplemental Figure 2). Subsequently, Partial Least Squares Discrimination Analysis (PLS-DA) and Orthogonal Partial Least Squares Discrimination Analysis (OPLS-DA) were performed to further screen out differential metabolites associated with sample groupings (Supplemental Figures 3 and 4). Meanwhile, the permutation test was used to ensure the validity of the PLS-DA and OPLS-DA models. The OPLS-DA variable importance in the projection (VIP) value of each metabolite was calculated and differential metabolites were further filtered with VIP value >1.
Results
Differential Metabolite Profiles of Bacterial and Cultured Supernatant
Summary statistics for the differential metabolite profiles of bacterial and cultured supernatant are presented in Supplementary Tables 1 and 2. Meanwhile, volcano plots were drawn to show the differential metabolite profiles, and the top 20 metabolites with the largest fold-change values were labeled (Figure 1).
In comparison with the DS-MTB, 74 metabolites were significantly increased, and 54 metabolites were significantly decreased in MDR-MTB isolate. Metabolites, including cysteic acid, guanine, and D-methionine, were significantly increased. Interestingly, D-alanyl-D-alanine, an essential component of the intracellular peptidoglycan precursor and an important target for the development of antibacterial drugs,11 was also increased in the MDR-MTB isolate (Supplemental Figure 5).
Compared to the DS-MTB, 48 metabolites were significantly increased, including L-tyrosine, D-tryptophan, 5’-S-methyl-5’-thioadenosine, nordihydroguaiaretic acid, and threonine, and 18 metabolites were significantly decreased in the MDR-MTB cultured supernatant.
Subsequently, metabolic pathway enrichment analysis of the differential metabolites was performed using MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/). The results showed that the differential metabolites of bacterial were mainly enriched in pyrimidine metabolism pathway, purine metabolism pathway, nicotinic acid and nicotinamide metabolism pathway, phenylalanine, tyrosine and tryptophan biosynthesis pathway, phenylalanine metabolism pathway, D-glutamine and D-glutamate metabolism pathway and arginine biosynthesis pathway (Figure 2A, Supplementary Table 3). And the differential metabolites of cultured supernatant were mainly enriched with aminoacyl-tRNA biosynthesis pathway, arginine biosynthesis pathway, cysteine and methionine metabolism pathway, and lysine biosynthesis pathway (Figure 2B, Supplementary Table 4).
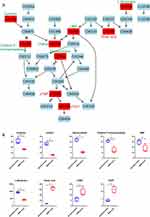
Pyrimidine and Purine Metabolism
Pyrimidine metabolism pathway is the pathway enriched with the most identified differential metabolites, containing three increased and six decreased metabolites in MDR-MTB isolate. As shown in the metabolic pathway diagram (Figure 3), important substrates and intermediate metabolites, such as cytosine, uridine, cytidine 5’-monophosphate, L-glutamine, deoxycytidine and uridine monophosphate (UMP) were significantly decreased in MDR-MTB isolate, while synthetic products, such as thymidine monophosphate (dTMP) and thymidine diphosphate (dTDP) and orotic acid, were significantly increased. Additionally, metabolomic results of the cultured supernatant also showed that uridine and deoxycytidine was also decreased in MDR-MTB isolate (Supplemental Figure 6). These results suggest that more uridine and deoxycytidine were consumed to produce more dTMP and dTDP in MDR-MTB isolate.
Purine metabolic pathway was also enriched with nine identified differential metabolites, including six increased and three decreased in MDR-MTB isolate. Through the metabolic pathway map, a phenomenon similar to the pyrimidine metabolism pathway can be observed. Compared with DS-MTB, more important substrates and intermediate metabolites, such as adenosine and adenosine 5’-monophosphate are consumed, producing more adenine and deoxyadenosine diphosphate (dADP) in MDR-MTB isolate (Figure 4). In addition, deoxyinosine monophosphate (dIMP), deoxyinosine and guanine were increased, and deoxyguanosine monophosphate (dGMP) was decreased in MDR-MTB isolate.
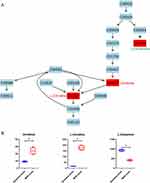
Nicotinate and Nicotinamide Metabolism Pathway
Nicotinate and nicotinamide metabolism pathways were enriched with four identified differential metabolites, including three increased and one decreased in MDR-MTB isolate. The results showed that nicotinic acid mononucleotide and nicotinamide adenine dinucleotide (NAD) were increased while nicotinic acid was decreased (Figure 5). Therefore, it might be assumed that the conversion of nicotinic acid mononucleotide to nicotinic acid was inhibited and the conversion to NAD was enhanced in MDR-MTB isolate. In addition, nicotinamide was also increased in the MDR-MTB isolate.
