Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 17
Investigation of Monogenic Diabetes Genes in Thai Children with Autoantibody Negative Diabetes Requiring Insulin
Authors Teerawattanapong N, Tangjarusritaratorn T, Narkdontri T , Santiprabhob J , Tangjittipokin W
Received 21 November 2023
Accepted for publication 3 February 2024
Published 14 February 2024 Volume 2024:17 Pages 795—808
DOI https://doi.org/10.2147/DMSO.S409713
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Konstantinos Tziomalos
Nipaporn Teerawattanapong,1– 3 Thanida Tangjarusritaratorn,1 Tassanee Narkdontri,1– 3 Jeerunda Santiprabhob,4,5 Watip Tangjittipokin1,2
1Department of Immunology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, 10700, Thailand; 2Siriraj Center of Research Excellence for Diabetes and Obesity, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, 10700, Thailand; 3Research Department, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, 10700, Thailand; 4Siriraj Diabetes Center of Excellence, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, 10700, Thailand; 5Division of Endocrinology & Metabolism, Department of Pediatrics, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, 10700, Thailand
Correspondence: Watip Tangjittipokin, Department of Immunology Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, 10700, Thailand, Tel +66 2-419-6635, Fax +66 2-418-1636, Email [email protected]
Purpose: The objective of this study was to clarify the phenotypic characteristics of monogenic diabetes abnormalities in Thai children with autoantibody-negative insulin.
Patients and Methods: Two hundred and thirty-one Thai type 1 diabetes (T1D) patients out of 300 participants with recent-onset diabetes were analyzed for GAD65 and IA2 pancreatic autoantibodies. A total of 30 individuals with T1D patients with negative autoantibody were screened for 32 monogenic diabetes genes by whole-exome sequencing (WES).
Results: All participants were ten men and twenty women. The median age to onset of diabetes was 8 years and 3 months. A total of 20 people with monogenic diabetes carried genes related to monogenic diabetes. The PAX4 (rs2233580) in ten patients with monogenic diabetes was found. Seven variants of WFS1 (Val412Ala, Glu737Lys, Gly576Ser, Cys673Tyr, Arg456His, Lys424Glu, and Gly736fs) were investigated in patients in this study. Furthermore, the pathogenic variant, rs115099192 (Pro407Gln) in the GATA4 gene was found. Most patients who carried PAX4 (c.575G>A, rs2233580) did not have a history of DKA. The pathogenic variant GATA4 variant (c.1220C>A, rs115099192) was found in a patient with a history of DKA.
Conclusion: This study demonstrated significant genetic overlap between autoantibody-negative diabetes and monogenic diabetes using WES. All candidate variants were considered disease risk with clinically significant variants. WES screening was the first implemented to diagnose monogenic diabetes in Thai children, and fourteen novel variants were identified in this study and need to be investigated in the future.
Keywords: monogenic diabetes, whole‑exome sequencing, autoantibody
Introduction
Diabetes is a condition in which the body cannot make enough insulin or cannot use insulin normally. Type 1 diabetes (T1D) is subclassified as type 1A associated with autoantibodies to pancreatic beta cells and type 1B which occurs independently.1 Type 1A diabetes is characterized by chronic immune-mediated destruction of pancreatic beta cells, leading to partial or in most cases absolute insulin deficiency. Currently, the islet autoantibody testing of glutamic acid decarboxylase (GAD), the tyrosine phosphatase-like insulinoma antigen 2 (IA2), insulin autoantibodies (IAA), and/or zinc transporter 8 (ZnT8) are used to diagnose type 1A diabetes. More than 90% of patients with early fasting hyperglycemia can have these autoantibodies detected.2 The prevalence of T1D in Thai was 62.6% of patients with diabetes onset before 30 years of age.3 The possibility of other types of diabetes should be considered clinically relevant in a child who does not have autoantibodies.
Monogenic forms of diabetes are caused by mutations in a single gene and account for 1% to 6% of pediatric diabetes patients.4 Maturity-onset diabetes of the young (MODY) is the most common type of monogenic diabetes and can be caused by a mutation in 15 genes.5 Monogenic diabetes also includes transient or permanent neonatal forms occurring under 6 months of age. More than 20 genes are known to be related to congenital neonatal diabetes.4 Clinically, monogenic diabetes may be difficult to classify as type 1 and type 2 diabetes, so it can be misdiagnosed and treated incorrectly. The previous study of known MODY genes was found in Thai patients, including HNF1A6 (R203C) and PAX47 (R192H). A molecular diagnosis of monogenic diabetes is essential for optimal treatment, prognosis, and genetic counseling. We used targeted whole-exome sequencing (WES) to detect genetic variants causative of monogenic diabetes. However, many cases of monogenic diabetes remain underestimated due to the lack of genetic information. In this study, we aimed to identify the spectrum of genetic variants associated with monogenic genes related to diabetes and possibly identify novel variants related to autoantibody-negative insulin-requiring diabetes mellitus.
