Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 17
Identification and Validation of CDKN1A and HDAC1 as Senescence-Related Hub Genes in Chronic Obstructive Pulmonary Disease
Authors Yang J, Zhang MY, Du YM, Ji XL, Qu YQ
Received 14 May 2022
Accepted for publication 31 July 2022
Published 10 August 2022 Volume 2022:17 Pages 1811—1825
DOI https://doi.org/10.2147/COPD.S374684
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Min Zhang
Jie Yang,1,2 Meng-Yu Zhang,1 Yi-Ming Du,2 Xiu-Li Ji,3 Yi-Qing Qu1
1Department of Pulmonary and Critical Care Medicine, Qilu Hospital of Shandong University, Shandong Key Laboratory of Infectious Respiratory Diseases, Jinan, People’s Republic of China; 2Department of Gerontology, The First Affiliated Hospital of Shandong First Medical University & Shandong Provincial Qianfoshan Hospital, Jinan, People’s Republic of China; 3Department of Pulmonary Disease, Jinan Traditional Chinese Medicine Hospital, Jinan, People’s Republic of China
Correspondence: Yi-Qing Qu, Department of Pulmonary and Critical Care Medicine, Qilu Hospital of Shandong University; Shandong Key Laboratory of Infectious Respiratory Diseases, Wenhuaxi Road 107#, Jinan, 250012, People’s Republic of China, Tel +86 531 8216 9335, Fax +86 531 8296 7544, Email [email protected]
Purpose: Cellular senescence participates in the occurrence and development of chronic obstructive pulmonary disease (COPD). This study aimed to identify senescence-related hub genes and explore effective diagnostic markers and therapeutic targets for COPD.
Methods: The microarray data from the GSE38974 dataset was downloaded from the Gene Expression Omnibus (GEO) database. The overlapping genes between genes from the GSE38974 dataset and CellAge database were considered differentially expressed senescence-related genes (DESRGs). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using R software. Protein-protein interaction (PPI), miRNA-mRNA network, and competitive endogenous RNA (ceRNA) network were constructed and visualized by Cytoscape software. GSE100281 and GSE103174 datasets were employed to validate the expression and diagnostic value of hub genes. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was used to measure the mRNA levels of hub genes in peripheral blood mononuclear cells (PBMCs) from COPD and control samples.
Results: A total of 23 DESRGs were identified between COPD samples and healthy controls. Enrichment analysis revealed that DESRGs were mainly related to apoptosis and senescence. Moreover, four hub genes and two key clusters were acquired by Cytohubba and MCODE plugin, respectively. CDKN1A and HDAC1 were verified as final hub genes based on GSE100281 and GSE103174 datasets validation. The mRNA expression level of CDKN1A was negatively related to forced expiratory volume in 1 second/forced vital capacity (FEV1/FVC), and HDAC1 expression had the opposite correlation. Finally, an HDAC1-based ceRNA network, including 6 miRNAs and 11 lncRNAs, was constructed.
Conclusion: We identified two senescence-related hub genes, CDKN1A and HDAC1, which may be effective biomarkers for COPD diagnosis and treatment. An HDAC1-related ceRNA network was constructed to clarify the role of senescence in COPD pathogenesis.
Keywords: chronic obstructive pulmonary disease, senescence, bioinformatics, CDKN1A, HDAC1
Introduction
Chronic obstructive pulmonary disease (COPD), ranking the third leading cause of death all over the world, is the fifth disease burden around the world.1 A recent epidemiological survey has shown that 13.6% of Chinese adults suffer from COPD.2 Early diagnosis and effective treatment are crucial to reducing COPD mortality. Currently, the confirmed diagnostic method is the lung function test. The major pharmacological treatment is bronchodilators and inhaled corticosteroids (ICSs). However, the early diagnosis rate and effect of treatment are unsatisfactory. Thus, more effective diagnostic markers and treatment targets need to be further developed.
Cell senescence characterized by permanent cell cycle arrest can be induced by several signals, including oxidative stress, mitochondrial dysfunction, and telomere dysfunction.3,4 As an aging-related disease, COPD is closely related to cellular senescence. Recent studies have revealed that p16, a marker of cellular senescence, was up-regulated in cigarette smoke extract (CSE) treated endothelial progenitor cells (EPCs) and COPD patients.5,6 Senescence-associated secretory phenotype (SASP), characterized by secretion of proinflammatory cytokines, chemokines, and matrix metalloproteinases (MMPs), is increased in COPD.7 SIRT6, a member of the sirtuin family, class III histone deacetylases (HDACs), is an anti-aging factor.8 A study has indicated that the expression level of SIRT6 was decreased in CSE-stimulated human bronchial epithelial cells (HBECs), and overexpression of SIRT6 could reverse cellular senescence induced by CSE.9 Therefore, we predicted that senescence-related genes (SRGs) would be effective diagnostic markers or therapeutic targets in COPD.