Amino Acid Metabolism
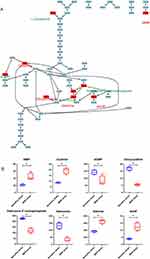
Arginine biosynthesis pathway was enriched with two increased metabolites, ornithine and L-citrulline, and one decreased metabolite, L-glutamine (Figure 6). Through the metabolic pathway map, it was apparent that the production of L-citrulline and its intermediate metabolite, ornithine was increased in MDR-MTB isolate. And it is interestingly to note that arginine biosynthetic pathway was also significantly enriched with the differential metabolites of the cultured supernatant (Supplementary Table 4). L-citrulline was also significantly increased in MDR-MTB cultured supernatant, along with the precursors of L-glutamine, L-glutamic acid (KEGG ID: C00025), and L-aspartic acid (KEGG ID: C00049, Supplemental Figure 7). In addition, an important precursor of L-citrulline, L-arginine (KEGG ID: C00062), was significantly decreased in the supernatant of MDR-MTB isolate, implying that more L-arginine was consumed to produce L-citrulline in MDR-MTB isolate.
L-phenylalanine and its key precursor metabolite, phenylpyruvate were the only two metabolites enriched in the phenylalanine metabolism pathway. Through the metabolic pathway map, it was apparent that increased production of L-phenylalanine and phenylpyruvate were increased in MDR-MTB isolate (Supplemental Figure 8).
Amino Acid Consumption of the Cultured Supernatant
The results of metabolic profile showed that the differential metabolites in the cultured supernatant were mostly amino acids, dipeptides, and their derivatives. Therefore, it is reasonable to believe that the type and quantity of amino acids intake by MDR-MTB isolate are significantly different from those of the DS-MTB isolate in which resistance mechanisms may be latent.
Through heatmap and boxplots, only two amino acids, L-lysine and L-arginine, were significantly decreased in MDR-MTB cultured supernatant, indicating that more of these two amino acids were consumed in MDR-MTB isolate (Figure 7). And the other ten amino acids (L-citrulline, L-glutamic acid, L-aspartic acid, L-norleucine, L-phenylalanine, L-methionine, L-tyrosine, D-tryptophan, valylproline, and D-methionine) were significantly increased, suggesting that MDR-MTB isolate consumed less of these ten amino acids. These results suggested that MDR-MTB isolate preferred to metabolize L-lysine and L-arginine rather than the other ten amino acids, and this amino acid metabolic signature might also be related to its drug resistance.
Discussion
In the present study, metabolomics analysis was performed on one DS-MTB clinical isolate and one MDR-MTB clinical isolate, as well as their cultured supernatant. The differential metabolites were mainly enriched in pyrimidine metabolism pathway, purine metabolism pathway, nicotinic acid and nicotinamide metabolism pathway, arginine biosynthesis, and phenylalanine metabolism pathway. In a recent study, a similar phenomenon was observed by metabolomic study of the H37Rv strain with or without rifampicin, which significantly altered metabolites associated with pyrimidine, purine, arginine, phenylalanine, tyrosine and tryptophan metabolic pathways.12 The above study was a complementation to the present study, and collectively indicated that drug resistance was related to pyrimidine, purine, arginine, and phenylalanine metabolic pathways.
D-alanyl-D-alanine is a dipeptide comprising D-alanine with a D-alanyl residue attached to the alpha-nitrogen.13 It is an essential component of the intracellular peptidoglycan precursor, uridine diphosphate-N-acetylcarbamate-pentapeptide,14 which is vital for the survival of pathogens by maintaining the integrity of bacterial cell walls15,16 through cross-linking peptidoglycan chains.17 Inhibition of D-alanine-D-alanine ligase (Ddl) would directly lead to a dramatic decrease of the strength of the bacterial cell wall, potentially leading to bacterial cell rupture.18 D-alanyl-D-alanine and Ddl might be important targets for the development of antibacterial drugs.11 There have also been several research based on Ddl as a potential anti-MTB drug target. Fakhar et al16 reported the biochemical function and homology modeling of Ddl and assessed the structural quality of the obtained homology modeled target. Yang et al19 developed a colorimetric method for high-throughput screening of Ddl inhibitors for TB-Ddl activity and identified eight potential interacting proteins of TB-Ddl. It is most likely that the strength of the cell wall was enhanced by increasing D-alanyl-D-alanine in MDR-MTB isolate, and thus became more resistant to the drugs.