Materials and Methods
Subjects and Study Design
A total of 300 participants diagnosed with diabetes mellitus were investigated in the project “Pediatric diabetes registry and a study on etiology, glycemic control, and complication in children and adolescents with diabetes” according to the criteria of the International Society for Pediatric and Adolescent Diabetes (ISPAD). This study was conducted at the Division of Endocrinology & Metabolism, Department of Pediatrics, Faculty of Medicine Siriraj Hospital, Thailand, during 2016–2018. Subjects were categorized into 4 groups including type 1 diabetes (T1D), type 2 diabetes (T2D), neonatal diabetes mellitus (NDM), and other types of diabetes mellitus (Figure 1). They underwent a thorough clinical examination and their medical records were retrospectively reviewed from their electronic and written medical records. Islet beta-cell autoantibodies (anti-GAD and IA-2) were determined.
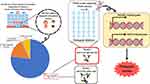 |
Figure 1 Classification of diabetes in Thai children. |
Participants who met the following criteria (i) diagnosed with diabetes according to the 2018 American Diabetes Association (ADA),8 (ii) diagnosed with diabetes mellitus before the age of 18 years, (iii) showed negative results for both GAD and IA-2 antibodies at the beginning of diabetes and (iv) required insulin therapy were enrolled in this study.
The control group included 16 individuals aged 49 to 56 and normal fasting glucose was tested. They had an annual check-up at Siriraj Hospital (Supplemental Table 1).
Ethics Statement
All parents of subjects were informed of the purpose of the study before signing an informed consent form for the genetic testing of their child. The research was carried out following the Declaration of Helsinki, and informed consent was obtained from patients where appropriate. The entire study was approved by the Siriraj Institutional Review Board (SIRB), Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand (COA no. Si145/2016).
Autoantibody Analyzes
Pancreatic autoantibodies (GAD65 and IA2) were determined in serum from participants at the recruitment step, using previously described standardized radio assays.9 GAD65 and IA2 autoantibodies were determined using standardized radio-binding assays using in vitro transcribed and translated [35S]-labeled recombinant human full-length GAD65, IA2ic (amino acids 605–979). The results of the antibodies for GAD65 and IA2 are expressed as a semiquantitative index calculated using a dilution curve of a positive sample. All cut-off values were set at the 99th percentile of the control population. Specificity was 100% for the two antibody assays, and sensitivity was 68% for GAD65 and 62% for IA2.
DNA Extraction and Whole-Exome Sequencing (WES) Analysis
A total of 5 mL of venous blood were collected from each subject, including both patients and controls. Genomic DNA was extracted from the buffy coat using Flexigene® DNA (Qiagen, Valencia, CA, USA). DNA purity determination was quantified using the NanoDrop-2000c (Scientific, USA).
Exome capture was performed using an Agilent SureSelect Human All Exon 50 Mb kit (Agilent Technologies, Inc., Santa Clara, CA, USA) according to the manufacturer’s protocol. The captured library was then loaded onto an Illumina HiSeq 2000 platform (Illumina, Inc., San Diego, CA, USA) for amplification and sequencing. Sequence reads were assigned to the reference human genome (UCSC NCBI37/hg19). Variant detections and annotations were analyzed using SAMtools10 and Genome Analysis Toolkit 11, respectively.
Variant Filtering and Bioinformatics Analysis
All variants detected in whole-exome sequencing analysis were selected missense variants with an allele frequency less than 0.05 from the 1000 Genome Project (http://www.1000genomes.org/) and the Exome Aggregation Consortium (ExAC) (Figure 2). Bioinformatic analysis of WES data was performed using VARCARDs online software (http://varcards.biols.ac.cn/).10 To identify genetic variants causative of monogenic diabetes in this study, pathogenicity prediction was tested using a damaging score >0.5 using VARCARDs as possibly pathogenic variants.10
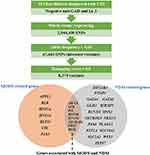 |
Figure 2 Variants filtering in MODY and/or NDM known genes. |
The obtained potential pathogenic variants were screened for 32 genes known to cause monogenic diabetes, MODY or NDM, and both MODY and NDM (APPL1, ABCC8, BLK, CEL, EIF2AK3, FOXP3, GATA4, GATA6, GCK, GLIS3, HNF1A, HNF1B, HNF4A, IER3IP1, INS, KCNJ11, KCNJ11, KLF11, MNX1, NEUROD1, NEUROG3, NKX2-2, PAX4, PAX6, PDX1, PLAGL1, PTF1A, RFX6, SLC19A2, SLC2A2, WFS1, ZFP57).4 All genetic variants of interest in the patient’s group were not identified in the Thai healthy control group (n = 16).
Statistical Analysis
Statistical analysis was carried out using SPSS software (version 21; SPSS Inc., Chicago, IL). Quantitative variables were expressed as means and standard deviations and qualitative variables as frequencies and percentages. The Mann–Whitney U-test was used to compare quantitative variables. Frequencies were compared using Pearson’s chi-square analysis and Fisher’s exact test when necessary. The significance level was defined as p < 0.05.
Results
Demographic Data and Clinical Characteristics
As shown in Table 1, the prevalence of type 1 diabetes and type 2 diabetes in the 300 patients with new-onset pediatric diabetes between March 2016 and August 2018 was 77% and 15.3%, respectively. The average age, age at diagnosis, duration of diabetes, height, weight, body mass index (BMI), waist circumference, systolic blood pressure, diastolic blood pressure, and hemoglobin A1c (HbA1c) were significantly different between types of diabetes (type 1 diabetes (T1D), type 2 diabetes (T2D), neonatal diabetes mellitus (NDM) and other types) (all p<0.001).
 |
Table 1 Demographic, Anthropometric, and Clinical Characteristics of Pediatric Diabetes |
An autoimmune response against pancreatic β-cells (anti-GAD) was detected in 60.6% of the patients to diagnose T1D. IA2-positive autoantibody results were present in 65.5% of patients with T1D. Forty-three percent (43.3%) of patients with a clinical diagnosis of T1D had positive autoantibodies (GAD65 and IA2). All negative autoantibodies (GAD65 and IA2) were found in 36 pediatric patients (36/201, 17.9%) with a clinical T1D diagnosis (Table 1).