Non-coding RNAs, including miRNAs and lncRNAs have been proved to be associated with COPD pathogenesis.10,11 LncRNAs act as miRNAs sponges that can regulate gene expression by competitively binding miRNAs, and this interaction among mRNAs, lncRNAs and miRNAs is also called competing endogenous RNA (ceRNA).12 Previous studies have revealed that ceRNA plays a pivotal role in COPD. The expression of lncRNA OIP5-AS1 is elevated in CSE-stimulated 16HBE cells and can enhance the inflammation and cell apoptosis by modulating the miR-410-3p/IL-13 signaling.13 LncRNA GAS5 functions as a ceRNA and regulates the miR-223-3p/NLRP3 axis to promote cell pyroptosis in COPD.14 However, the role of SRGs-based ceRNA in COPD has not been investigated.
In this study, for the first time, we elucidated the relationship between SRGs and COPD by the bioinformatics method. We screened differentially expressed senescence-related genes (DESRGs) based on the Gene Expression Omnibus (GEO) and CellAge database. Then, we performed enrichment analysis, protein-protein interaction (PPI) network analysis, and correlation analysis. Furthermore, hub genes were acquired, and another two separate datasets were used for validation. The expression levels of hub genes in peripheral blood mononuclear cells (PBMCs) of COPD and control samples were further measured by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). ceRNA among hub genes, miRNAs, and lncRNAs was also constructed. The flow chart of this study is shown in Figure 1. We believe that these results will provide a novel platform for exploring COPD pathogenesis and improving the prognosis of COPD patients.
Materials and Methods
Data Acquisition
In this study, three mRNA expression data of lung tissues were downloaded from the GEO datasets (https://www.ncbi.nlm.nih.gov/gds/). The keywords for searching were “chronic obstructive pulmonary disease”, “expression profiling by array”, “Homo sapiens”, and “tissue”. A GPL4133-platform dataset, GSE38974, which contained 9 healthy samples and 23 COPD samples, was identified as the test dataset.15 GSE103174 and GSE100281 were selected as validation datasets. GSE103174, which contained 11 healthy samples and 23 COPD samples, was acquired from the GPL13667 platform, and GSE100281, which included 16 healthy samples and 79 COPD samples, was acquired from the GPL11532 platform.16 A total of 279 senescence-related genes (SRGs) were obtained from the CellAge database (https://genomics.senescence.info/cells/).17
Differentially Expressed SRGs in COPD and Correlation Analysis
GEO2R was employed to analyze the gene expression between the control and COPD groups in the GSE38974 dataset. SRGs between the control and COPD groups were acquired by intersecting genes from the GSE38974 dataset and CellAge database. The cut-off criteria of DESRGs was an adjusted p-value of <0.050 and log fold change (FC) of >1. Spearman correlation analysis was used to conduct the correlation analysis of DESRGs.
Functional Enrichment Analysis of DESRGs in COPD
We used the “cluster Profiler R” package (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and the “GOplot R” package (https://cran.r-project.org/web/packages/GOplot/citation.html) for visualization. An adjusted p-value of < 0.050 indicated statistical significance.
PPI Network Analysis
The PPI network was predicted by the STRING database (https://cn.string-db.org/) by using an interaction score of > 0.4.18 Then, the interaction data were downloaded and visualized using Cytoscape software (version 3.9.1). The key clusters were obtained by Molecular Complex Detection (MCODE).19 Cytohubba was employed to screen important genes by several topological algorithms.20 In this study, the top six hub genes were obtained by five algorithms, including Degree, Maximal Clique Centrality (MCC), Maximum Neighborhood Component (MNC), Stress and EcCentricity. Then, the genes obtained from the above five algorithms were intersected to gain the final hub genes.
mRNA-miRNA Co-Expressed Network Construction
The Encyclopedia of RNA Interactomes (ENCORI, https://starbase.sysu.edu.cn) and miRWalk database (http://mirwalk.umm.uni-heidelberg.de) were used to predict the target miRNAs of 4 hub genes.21,22 A co-expressed network of mRNAs and miRNAs was constructed by Cytoscape software.