Purine metabolic pathway plays an important role in mycobacterial physiology and is a target for MTB drug development. MTB has the ability to adopt a growth, slow growth or even non-growth state within the host to survive in the face of prolonged multidrug therapy, and it has been revealed that the pyrimidine metabolism is a metabolic checkpoint during MTB adaptation to non-growing state.20 Furthermore, the pyrimidine salvage pathway has an important role in the latent state of MTB, as MTB must recover bases and/or nucleosides in order to survive in the harsh environment imposed by the host.21 Furthermore, TMPK, the enzyme that catalyzes the phosphorylation of dTMP to dTDP, is indispensable for growth and survival.Therefore it represents a potential target for developing new anti-TB drugs.22
Purine metabolism pathway plays an essential role in the physiology of MTB and other mycobacteria owing to the critical importance of the purine salvage enzyme hypoxanthine-guanine phosphate nucleotidyl transferase (HGPRT) for the growth of MTB in vitro.23 Similar to the pyrimidine metabolism, purine metabolism is also a target for drug development in MTB, and a large number of potent purine analogs have been designed.24
Nicotinate and nicotinamide metabolism pathways are vital pathways for the biosynthesis of NAD. It is reported that nicotinamide nucleotide adenosyltransferase, the enzyme that catalyzes nicotinic acid mononucleotide with ATP to form NAD, was selected as a potential drug target as it has been proven in vitro to be essential for the growth of MTB.25 This founding partly explained the result of our study that more nicotinic acid mononucleotide was converted to more NAD in MDR-MTB isolate. We assumed that MDR-MTB isolate requires more NAD than DS-MTB isolate, so more nicotinic acid mononucleotide was consumed to synthesize NAD. In addition, nicotinamide was reported to be able to limit the replication of MTB within macrophages.26 In this study, we found that nicotinamide was also increased in MDR-MTB isolate. Hence, resistance of MDR-MTB isolate might also be related to its resistance to higher concentrations of nicotinamide.
The DNA replication mechanisms, protein synthesis and cell wall biogenesis of MTB have been targeted for the development of anti-tuberculosis drugs, but some essential metabolic pathways that are critical for MTB survival have received little attention. Currently, many amino acid biosynthetic pathways have recently been shown to be critical for the survival and pathogenesis of MTB,27 and these amino acid metabolism pathways might be critical in the development of drug-resistance in MDR-MTB isolate.
Arginine metabolism pathway has been reported to serve as one of the metabolic pathways of drug targets for the control of TB.27 The arginine biosynthetic enzyme argB (acetylglutamate kinase) is essential for MTB and its homologous gene is missing in humans, therefore this enzyme could be a potential target for drugs.27 The MTB argB and argF (ornithine carbamoyltransferase) mutants were unable to grow in arginine-deprived medium and can be restored by supplementation with 1 mM of arginine or by gene complementation. It is reported that arginine deprivation rapidly sterilizes the MTB through accumulation of reactive oxygen species and DNA damage, and arginine starvation does not affect the survival of immunodeficient, or immunocompromised mice infected with MTB argB and argF mutants.28,29 Combined with the results that arginine consumption was significantly higher in MDR-MTB isolate, it is speculated that arginine metabolism might be involved in the development of drug resistance.
One highlight of this study is that instead of using the standard strain, the clinical isolates were used, which could more realistically reflect the metabolic status of MTB in the TB patients. Another highlight of this study is that, unlike other studies that only analyzed the bacterial organism, the cultured supernatant was also analyzed in this study so that it could (1) enhance the robustness of the study and (2) reveal the intake and consumption of metabolites in MTB.
There are certainly many limitations in the study. Firstly, standard control strains, such as H37Rv, were not set. Secondly, only one DS-MTB and one MDR-MTB isolate were analysis, large numbers of samples are needed to demonstrate the universality of the results. Thirdly, only preliminary screen study was performed, more experiments such verification are needed to be done to get a more robust and convincing conclusion.
Conclusion
Through high-throughput metabolomics, the metabolic profiles of DS-MTB clinical isolate and MDR-MTB clinical isolate and their cultured supernatants were revealed. The differential metabolites were associated with pyrimidine metabolism, purine metabolism, nicotinate and nicotinamide metabolism, arginine acid metabolism, and phenylalanine metabolism. Furthermore, drug-resistance might relate to the type and quantity of amino acids taken from cultured supernatant.