This study recruited only 30 out of 36 monogenic diabetes subjects with negative autoantibody (GAD65 and IA2) and clinical characteristics were shown (Table 2). Ten men and twenty women are among the participants. The median age to the onset of diabetes was 8 years and 3 months. The median age at the time of diagnosis of diabetes was 13 years and 2 months. Family history information on diabetes included 25 patients (80%). The median BMI at diagnosis was 19.3 kg/m2. The median HbA1C at the time of diagnosis was 9.1%. The median birth weight was 3115 grams. The average systolic and diastolic blood pressures were 110.3 and 68.2 mmHg, respectively. The average pulse rate was 89.4 bpm. The average waist circumference was 67.7 cm. The average amount of insulin required per day was 1.1 units/kg. The insulin regimen included 13 patients with modified conventional, 16 patients with basal-bolus, and 1 patient with mixed twice. No subjects had monogenic diabetes phenotypes that present with diabetes before 6 months of age or had a family history of diabetes in one parent or other first‐degree relatives, and all islet autoantibodies were positive.
 |
Table 2 Characteristics of Participants with Autoantibody-Negative Insulin-Requiring Diabetes Mellitus |
Genetic Screening
This present study utilized a targeted whole-exome sequencing for the screening of pathogenic variants in 32 genes causative of monogenic diabetes, including 7 genes that have been associated with MODY but not NDM (HNF4A, HNF1A, KLF11, CEL, PAX4, BLK, APPL1) and 17 genes that have been associated with NDM but not MODY (EIF2AK3, FOXP3, GATA6, IER3IP1, MNX1, NKX2-2, PLAGL1, SLC19A2, WFS1, GATA4, GLIS3, KCNJ11, NEUROG3, PAX6, PTF1A, SLC2A2, ZFP57), and 8 monogenic diabetes genes that have been identified to cause both MODY and NDM (ABCC8, GCK, INS, HNF1B, KCNJ11, NEUROD1, PDX1, RFX6).4,11 The 2,546,430 variants were called using the Genome analysis toolkit. The WES data was then analyzed using VARCARDs based on allele frequencies and damaging scores.10 The 47,661 variants with allele frequencies <0.05 were included. After that, the variants were filtered for further analysis if the predicted damaging score >0.5. Using these criteria, 8279 variants were suspected as pathogenic variants and screened for causative genes of MODY or NDM.
The results showed that only 25 variants were identified in known genes that cause MODY and/or NDM (Figure 2). Twenty out of 30 patients (67%) had genetic variants in the target genes (Table 3). Genetic variants in different target genes were found in 12 monogenic diabetes patients (patients # 3, 9, 11, 12, 13, 15, 17, 18, 20, 24, 26, 27). Five patients had genetic variants in only five genes (PAX4, HNF1A, HNF4A, KLF11, and CEL) associated with MODY (patients # 3, 8, 25, 28, and 29). There are 5 patients (patients # 1, 4, 7, 18, and 19) who had gene mutations in only four NDM-related genes (WFS1, EIF2AK3, GATA6, and KCNJ11). On the other hand, eight patients (patients # 12, 13, 15, 17, 20, 24, 26, and 27) carried multiple genetic variants in genes associated with MODY and genes associated with NDM. Two patients (patients # 9 and 11) detected mutations in genes that were previously reported as causative genes of MODY and NDM (RFX6 and PDX1).
 |
Table 3 Characteristics of Patients Diagnosed with Monogenic Diabetes |
MODY-Related Genes
Half of the patients who carried variants in known pathogenic genes (10 out of 20: patient# 3, 8, 9, 11, 12, 13, 15, 24, 25, and 26) had a missense mutation (rs2233580) in PAX4 (MODY9). Missense mutations (patient# 17: rs1800961 and patient# 3: rs150776703) in HNF4A (MODY1) were identified in two patients. In addition, two cases of a missense mutation (patient# 27: c.1040T>C and patient # 28: rs62576769) were detected in CEL (MODY8). Other MODY-related genetic variants, including missense mutations in rs1800574 HNF1A (MODY3) (patient # 29) and rs150096859 KLF11 (MODY7) (patient # 20) were found in each patient. In addition to diabetes patients, a case mutation in KLF11 (patient # 20: rs150096859) was also found in a control subject (Control# 14) (Table 4).
 |
Table 4 Genetic Variants Identified in 20 Monogenic Diabetes Patients |
NDM-Related Genes
For genetic variants in NDM-related genes, most of them had genetic variants in WFS1, which included six missense mutations and one frameshift insertion. Four of these variants have not been reported. These included a missense mutation (c.1726G>A) that was found in two patients (patients # 15 and 20). In addition, no case of a missense and frameshift mutation presented in WFS1 (c.1270A>G and c.2204_2205insT) has previously been identified (patient # 27). The other genetic variants causative of NDM were found in 11 patients. These included a missense mutation in EIF2AK3, GATA6, and SLC19A2 that were found in two patients each, three cases of a missense mutation in GATA4, and one case each for a missense mutation in GLIS3 and KCNJ11. Of these, four genetic variants were previously reported, and five genetic variants were novel. Only the mutation in GATA4 (rs115099192) was classified as pathogenic (patient # 9 and 27). Additional control subjects in this study showed the presence of a genetic variant in WFS1 (Control# 14: c.1367G>A), GATA6 (Control# 8: c.43G>C), GLIS3 (Control# 12: c.2678C>T) and SLC19A2 (Control# 1, 2 and 9: c.1322T>C). It is noticeable that just under a fifth of the control (3 out of 16) also had a missense mutation in SLC19A2.