Clinical Samples Collecting and Preparation
Briefly, 5-mL venous blood samples were collected from 20 healthy controls and 24 COPD samples at Qilu Hospital of Shandong University. Thereafter, PBMCs were separated using isolation fluids (TBD sciences, Tianjin, China). The samples meeting the following criteria were included in the COPD group: the value of forced expiratory volume in 1 second/forced vital capacity (FEV1/ FVC) of < 0.7 after bronchodilator treatment and the age of 40 to 80 years. Healthy controls matched COPD samples in age and gender. The Medical Ethics Committee of Qilu Hospital of Shandong University approved the study procedure, which complied with the Declaration of Helsinki (as revised in 2013). All patients signed informed consent forms before the experiment.
RNA Isolation and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
The total RNA of PBMCs was extracted using RNAfast200 Reagent (Fastegen, Shanghai, China). Then, cDNA was synthesized using the PrimeScript RT reagent kit (Takara, Japan). RT-qPCR was performed using the TB Green Premix Ex Taq II (TaKaRa). ACTB was considered an endogenous control. The gene-specific primer sequences are listed in Supplementary Table S1. The 2−ΔCT method was employed to quantify the mRNA levels of the genes.
ceRNA Construction
An HDCA1-related ceRNA network was constructed. ENCORI was employed to predict the target lncRNA of six miRNAs. The predicted results were intersected using the “UpSetR” package.23 Then, the ceRNA network among mRNA, miRNA, and lncRNA was visualized by Cytoscape software.
Statistics
Statistical analysis was performed using the R language (version 3.6.3), GraphPad prism 7.0 (GraphPad Software, CA, USA) and IBM SPSS Statistics 28 (USA). Student’s t-test and Mann–Whitney U-test were used to compare gene expression levels between the control and COPD groups. Fisher-Freeman-Halton exact probability test was used to analyze the correlation between the Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage and other clinical characteristics. A p value of < 0.050 was considered statistically significant.
Results
Gain of DESRGs and Correlation Analysis
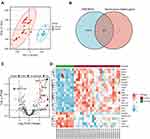
In this study, GSE38974 was used to analyze gene expression levels between the control and COPD groups. The principal component analysis (PCA) revealed that data of GSE38974 can be well repeated (Figure 2A). After GEO2R analysis, 19,287 genes were acquired. Figure 2B shows that 272 genes were obtained after the interaction with SRGs from the CellAge database. With the thresholds of the adjusted p-value of < 0.050 and log fold change (FC) > 1, 23 DESRGs, including 17 upregulated genes and 6 downregulated genes, were obtained (Supplementary Table S2). Volcano plots and heatmaps were created to visualize DESRGs (Figure 2C and D). The top 5 significantly different genes were SNAI1, MMP9, ATM, YAP1, and NOTCH3. Meanwhile, expression levels of DESRGs between the control and COPD groups were explored and visualized in the form of a boxplot. Figure 3A shows that all the DESRGs were significantly different between the two groups in the GSE38974 dataset. Furthermore, Spearman correlation analysis was used to explore the correlation among DESRGs. The specific results are shown on the heatmap in Figure 3B. The red color represents positive correlation, and the blue color represents negative correlation. The value of p of < 0.050 was considered statistically significant.
GO and KEGG Enrichment Analyses
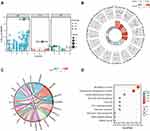
GO and KEGG analyses were performed to explore the biological functions of DESRGs. The top 5 GO biological processes of DESRGs were intrinsic apoptotic signaling pathway (p = 1.83e-06), intrinsic apoptotic signaling pathway in response to DNA damage (p = 1.15e-04), regulation of apoptotic signaling pathway (p = 1.79e-04), signal transduction in response to DNA damage (p = 1.79e-04) and response to gamma radiation (p = 1.79e-04) (Figure 4A and B; Table 1). According to the results of the KEGG analysis, DESRGs were mainly involved in transcriptional misregulation in cancer, microRNA in cancer, endocrine resistance, the hypoxia-inducible factor (HIF-1) signaling pathway and cell cycle (Figure 4C and D). The results indicated that DESRGs were closely related to senescence and apoptosis.