Abbreviations
TB, Tuberculosis; MTB, Mycobacterium tuberculosis; MDR-MTB, Multidrug-resistant Mycobacterium tuberculosis; DS-MTB, Drug-susceptible Mycobacterium tuberculosis; MIC, Minimum inhibition concentration; MDR-TB, Multidrug-resistant tuberculosis; RR-TB, Rifampicin-resistant tuberculosis; PLS-DA, Partial Least Squares Discrimination Analysis; OPLS-DA, Orthogonal Partial Least Squares Discrimination Analysis; VIP, Variable Importance in the Projection; SCID, Severe combined immunodeficiency.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was funded by: Shanghai 2020 “Science and technology innovation action plan” technological innovation fund:Clinical Study on New Short-Course treatment regimens and Host-Directed Therapy for MDR-TB (20Z11900500). Shanghai Clinical Research Center for infectious disease (tuberculosis) (Grant ID:19MC1910800). The three-year action plan for promoting clinical skills and clinical innovation in municipal hospitals of Shanghai Shenkang 2020–2022 (Grant ID: SHDC2020CR6024). National Natural Science Foundation of China (Nos.82102251). Shanghai key clinical specialty construction project - Tuberculosis department (Shslczdzc03001).
Disclosure
The authors declare that they have no conflict of interest.
References
1. Jeremiah C, Petersen E, Nantanda R, et al. The WHO Global Tuberculosis 2021 Report - not so good news and turning the tide back to End TB. Int J Infect Dis. 2022. doi:10.1016/j.ijid.2022.03.011
2. Ledesma JR, Ma J, Vongpradith A; Collaborators, G.B.D.T. Global, regional, and national sex differences in the global burden of tuberculosis by HIV status, 1990-2019: results from the Global Burden of Disease Study 2019. Lancet Infect Dis. 2022;22(2):222–241. doi:10.1016/S1473-3099(21)00449-7
3. World Health Organization. Global Tuberculosis Report 2021. Geneva: World Health Organization; 2021. Licence: CC BY-NC-SA 3.0 IGO.
4. Khawbung JL, Nath D, Chakraborty S. Drug resistant Tuberculosis: a review. Comp Immunol Microbiol Infect Dis. 2021;74:101574. doi:10.1016/j.cimid.2020.101574
5. Judge A, Dodd MS. Metabolism. Essays Biochem. 2020;64(4):607–647. doi:10.1042/EBC20190041
6. Planck KA, Rhee K. Metabolomics of Mycobacterium tuberculosis. Methods Mol Biol. 2021;2314:579–593. doi:10.1007/978-1-0716-1460-0_25
7. Xiao G, Zhang S, Zhang L, et al. Untargeted metabolomics analysis reveals Mycobacterium tuberculosis strain H37Rv specifically induces tryptophan metabolism in human macrophages. BMC Microbiol. 2022;22(1):249. doi:10.1186/s12866-022-02659-y
8. Jiang Q, Qiu Y, Kurland IJ, et al. Glutamine Is Required for M1-like Polarization of Macrophages in Response to Mycobacterium tuberculosis Infection. mBio. 2022;13(4):e0127422. doi:10.1128/mbio.01274-22
9. Caleffi-Ferracioli KR, Maltempe FG, Siqueira VL, et al. Fast detection of drug interaction in Mycobacterium tuberculosis by a checkerboard resazurin method. Tuberculosis. 2013;93(6):660–663. doi:10.1016/j.tube.2013.09.001
10. Ying R, Huang X, Gao Y, et al. In vitro Synergism of Six Antituberculosis Agents Against Drug-Resistant Mycobacterium tuberculosis Isolated from Retreatment Tuberculosis Patients. Infect Drug Resist. 2021;14:3729–3736. doi:10.2147/IDR.S322563
11. Qin Y, Xu L, Teng Y, et al. Discovery of novel antibacterial agents: recent developments in D-alanyl-D-alanine ligase inhibitors. Chem Biol Drug Des. 2021;98(3):305–322. doi:10.1111/cbdd.13899
12. Yelamanchi SD, Mishra A, Behra SK, et al. Rifampicin-Mediated Metabolic Changes in Mycobacterium tuberculosis. Metabolites. 2022;12(6):493. doi:10.3390/metabo12060493