MODY and NDM-Related Genes
In this study, a mutation in RFX6 and PDX1 was also found which has been reported as a causative gene for both MODY and NDM. A new variant of regulatory factor X 6 (RFX6) (c.523C>T, rs868202124) was identified in patient # 9. For PDX1 (pancreatic and duodenal homeobox-1), also known as IPF1 (insulin promoter factor-1), one missense variant (c.670G>A, rs137852787) of this gene was detected in patient #11.
Variants and Diabetic Phenotypes
The clinical presentation was also considered to examine the relationship between genetic variants and diabetic phenotypes as shown in Tables 3 and 4. Age at the time of onset, sex, birth weight, diabetic ketoacidosis (DKA) at first presentation, body mass index (BMI), average level of glycated hemoglobin (HbA1c), treatment regimen, and daily insulin dose used per kilogram and family history of diabetes mellitus were presented in each patient. The diagnosis of diabetic ketoacidosis (DKA) was based on the presence of acidosis symptoms and was combined with hyperglycemia acidosis ketone in blood or urine. The clinical significance of the variants obtained was represented in ClinVar and showed that most of them were reported as likely benign (Table 4 and Figure 3). However, the clinical and functional significance of clinical testing or experimental evidence was not provided.
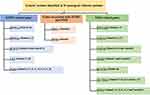 |
Figure 3 Flowchart of monogenic diabetes variants is identified in patients. [Gene(Patient number)]. |
A total of 30 patients with monogenic diabetes participated in this study. We found that 14 patients were diagnosed with DKA at the first clinical presentation of diabetes mellitus. Of these, only six patients (patient # 3, 9, 17, 19, 27, and 29) carried MODY and/or NDM-related variants in pathogenic genes such as PAX4, GATA4, GATA6, RFX6, HNF1A, HNF4A, SLC19A2, CEL and WFS1 (Table 4). While the largest number of patients in this study (10 out of 30: patient # 3, 8, 9, 11, 12, 13, 15, 24, 25, and 26) had genetic variants in PAX4 c.575G>A (rs2233580), which was not detected in the 16 controls.
Most PAX4 mutation-positive (c.575G>A, rs2233580) patients (8 out of 10: patient# 8, 11, 12, 13, 15, 24, 25, and 26) did not show the presence of DKA at the first diagnosis. Patients #25 and #26 carried the same PAX4 (c.575G>A, rs2233580) but lacked DKA. They were siblings, and only the brother (patient # 26) had another genetic variant in GATA6 (c.187C>T, rs559705145).
The clinical significance of the GATA4 variants (c.1220C>A, rs115099192) was represented as pathogenic in ClinVar, which was found in patients # 9 and 27 but not detected in the 16 controls (Table 4). The clinical presentation of patients # 9 and 27 were female, with a history of DKA at first diagnosis, basal-bolus treatment, and a family history of diabetes mellitus (Table 3). The clinical picture in some patients with multiple genetic variants was not different from that of a single variant mutation, including age at the time of onset, BMI, DKA at the time of onset, and insulin treatment.
Discussion
Our study provides a wide spectrum of variants in monogenic diabetes-related genes and 25 variants were identified from 30 patients who were diagnosed before 18 years of age and negative for GAD and IA-2 antibodies. In this study, 14 variants were not reported as pathogenic variants in the case of MODY and/ or NDM. It should be noted that some patients in this study did not have a family history of diabetes.
A recent study has shown that some genetic variants can be associated with de novo occurrence or incomplete penetration in MODY patients.22 Thus, our study could have benefits in establishing MODY and/or NDM-related genes among patients with a negative family history. In our cohort of patients with monogenic diabetes, de novo mutations in PAX4, WFS1, HNF4A, and SLC19A2 were present in three patients (patients # 1, 8, and 17) without a history of diabetes.