 |
Table 1 The Top 15 GO Enrichment Analysis of 23 DESRGs |
Identification of PPI Network and Hub Genes
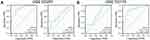
The PPI network was constructed using the STRING database to identify the interactive relationship between DESRGs, and the results, including 19 nodes and 47 edges, were further visualized by Cytoscape software. Figure 5A shows the PPI network of DESRGs. The interaction number of each DESRG is presented in Figure 5B. GAPDH, SNAI1, CDKN1A, HDAC1, and ATM were the top 5 genes with the maximum interaction numbers. Next, two significant clusters were obtained by the MCODE app. Cluster 1, which had the highest score, contained 5 nodes and 8 edges, including ATM, CDKN1A, GAPDH, NOTCH3, and SNAI1 (Figure 5C). Cluster 2 included 3 nodes and 3 edges (Figure 5D). Then, Cytohubba was employed to explore hub genes. Four hub genes, including ATM, CDKN1A, GAPDH, and HDAC1, were gained by intersecting the top 6 genes obtained by Degree, MCC, MNC, Stress and EcCentricity algorithms (Table 2). Furthermore, the diagnostic value of four hub genes in COPD was examined by receiver operating characteristic (ROC) curves. The best diagnostic genes were HDAC1 (area under the curve (AUC): 1.000) and ATM (AUC: 1.000), followed by GAPDH (AUC: 0.971) and CDKN1A (AUC: 0.874) (Figure 6). These results revealed that ATM, CDKN1A, GAPDH, and HDAC1 may be hub genes of COPD, and the above-mentioned genes had good diagnostic value.
 |
Table 2 Four Hub Genes Identified by Five Algorithms of CytoHubba |
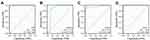 |
Figure 6 Diagnostic value of 4 hub genes. ROC curves of ATM (A), CDKN1A (B), GAPDH (C) and HDAC1 (D). Abbreviation: ROC, receiver operating characteristic. |
Prediction of Target miRNAs of Hub Genes and Construction of miRNA-mRNA Network
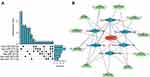
In this study, we used ENCORI and miRWalk databases to predict target miRNAs. After the intersection of the results predicted by the ENCORI and miRWalk databases, 31 miRNAs of ATM, 25 miRNAs of CDKN1A, 6 miRNAs of HDAC1, and 4 miRNAs of GAPDH were obtained. Then, a network between miRNAs and 4 hub genes, including 67 nodes and 66 edges, was visualized by Cytoscape software (Figure 7). The common target miRNA of ATM, CDKN1A, and GAPDH was has-miR-1224-5p.
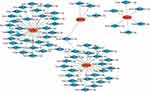 |
Figure 7 Co-expressed network of hub genes and target miRNAs. Red ellipses represent hub genes, and blue diamonds represent target miRNAs. |
Validation of Differentially Expressed Hub Genes in Two GSE Datasets
To further confirm differential expression levels of hub genes between the control and COPD groups, GSE103174, containing 11 healthy samples and 23 COPD samples, and GSE100281, including 16 healthy samples and 79 COPD samples, were employed. Figure 8A shows that the expression levels of CDKN1A and HDAC1 were significantly different between the two groups in the GSE100281 dataset. In the GSE103174 dataset, CDKN1A, GAPDH, and HDAC1 were differentially expressed between the control and COPD groups (Figure 8B). These results indicated that CDKN1A and HDAC1 might be the final hub genes of COPD.
Diagnostic Value Analysis of CDKN1A and HDAC1 in COPD Patients
In this part, the ROC curve was used to verify the diagnostic value of hub genes. The AUC can reflect both specificity and sensitivity. The criteria of AUC were 0.5–0.7 meaning moderate indicator, 0.7–0.9 on behalf of better indicator, and > 0.9 representing high indicator. The AUC of CDKN1A and HDAC1 in the GSE100281 dataset was 0.888 and 0.672, respectively (Figure 9A). In the GSE103174 dataset, the AUC of CDKN1A and HDAC1 was 0.678 and 0.839, respectively (Figure 9B). These findings suggested that CDKN1A and HDAC1 may be diagnostic markers for COPD.
The Expression of CDKN1A and HDAC1 in PBMCs of COPD Patients and Correlation Analysis with Lung Function
We measured the mRNA expression levels of CDKN1A and HDAC1 in PBMCs of COPD and healthy control samples. A total of 20 healthy controls and 24 COPD samples were screened. The clinical characteristics of cases are listed in Table 3. According to the correlation analysis of the GOLD stage and other clinical characteristics of the patients, we found that smoking exposure (p < 0.001) and body mass index (BMI) (p = 0.022) were related to the GOLD stage (Supplementary Table S3). The mRNA expression level of CDKN1A in PBMCs of COPD samples was higher than that in the healthy controls (p < 0.050) (Figure 10A), which was consistent with the results of the GSE100281 and GSE103174 datasets. The CDKN1A level was negatively related to FEV1/FVC value (r = −0.310, p = 0.041) (Figure 10B), while it was not related to FEV1% pred (Figure 10C). Compared to the healthy controls, the expression level of HDAC1 was decreased in PBMCs of COPD samples (p < 0.001) (Figure 10D). Correlation analysis revealed that the expression of HDAC1 was positively associated with FEV1/FVC value (r = 0.520, p < 0.001) and FEV1% pred (r = 0.457, p = 0.002) (Figure 10E and F).