13. Pederick JL, Thompson AP, Bell SG, et al. d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations. J Biol Chem. 2020;295(23):7894–7904. doi:10.1074/jbc.RA120.012936
14. Boudrioua A, Li Y, Hartke A, et al. Opposite effect of vancomycin and D-Cycloserine combination in both vancomycin resistant Staphylococcus aureus and enterococci. FEMS Microbiol Lett. 2020;367(8). doi:10.1093/femsle/fnaa062
15. Zhang S, Oh JH, Alexander LM, et al. d-Alanyl-d-Alanine Ligase as a Broad-Host-Range Counterselection Marker in Vancomycin-Resistant Lactic Acid Bacteria. J Bacteriol. 2018;200(13). doi:10.1128/JB.00607-17
16. Fakhar Z, Naiker S, Alves CN, et al. A comparative modeling and molecular docking study on Mycobacterium tuberculosis targets involved in peptidoglycan biosynthesis. J Biomol Struct Dyn. 2016;34(11):2399–2417. doi:10.1080/07391102.2015.1117397
17. Hrast M, Vehar B, Turk S, et al. Function of the d-Alanine: d-Alanine Ligase Lid Loop: a Molecular Modeling and Bioactivity Study. J Med Chem. 2012;55(15):6849–6856. doi:10.1021/jm3006965
18. Chen Y, Xu Y, Yang S, et al. Deficiency of D-alanyl-D-alanine ligase A attenuated cell division and greatly altered the proteome of Mycobacterium smegmatis. Microbiologyopen. 2019;8(9):e00819. doi:10.1002/mbo3.819
19. Yang S, Xu Y, Wang Y, et al. The Biological Properties and Potential Interacting Proteins of d-Alanyl-d-alanine Ligase A from Mycobacterium tuberculosis. Molecules. 2018;23(2). doi:10.3390/molecules23020324
20. Shi KX, Wu YK, Tang BK, et al. Housecleaning of pyrimidine nucleotide pool coordinates metabolic adaptation of nongrowing Mycobacterium tuberculosis. Emerg Microbes Infect. 2019;8(1):40–44. doi:10.1080/22221751.2018.1559706
21. Villela AD, Sanchez-Quitian ZA, Ducati RG, et al. Pyrimidine salvage pathway in Mycobacterium tuberculosis. Curr Med Chem. 2011;18(9):1286–1298. doi:10.2174/092986711795029555
22. Van Calenbergh S, Pochet S, Munier-Lehmann H. Drug design and identification of potent leads against mycobacterium tuberculosis thymidine monophosphate kinase. Curr Top Med Chem. 2012;12(7):694–705. doi:10.2174/156802612799984580
23. Knejzlik Z, Herkommerova K, Hockova D, et al. Hypoxanthine-Guanine Phosphoribosyltransferase Is Dispensable for Mycobacterium smegmatis Viability. J Bacteriol. 2020;202(5). doi:10.1128/JB.00710-19
24. Parker WB, Long MC. Purine metabolism in Mycobacterium tuberculosis as a target for drug development. Curr Pharm Des. 2007;13(6):599–608. doi:10.2174/138161207780162863
25. Cloete R, Shahbaaz M, Grobbelaar M, et al. In silico repurposing of a Novobiocin derivative for activity against latency associated Mycobacterium tuberculosis drug target nicotinate-nucleotide adenylyl transferase (Rv2421c). PLoS One. 2021;16(11):e0259348. doi:10.1371/journal.pone.0259348
26. Simmons JD, Peterson GJ, Campo M, et al. Nicotinamide Limits Replication of Mycobacterium tuberculosis and Bacille Calmette-Guerin Within Macrophages. J Infect Dis. 2020;221(6):989–999. doi:10.1093/infdis/jiz541
27. Yelamanchi SD, Surolia A. Targeting amino acid metabolism of Mycobacterium tuberculosis for developing inhibitors to curtail its survival. IUBMB Life. 2021;73(4):643–658. doi:10.1002/iub.2455
28. Tiwari S, van Tonder AJ, Vilcheze C, et al. Arginine-deprivation-induced oxidative damage sterilizes Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2018;115(39):9779–9784. doi:10.1073/pnas.1808874115
29. Mizrahi V, Warner DF. Death of Mycobacterium tuberculosis by l-arginine starvation. Proc Natl Acad Sci U S A. 2018;115(39):9658–9660. doi:10.1073/pnas.1813587115
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.