Several of these patients had genetic variants in PAX4 c.575G>A (rs2233580). This missense mutation led to a substitution of amino acid from arginine to histidine at codon 192 (R192H). The PAX4 (R192H) mutation had previously been identified as causing MODY9.7 The role of PAX4 is affected in the development, differentiation, and survival of insulin-producing beta cells.23 PAX4 rs2233580 (R192H) results in the change of an amino acid at a highly conserved position across different species, and the change of an amino acid in the homeodomain that is required for DNA binding to target gene promoters could contribute to a defect in transcription activity.24 A younger age at diagnosis of diabetes was related to the risk allele (T) of PAX4 rs2233580.24 PAX4 R192H can disrupt its binding to target DNA sequences and exert a reduction in the transcriptional activity of target genes.25 We found this PAX4 c.575G>A (rs2233580) in 10 out of 30 patients with monogenic diabetes but not in control subjects. This is consistent with a previous study showing a high prevalence of PAX4 c.575G>A (rs2233580) among Thai children with autoantibody-negative insulin-requiring diabetes mellitus. The study also found that PAX4 c.575G>A (R192H) increased in frequency in MODY probands compared to non-diabetic controls in the Thai population.7 Additionally, a previous study in Thailand and Singapore had shown that PAX4 rs2233580 (R192H) influences T2D risk.26 Ethnic differences might play an important role in determining the epidemiology of monogenic diabetes. This may suggest that PAX4 rs2233580 (R192H) is responsible for diabetes susceptibility in Thai populations through the mechanism of decreased beta-cell activities. The correlation between diabetic ketoacidosis at first presentation and PAX4 rs2233580 (R192H) was examined, and it was found that 8 of 10 patients with PAX4 R192H had not presented DKA at the first clinical presentation. In the same way, most subjects who were MODY did not have DKA at diagnosis.27 It should be noted that two patients in the PAX4-positive group were siblings. Thus, this variant could be inherited from their parents. However, the correlation between diabetic ketoacidosis at first presentation and PAX4 R192H has not been investigated.
A mutation in the Hepatocyte Nuclear Factor 1A (HNF1A) gene, which is the most common genetic cause of MODY, was found in a patient in this study (patient # 29). We detected a missense mutation in HNF1A that causes a substitution of amino acid from alanine to valine at codon 98 (A98V). This position is located in the DNA binding domain, and an in vivo study demonstrated that HNF1A (A98V) polymorphism was correlated with a decreased in insulin release when responding to glucose.28 A study in India reported that the A98V polymorphism of HNF1A is associated with MODY and an earlier age at the beginning of T2D.13
In addition to HNF1A, we detected two mutations in the HNF4A gene, including T139I and P437S. One case had an amino acid substitution from threonine to isoleucine at codon 139 (T139I) in the HNF4 gene. This variant has been reported and shows that it is located within the hinge/DNA binding domain within the gene, potentially leading to altering interactions with targeted promoter regions.29 Although mutations in the HNF4A gene cause type 1 MODY (MODY1), this variant of T139I variant is associated with T2D.29 Furthermore, this missense mutation c.416C>T in the HNF4A gene has been identified in one case among 43 Turkish children diagnosed with MODY.14 Another case in which we found a mutation in the HNF4A gene that leads to a substitution of amino acid from proline to serine at codon 437 (P437S, rs150776703) has not been studied in terms of its functions. This variant has been classified in ClinVar interpretation as conflicting interpretations of pathogenicity.
In this study, 16 mutations were identified in NDM-related genes. Seven variants in WFS1 represented most of the patients with NDM-related genes in the cohort. WFS1 is expressed in various types of tissues, including the pancreas, and is involved in islet β-cells.30 WFS1 c.1235T>C (V412A, rs144951440) and c.2209G>A (E737K, rs147834269) that caused an amino acid substitution from valine to alanine at codon 412 and glutamate to lysine at codon 737, respectively, have been reported as a pathogenic variant in low-frequency non-syndromic hearing loss.15,16 However, hearing defects in patients #1 and 15 had not been detected in medical records. Furthermore, we identified WFS1 c.1367G>A that led to a substitution of amino acid from arginine to histidine at codon 456 (R456H) and was involved in the development of common T1D in the Japanese population.31 This missense mutation was detected in only one patient (patient #24) who carried another mutation in PAX4 of the MODY-related gene. This patient was a preterm female infant with a low birth weight (800 g) and a family history of diabetes. She was diagnosed with diabetes at 13 years of age and did not have DKA at the first presentation. At the moment, she has controlled her blood sugar with insulin premixed twice daily. However, this WFS1 R456H mutation was also identified in a Thai non-diabetic control subject. WFS1 R456H may be only a polymorphism of the WFS1 gene. The presence of DKA at the first diagnosis was considered in these WFS1 mutation-positive participants. Interestingly, 5 out of 6 patients who had WFS1 genetic variants did not present with DKA at the first diagnosis. This is consistent with a previous report showing that WFS1 mutations in Wolfram syndrome are associated with a lower prevalence of ketoacidosis and fewer positive autoantibodies.32
The GATA family of transcription factors, including GATA4 and GATA6, plays an important role in the development of the pancreas.33 All variants in the GATA family found in this study have not been studied in diabetic patients, except for a single variant in the GATA4 gene that caused a substitution of amino acid from proline to glutamine at codon 407 (P407Q, rs115099192). The mutation at codon 407 has been identified as a pathogenic variant of monogenic diabetes in patients with early-onset diabetes in South Korea.17 Furthermore, this GATA4 (P407Q) mutation is found to be associated with congenital heart disease.34 Another missense mutation in GATA4, valine is mutated to methionine at codon 267 (V267M) and is known to affect cardiac function on the transcriptional properties of GATA4.18 Two variants in GATA6 (G15R, rs116262672, and P63S, rs559705145) identified in this study are not associated with diabetes mellitus. The missense mutation in GATA6 (G15R, rs116262672) has been reported to be associated with nonsyndromic congenital heart disease.35