 |
Table 3 The Clinical Characteristics of Patients |
Construction of ceRNA Between HDAC1 and Its Target Non-Coding RNAs
The expression level of HDAC1 was decreased in COPD samples. Meanwhile, the expression of HDAC1 was related to FEV1/FVC and FEV1% pred value. Increasing HDAC1 expression levels has been proved to alleviate the release of inflammatory cytokines.24 We proposed that HDAC1 was a promising therapeutic target for COPD patients. Therefore, we constructed an HDAC1-related ceRNA network. The target miRNAs of HDAC1 were has-miR-1271-5p, has-miR-3614-5p, has-miR-28-3p, has-miR-7-5p, has-miR-214-3p, and has-miR-330-3p. We predicted 7 target lncRNAs of has-miR-1271-5p, 10 target lncRNAs of has-miR-3614-5p, 11 target lncRNAs of has-miR-28-3p, 13 target lncRNAs of has-miR-7-5p, 19 target lncRNAs of has-miR-214-3p, and 14 target lncRNAs of has-miR-330-3p using the ENCORI database. LncRNA, which was the predicted target of at least two miRNAs, was selected as the final target lncRNA. Finally, 11 lncRNAs were obtained based on the above criteria (Figure 11A). The HDAC1-related ceRNA network was constructed and visualized by Cytoscape software (Figure 11B).
Discussion
COPD is one of the leading mortal diseases worldwide.25 In the past decades, various biological processes, including inflammation, autophagy, and pyroptosis, have been considered to be involved in COPD pathogenesis.26–28 Recently, numerous studies have indicated that cellular senescence was associated with COPD. However, the value of SRGs and their mechanism of action in COPD have remained unclear. In this paper, for the first time, we identified key SRGs and explored the function of SRGs in COPD pathogenesis by bioinformatics analysis.
We screened 23 DESRGs of COPD from the GEO dataset and CellAge database. GO analysis revealed that DESRGs were enriched in apoptosis and response to damage. KEGG pathway analysis showed that DESRGs were involved in cell rest and HIF-1 signaling pathway. Previous studies have shown that DNA damage induces cellular senescence via several signaling pathways.4 HIF-1α prevents cellular senescence and is involved in the pathogenesis of aging-related chronic diseases. HIF-1 signaling pathway can interact with multiple pathways involved in cell senescence, including the SIRT1 pathway.29 Hence, we predict that SRGs may participate COPD pathogenesis via the HIF-1 signaling pathway.
After the construction of the PPI network, four hub genes (ATM, CDKN1A, GAPDH, and HDAC1) were acquired. CDKN1A and HDAC1 were identified as final hub genes based on validation by another two independent GEO datasets. CDKN1A, also known as P21 or CIP1/WAF1, a cyclin-dependent kinase (CDK) inhibitor, has been found to play a key role in controlling cell cycle progression.30 Several studies have indicated that the expression of CDKN1A is upregulated in the COPD group.31,32 In three GEO datasets and the PBMCs of our clinical cases, the expression level of CDKN1A in COPD samples was higher than that in control samples, which is consistent with the above-mentioned findings. ROC curve analysis implied that CDKN1A had a better prognostic value. HDAC1, a member of the class I HDAC family, plays an important role in controlling inflammation in COPD in vivo and in vitro.24,33 Furthermore, HADC1 is a key regulatory factor in cellular senescence.34,35 Previous studies had shown that the expression level of HADC1 in COPD is decreased compared to the control group, which is consistent with our findings.36 A study has observed that erythromycin reverses the declined expression of HDAC1 induced by cigarette smoke extract (CSE) to alleviate the release of inflammatory cytokines in human macrophages.24 These results suggested that CDKN1A may be an effective diagnostic marker, and an HADC1 inducer may be a promising therapeutic agent for COPD patients.
Protein-coding RNA and non-coding RNA, acting as endogenous miRNA sponges, have been found to interact with each other, and the crosstalk between RNAs is called ceRNA.12 Studies have demonstrated that ceRNA is an important regular in hypoxia-induced pulmonary hypertension (HPH),37 interstitial lung disease (ILD),38 and COPD.39,40 However, the role of the senescence-related ceRNA network in COPD pathogenesis has not been explored yet. In this paper, we confirmed the downregulated HDAC1 in COPD samples. Meanwhile, we found that the expression of HDAC1 was positively correlated with FEV1/FVC and FEV1% pred. Thus, HDAC1, a senescence-related gene, was selected for analysis. An HDAC1-related ceRNA network, including 11 lncRNAs (XIST, NEAT1, ZFAS1, OIP5-AS1, LINC02381, MIR17HG, FGD5-AS1, AL035425.3, AL078639.1, AC079781.5 and AC016876.2) and 6 miRNAs was constructed. Among the target lncRNAs of miRNAs, several lncRNAs have been reported. A previous study has implied that NEAT1 is upregulated in aged bone marrow mesenchymal stem cells (BMSCs) and regulates mitochondrial function by sponging miR-27b-3p.41 Additionally, NEAT1 is a marker of COPD susceptibility, and NEAT1/miR-193a affects the development of COPD.42 In CSE-exposed 16HBE cells, XIST/miR-200c-3p/EGR3 can intensify inflammation action and apoptosis.43 OIP5-AS1 is a diagnostic marker for COPD, and the OIP5-AS1/miR-410-3p/IL-13 axis plays a crucial role in regulating COPD.13 Hence, we predicted that the HDAC1-related ceRNA might contribute to clarifying the effect of HDAC1 on senescence and the role of senescence in the development of COPD. The specific role and mechanism of ceRNA in COPD need to be further verified.
Although important discoveries were revealed by this study, there are also limitations. First, due to the lack of demographic and clinical information about patients in the three datasets, the correlation analysis between this information and hub genes was not performed. Second, we only investigated the mRNA expression levels of hub genes in peripheral blood, and further verification, including protein levels of genes, and the mechanism of SRGs in COPD pathogenesis needs to be conducted both in vivo and in vitro. Additionally, we did not confirm the interplay between RNAs in the ceRNA network, and it needs to be further studied and experimentally validated.
Conclusions
In the present study, we identified 23 DESRGs between the control samples and COPD samples via bioinformatics analysis. Based on our analysis and validation, CDKN1A and HDAC1 were verified as senescence-related hub genes of COPD. CDKN1A may be a promising diagnostic marker for COPD. The HDAC1-related ceRNA may contribute to elucidating the role of cellular senescence in COPD pathogenesis. HDAC1 seemed to be a crucial therapeutic target for COPD patients.
Abbreviations
BMSC, bone marrow mesenchymal stem cells; COPD, chronic obstructive pulmonary disease; ceRNA, competitive endogenous RNA; GOLD, global initiative for chronic obstructive lung disease; CSE, cigarette smoke extract; DESRGs, differentially expressed senescence-related genes; ENCORI, encyclopedia of RNA interactomes; EPCs, endothelial progenitor cells; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; GEO, gene expression omnibus; GO, gene ontology; HBECs, human bronchial epithelial cells; HDACs, histone deacetylases; HBECs, human bronchial epithelial cells; HPH, hypoxia-induced pulmonary hypertension; ICSs, inhaled corticosteroids; ILD, interstitial lung disease; KEGG, Kyoto encyclopedia of genes and genomes; MMPs, matrix metalloproteinases; PBMCs, peripheral blood mononuclear cells; PCA, principal component analysis; PPI, protein-protein interaction; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SASP, senescence-associated secretory phenotype; SRGs, senescence-related genes.
Data Sharing Statement
The datasets analyzed in this paper can be found in the GEO database. Senescence-related genes are downloaded from the CellAge database.
Ethics Approval
The trial was conducted according to the Declaration of Helsinki (as revised in 2013). The experimental procedure was approved by the Medical Ethics Committee of Qilu Hospital of Shandong University. All participants signed informed consent forms prior to study.