Mutations in the GLIS3, SLC19A2, and KCNJ11 genes that cause NDM were also identified. Although variants in GLIS3 c.2678C>T (p.Ser893Phe) that cause a substitution of amino acid at codon 893 from serine to phenylalanine have not been discovered, mutations in SLC19A2 c.1322T>C (p.Ile441Thr, rs17847484)19 and KCNJ11 c.1154C>G (p.Ser385Cys, rs41282930)36 have previously been studied. In this study, a variant detected in SLC19A2 c.1322T>C (p.Ile441Thr, rs17847484) has been implicated in the pathogenesis of thiamine responsive megaloblastic anemia syndrome (TRMAS).19 TRMAS is a rare autosomal recessive disorder with a triad of disease characteristics: megaloblastic anemia, early-onset deafness, and non-type I diabetes.37 Thus, it would be assumed that patients # 17 and 20 had diabetes with negative autoantibodies, which could be non-T1D. However, no signs and symptoms of TRMAS, including megaloblastic anemia and deafness, had been shown. This variant was also identified in three healthy control subjects. Neonatal diabetes mellitus (NDM) is mainly caused by mutations in the KCNJ11 gene.20 The Kir 6.2 subunit of the ATP-sensitive potassium channel, which controls pancreatic beta cell insulin secretion, is encoded by the KCNJ11 gene.20 KCNJ11 S385C mutations were found as polymorphisms in patients with NDM, but they did not respond with sulfonylurea.20
In this present study, two variants of the RFX6 and PDX1 genes were found that are associated with MODY and NDM. RFX6 c.523C>T (p.Pro175Ser, rs868202124) has not been reported, while PDX c.670G>A (p.Glu224Lys, rs137852787) has previously been studied. The mutation in the regulatory factor X 6 (RFX6) gene was known to be associated with beta cell dysfunction.38 Previously, variants in RFX6 associated with NDM were discovered in neonatal diabetes patients with other digestive system defects (known as Mitchell-Riley syndrome).39 Furthermore, variants truncated with the RFX6 protein, ie p.His293Leufs are found to be a cause of MODY in European patients.40
Pancreatic and duodenal homeobox 1 (PDX-1) plays an important role in regulating the function of the pancreatic β-cell. PDX-1 mutations cause pancreatic agenesis and neonatal diabetes.41 The variants that caused an amino acid substitution mutation at codon 224 from glutamate to lysine (E224K, rs137852787) were found in patient # 11 and were not detected in 16 controls. Residue 224 of PDX1 is conserved between different species and the E224K mutation has been shown to cosegregate with early-onset diabetes or impaired glucose tolerance in an Indo-Trinidadian family.21 The E224K mutation of PDX1 has significantly reduced transactivation properties, suggesting that this mutation is associated with MODY4 rather than type 2 diabetes.21
By targeting WES, we enabled the identification of novel variants in known MODY- and/or NDM-related genes in Thai children diagnosed with diabetes mellitus and negative for both GAD and IA-2 antibodies. This study provides evidence for an association of genetic polymorphism with autoantibody-negative diabetes and monogenic diabetes, although considerable limitations of our study should be acknowledged. First, this data was collected from only 30 cases (28 when excluding two siblings). We may have missed some rare variants for evaluation. Second, we did not test for segregation within their pedigree. Third, we tested only 2 autoantibodies GAD and IA2, including ZnT8 and insulin, that will be missing some positivity. Fourth, the control subjects are older than the cases, some parameters will not compare even the genetic factors will not be affected. Last, it could not be interpreted that all of these variants were pathogenic. It can be difficult to classify novel, uncommon variations as pathogenic or benign; as a result, several variants have been classified as either uncertainly significant or likely benign. Although this is still inconclusive, we cannot exclude the possibility that some novel genes remain unidentified and uninvestigated. The basic history, signs, symptoms, and clinical characteristics are still essential and should be included in the diagnosis. To reach a strong conclusion about the pathogenicity of each variant, further studies are required to provide more information, including functional studies to reveal the impact of these variants as a causative of diabetes.
Conclusions
The prevalence of monogenic diabetes with negative autoantibody was analyzed using WES. All candidate variants were considered disease risk with clinically significant variants from the ClinVar database. We found 5 MODY-related genes (PAX4, HNF1A, HNF4A, KLF11, and CEL), 7 NDM-related genes (WFS1, EIF2AK3, GATA4, GATA6, GLIS3, SLC19A2, KCNJ11) and 2 MODY and NDM related genes (RFX6 and PDX1) in Thai children with autoantibody negative insulin-requiring diabetes mellitus. Therefore, all candidate variants of monogenic diabetes could be used for appropriate treatment in each patient. Fourteen novel variants were identified in this study and need to be investigated in the future.
Data Sharing Statement
Data will be made available by the corresponding author upon reasonable request from the journal editor.
Ethics Approval
This study involves human participants, and the study protocol was approved by the Siriraj Institutional Review Board (SIRB) of the Siriraj Hospital, Faculty of Medicine, Mahidol University, Bangkok, Thailand (COA no. Si145/2016).
Acknowledgments
The authors gratefully acknowledge the diabetes patients who graciously agreed to participate in this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by a Siriraj Research Grant for Research and Development to WT (grant no. R015934015) from Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand.
Disclosure
All authors declare no personal or professional conflicts of interest, and no financial support from the companies that produce and/or distribute the drugs, devices, or materials described in this report.