Author Contributions
All authors made a significant contribution to the work reported, whether it was in the conception, study design, execution, acquisition of data, analysis and interpretation, or all of these; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the National Natural Science Foundation of China (grant no. 81800039) and the Jinan Clinical Research Center for Prevention and Control Project of Major Respiratory Diseases (grant no. 201912011).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Barnes PJ, Burney PG, Silverman EK, et al. Chronic obstructive pulmonary disease. Nat Rev Dis Primers. 2015;1(1):15076. doi:10.1038/nrdp.2015.76
2. Fang L, Gao P, Bao H, et al. Chronic obstructive pulmonary disease in China: a nationwide prevalence study. Lancet Respir Med. 2018;6(6):421–430. doi:10.1016/s2213-2600(18)30103-6
3. Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med. 2015;21(12):1424–1435. doi:10.1038/nm.4000
4. He S, Sharpless NE. Senescence in health and disease. Cell. 2017;169(6):1000–1011. doi:10.1016/j.cell.2017.05.015
5. Adnot S, Amsellem V, Boyer L, et al. Telomere dysfunction and cell senescence in chronic lung diseases: therapeutic potential. Pharmacol Ther. 2015;153:125–134. doi:10.1016/j.pharmthera.2015.06.007
6. He Z, Peng H, Gao M, Liang G, Zeng M, Zhang X. p300/Sp1-mediated high expression of p16 promotes endothelial progenitor cell senescence leading to the occurrence of chronic obstructive pulmonary disease. Mediators Inflamm. 2021;2021:5599364. doi:10.1155/2021/5599364
7. Barnes PJ. Senescence in COPD and Its Comorbidities. Annu Rev Physiol. 2017;79(1):517–539. doi:10.1146/annurev-physiol-022516-034314
8. Kuwano K, Araya J, Hara H, et al. Cellular senescence and autophagy in the pathogenesis of chronic obstructive pulmonary disease (COPD) and idiopathic pulmonary fibrosis (IPF). Respir Investig. 2016;54(6):397–406. doi:10.1016/j.resinv.2016.03.010
9. Takasaka N, Araya J, Hara H, et al. Autophagy induction by SIRT6 through attenuation of insulin-like growth factor signaling is involved in the regulation of human bronchial epithelial cell senescence. J Immunol. 2014;192(3):958–968. doi:10.4049/jimmunol.1302341
10. Huang X, Zhu Z, Guo X, Kong X. The roles of microRNAs in the pathogenesis of chronic obstructive pulmonary disease. Int Immunopharmacol. 2019;67:12. doi:10.1016/j.intimp.2018.12.013
11. Devadoss D, Long C, Langley RJ, et al. Long noncoding transcriptome in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2019;61(6):10. doi:10.1165/rcmb.2019-0184TR
12. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
13. Hao W, Lin F, Shi H, Guan Z, Jiang Y. Long non-coding RNA OIP5-AS1 regulates smoke-related chronic obstructive pulmonary disease via targeting micro RNA −410-3p/IL-13. Bioengineered. 2021;12(2):11664–11676. doi:10.1080/21655979.2021.2000199
14. Mo R, Li J, Chen Y, Ding Y. lncRNA GAS5 promotes pyroptosis in COPD by functioning as a ceRNA to regulate the miR2233p/NLRP3 axis. Mol Med Rep. 2022;26(1). doi:10.3892/mmr.2022.12735
15. Ezzie ME, Crawford M, Cho JH, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67(2):122–131. doi:10.1136/thoraxjnl-2011-200089
16. SAG Willis-Owen, Thompson A, Kemp PR, et al. COPD is accompanied by co-ordinated transcriptional perturbation in the quadriceps affecting the mitochondria and extracellular matrix. Sci Rep. 2018;8(1):12165. doi:10.1038/s41598-018-29789-6
17. Avelar RA, Ortega JG, Tacutu R, et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 2020;21(1):91. doi:10.1186/s13059-020-01990-9
18. Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi:10.1093/nar/gkaa1074
19. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4(1):2. doi:10.1186/1471-2105-4-2
20. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11
21. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. doi:10.1093/nar/gkt1248
22. Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018;13(10):e0206239. doi:10.1371/journal.pone.0206239
23. Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–2940. doi:10.1093/bioinformatics/btx364
24. Li M, Zhong X, He Z, et al. Effect of erythromycin on cigarette-induced histone deacetylase protein expression and nuclear factor-kappaB activity in human macrophages in vitro. Int Immunopharmacol. 2012;12(4):643–650. doi:10.1016/j.intimp.2011.12.022
25. Rabe KF, Watz H. Chronic obstructive pulmonary disease. Lancet. 2017;389(10082):1931–1940. doi:10.1016/s0140-6736(17)31222-9.