References
1. The Expert Committee on the D, Classification of Diabetes M. Report of the Expert Committee on the diagnosis and classification of diabetes mellitus. Diabetes Care. 1997;20(7):1183–1197. doi:10.2337/diacare.20.7.1183
2. Watkins RA, Evans-Molina C, Blum JS, DiMeglio LA. Established and emerging biomarkers for the prediction of type 1 diabetes: a systematic review. Transl Res. 2014;164(2):110–121. doi:10.1016/j.trsl.2014.02.004
3. Dejkhamron P, Santiprabhob J, Likitmaskul S, et al. Young-onset diabetes patients in Thailand: data from Thai type 1 diabetes and diabetes diagnosed age before 30 years Registry, Care and Network (T1DDAR CN). J Diabetes Investig. 2022;13(5):796–809. doi:10.1111/jdi.13732
4. Hattersley AT, Greeley SAW, Polak M, et al. ISPAD clinical practice consensus guidelines 2018: the diagnosis and management of monogenic diabetes in children and adolescents. Pediatr Diabetes. 2018;19(Suppl 27):47–63. doi:10.1111/pedi.12772
5. Bansal V, Gassenhuber J, Phillips T, et al. Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals. BMC Med. 2017;15(1):213. doi:10.1186/s12916-017-0977-3
6. Plengvidhya N, Tangjittipokin W, Teerawattanapong N, Narkdontri T, Yenchitsomanus P-T. HNF1A mutation in a Thai patient with maturity-onset diabetes of the young: a case report. World J Diabetes. 2019;10(7):414–420. doi:10.4239/wjd.v10.i7.414
7. Plengvidhya N, Kooptiwut S, Songtawee N, et al. PAX4 mutations in Thais with maturity onset diabetes of the young. J Clin Endocrinol Metab. 2007;92(7):2821–2826. doi:10.1210/jc.2006-1927
8. American Diabetes Association. Standards of Medical Care in Diabetes—2018 abridged for primary care providers. Clin Diabetes. 2018;36(1):14–37. doi:10.2337/cd17-0119
9. Bingley PJ, Bonifacio E, Mueller PW. Diabetes antibody standardization program: first assay proficiency evaluation. Diabetes. 2003;52(5):1128–1136. doi:10.2337/diabetes.52.5.1128
10. Li J, Shi L, Zhang K, et al. VarCards: an integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 2018;46:D1039–D1048.
11. Firdous P, Nissar K, Ali S, et al. Genetic testing of maturity-onset diabetes of the young current status and future perspectives. Front Endocrinol. 2018;9:253. doi:10.3389/fendo.2018.00253
12. Sujjitjoon J, Kooptiwut S, Chongjaroen N, et al. PAX4 R192H and P321H polymorphisms in type 2 diabetes and their functional defects. J Hum Genet. 2016;61(11):943–949. doi:10.1038/jhg.2016.80
13. Anuradha S, Radha V, Deepa R, et al. A prevalent amino acid polymorphism at codon 98 (Ala98Val) of the hepatocyte nuclear factor-1 alpha is associated with maturity-onset diabetes of the young and younger age at onset of type 2 diabetes in Asian Indians. Diabetes Care. 2005;28(10):2430–2435. doi:10.2337/diacare.28.10.2430
14. Ağladıoğlu SY, Aycan Z, Çetinkaya S, et al. Maturity onset diabetes of youth (MODY) in Turkish children: sequence analysis of 11 causative genes by next generation sequencing. J Pediatr Endocrinol Metab. 2016;29(4):487–496. doi:10.1515/jpem-2015-0039
15. Choi HJ, Lee JS, Yu S, et al. Whole-exome sequencing identified a missense mutation in WFS1 causing low-frequency hearing loss: a case report. BMC Med Genet. 2017;18(1):151. doi:10.1186/s12881-017-0511-7
16. Baek J-I, Oh S-K, Kim D-B, et al. Targeted massive parallel sequencing: the effective detection of novel causative mutations associated with hearing loss in small families. Orphanet J Rare Dis. 2012;7(1):60. doi:10.1186/1750-1172-7-60
17. Kwak SH, Jung C-H, Ahn CH, et al. Clinical whole exome sequencing in early onset diabetes patients. Diabetes Res Clin Pract. 2016;122:71–77. doi:10.1016/j.diabres.2016.10.005
18. Riching AS, Danis E, Zhao Y, et al. Suppression of canonical TGF-β signaling enables GATA4 to interact with H3K27me3 demethylase JMJD3 to promote cardiomyogenesis. J Mol Cell Cardiol. 2021;153:44–59. doi:10.1016/j.yjmcc.2020.12.005
19. Liu G, Yang F, Han B, Liu J, Nie G. Identification of four SLC19A2 mutations in four Chinese thiamine responsive megaloblastic anemia patients without diabetes. Blood Cells Mol Dis. 2014;52(4):203–204. doi:10.1016/j.bcmd.2013.11.002
20. Madani HA, Fawzy N, Afif A, Abdelghaffar S, Gohar N. Study of KCNJ11 gene mutations in association with monogenic diabetes of infancy and response to sulfonylurea treatment in a cohort study in Egypt. Acta endocrinologica. 2016;12(2):157–160. doi:10.4183/aeb.2016.157
21. Cockburn BN, Bermano G, Boodram L-LG, et al. Insulin promoter factor-1 mutations and diabetes in Trinidad: identification of a novel diabetes-associated mutation (E224K) in an Indo-Trinidadian family. J Clin Endocrinol Metab. 2004;89(2):971–978. doi:10.1210/jc.2003-031282
22. Stanik J, Dusatkova P, Cinek O, et al. De novo mutations of GCK, HNF1A and HNF4A may be more frequent in MODY than previously assumed. Diabetologia. 2014;57(3):480–484. doi:10.1007/s00125-013-3119-2
23. Lorenzo PI, Juárez-Vicente F, Cobo-Vuilleumier N, García-Domínguez M, Gauthier BR. The diabetes-linked transcription factor PAX4: from gene to functional consequences. Genes. 2017;9(1):8. doi:10.3390/genes9010008
24. Cheung CY, Tang CS, Xu A, et al. Exome-chip association analysis reveals an Asian-specific missense variant in PAX4 associated with type 2 diabetes in Chinese individuals. Diabetologia. 2017;60(1):107–115. doi:10.1007/s00125-016-4132-z
25. Kooptiwut S, Plengvidhya N, Chukijrungroat T, et al. Defective PAX4 R192H transcriptional repressor activities associated with maturity onset diabetes of the young and early onset-age of type 2 diabetes. J Diabet Complicat. 2012;26(4):343–347. doi:10.1016/j.jdiacomp.2012.03.025
26. Ang SF, Tan CSH, Wang L, et al. PAX4 R192H is associated with younger onset of Type 2 diabetes in East Asians in Singapore. J Diabet Complicat. 2019;33:53–58. doi:10.1016/j.jdiacomp.2018.10.002
27. Chambers C, Fouts A, Dong F, et al. Characteristics of maturity onset diabetes of the young in a large diabetes center. Pediatr Diabetes. 2016;17(5):360–367. doi:10.1111/pedi.12289
28. Bonatto N, Nogaroto V, Svidnicki PV, et al. Variants of the HNF1alpha gene: a molecular approach concerning diabetic patients from southern Brazil. Genet Mol Biol. 2012;35(4):737–740. doi:10.1590/S1415-47572012005000061
29. Tin A, Marten J, Halperin Kuhns VL, et al. Target genes, variants, tissues and transcriptional pathways influencing human serum urate levels. Nat Genet. 2019;51(10):1459–1474. doi:10.1038/s41588-019-0504-x
30. Abreu D, Asada R, Revilla JMP, et al. Wolfram syndrome 1 gene regulates pathways maintaining beta-cell health and survival. Lab Invest. 2020;100(6):849–862. doi:10.1038/s41374-020-0408-5
31. Awata T, Inoue K, Kurihara S, et al. Missense variations of the gene responsible for Wolfram syndrome (WFS1/wolframin) in Japanese: possible contribution of the Arg456His mutation to type 1 diabetes as a nonautoimmune genetic basis. Biochem Biophys Res Commun. 2000;268(2):612–616. doi:10.1006/bbrc.2000.2169
32. Rohayem J, Ehlers C, Wiedemann B, et al. Diabetes and neurodegeneration in Wolfram syndrome: a multicenter study of phenotype and genotype. Diabetes Care. 2011;34(7):1503–1510. doi:10.2337/dc10-1937
33. Xuan S, Borok MJ, Decker KJ, et al. Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. J Clin Invest. 2012;122(10):3516–3528. doi:10.1172/JCI63352
34. Kalayinia S, Maleki M, Rokni-Zadeh H, et al. GATA4 screening in Iranian patients of various ethnicities affected with congenital heart disease: co-occurrence of a novel de novo translocation (5;7) and a likely pathogenic heterozygous GATA4 mutation in a family with autosomal dominant congenital heart disease. J Clin Lab Anal. 2019;33:e22923.
35. Lin X, Huo Z, Liu X, et al. A novel GATA6 mutation in patients with tetralogy of Fallot or atrial septal defect. J Hum Genet. 2010;55(10):662–667. doi:10.1038/jhg.2010.84
36. Phani NM, Guddattu V, Bellampalli R, et al. Population specific impact of genetic variants in KCNJ11 gene to type 2 diabetes: a case-control and meta-analysis study. PLoS One. 2014;9:e107021.
37. Olety SS, Vellakampadi D. TRMA syndrome (thiamine-responsive megaloblastic anaemia): an example of rare monogenic diabetes: is thiamine a magic pill for anaemia and diabetes? Int j Diabetes Dev Countries. 2016;36(4):389–392. doi:10.1007/s13410-016-0478-5
38. Piccand J, Strasser P, Hodson DJ, et al. Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Rep. 2014;9(6):2219–2232. doi:10.1016/j.celrep.2014.11.033
39. Chandra V, Albagli-Curiel O, Hastoy B, et al. RFX6 regulates insulin secretion by modulating Ca2+ homeostasis in human β cells. Cell Rep. 2014;9(6):2206–2218. doi:10.1016/j.celrep.2014.11.010
40. Patel KA, Kettunen J, Laakso M, et al. Heterozygous RFX6 protein truncating variants are associated with MODY with reduced penetrance. Nat Commun. 2017;8(1):888. doi:10.1038/s41467-017-00895-9
41. De Franco E, Shaw‐Smith C, Flanagan SE, et al. Biallelic PDX1 (insulin promoter factor 1) mutations causing neonatal diabetes without exocrine pancreatic insufficiency. Diabetic Med. 2013;30(5):e197–200. doi:10.1111/dme.12122
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