26. Vij N, Chandramani-Shivalingappa P, Van Westphal C, Hole R, Bodas M. Cigarette smoke-induced autophagy impairment accelerates lung aging, COPD-emphysema exacerbations and pathogenesis. Am J Physiol Cell Physiol. 2018;314(1):C73–C87. doi:10.1152/ajpcell.00110.2016
27. Zhang MY, Jiang YX, Yang YC, et al. Cigarette smoke extract induces pyroptosis in human bronchial epithelial cells through the ROS/NLRP3/caspase-1 pathway. Life Sci. 2021;269:119090. doi:10.1016/j.lfs.2021.119090
28. Wu Y, Li Y, Wu B, et al. beta-Arrestin2 inhibits expression of inflammatory cytokines in BEAS-2B lung epithelial cells treated with cigarette smoke condensate via inhibition of autophagy. Cell Physiol Biochem. 2018;50(4):1270–1285. doi:10.1159/000494586
29. Yeo EJ. Hypoxia and aging. Exp Mol Med. 2019;51(6):1–15. doi:10.1038/s12276-019-0233-3
30. López-Domínguez JA, Rodríguez-López S, Ahumada-Castro U, et al. Cdkn1a transcript variant 2 is a marker of aging and cellular senescence. Aging. 2021;13(10):12. doi:10.18632/aging.203110
31. Yang D, Yan Y, Hu F, Wang T. CYP1B1, VEGFA, BCL2, and CDKN1A affect the development of chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2020;15:167–175. doi:10.2147/COPD.S220675
32. Sun S, Shen Y, Wang J, Li J, Cao J, Zhang J. Identification and validation of autophagy-related genes in chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2021;16:67–78. doi:10.2147/COPD.S288428
33. Chen X, Guan XJ, Peng XH, Cui ZL, Luan CY, Guo XJ. Acetylation of lysine 9 on histone H3 is associated with increased pro-inflammatory cytokine release in a cigarette smoke-induced rat model through HDAC1 depression. Inflamm Res. 2015;64(7):513–526. doi:10.1007/s00011-015-0832-y
34. Pao PC, Patnaik D, Watson LA, et al. HDAC1 modulates OGG1-initiated oxidative DNA damage repair in the aging brain and Alzheimer’s disease. Nat Commun. 2020;11(1):2484. doi:10.1038/s41467-020-16361-y
35. Willis-Martinez D, Richards HW, Timchenko NA, Medrano EE. Role of HDAC1 in senescence, aging, and cancer. Exp Gerontol. 2010;45(4):279–285. doi:10.1016/j.exger.2009.10.001
36. Leus NG, van den Bosch T, van der Wouden PE, et al. HDAC1-3 inhibitor MS-275 enhances IL10 expression in RAW264.7 macrophages and reduces cigarette smoke-induced airway inflammation in mice. Sci Rep. 2017;7(1):45047. doi:10.1038/srep45047
37. Wang J, Niu Y, Luo L, et al. Decoding ceRNA regulatory network in the pulmonary artery of hypoxia-induced pulmonary hypertension (HPH) rat model. Cell Biosci. 2022;12(1):27. doi:10.1186/s13578-022-00762-1
38. Wu Q, Liu Y, Xie Y, Wei S, Liu Y. Identification of potential ceRNA network and patterns of immune cell infiltration in systemic sclerosis-associated interstitial lung disease. Front Cell Dev Biol. 2021;9:622021. doi:10.3389/fcell.2021.622021
39. Duan R, Niu H, Yu T, et al. Identification and bioinformatic analysis of circular RNA expression in peripheral blood mononuclear cells from patients with chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2020;15:1391–1401. doi:10.2147/COPD.S252896
40. Shi ZE, Zhang MY, Liu JY, et al. Autophagy induced by BCL2-Related ceRNA network participates in the occurrence of COPD. Int J Chron Obstruct Pulmon Dis. 2022;17:791–808. doi:10.2147/COPD.S347733
41. Zhang H, Xu R, Li B, et al. LncRNA NEAT1 controls the lineage fates of BMSCs during skeletal aging by impairing mitochondrial function and pluripotency maintenance. Cell Death Differ. 2022;29(2):351–365. doi:10.1038/s41418-021-00858-0
42. Ming X, Duan W, Yi W. Long non-coding RNA NEAT1 predicts elevated chronic obstructive pulmonary disease (COPD) susceptibility and acute exacerbation risk, and correlates with higher disease severity, inflammation, and lower miR-193a in COPD patients. Int J Clin Exp Pathol. 2019;12(8):11.
43. Chen P, Jiang P, Chen J, Yang Y, Guo X. XIST promotes apoptosis and the inflammatory response in CSE-stimulated cells via the miR-200c-3p/EGR3 axis. BMC Pulm Med. 2021;21(1):215. doi:10.1186/s12890-021-01582-8
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.