Back to Journals » Infection and Drug Resistance » Volume 16
Genomic Determinants of Pathogenicity and Antimicrobial Resistance of Nosocomial Acinetobacter baumannii Clinical Isolates of Hospitalized Patients (2019–2021) from a Sentinel Hospital in Hangzhou, China
Authors Wei C, Chen J, Anwar TM, Huang L, Yang W, Dong X, Chen Q, Yue M , Yu D
Received 21 February 2023
Accepted for publication 29 April 2023
Published 12 May 2023 Volume 2023:16 Pages 2939—2952
DOI https://doi.org/10.2147/IDR.S407577
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Chenxing Wei,1,* Jian Chen,1,* Tanveer Muhammad Anwar,2,* Lingling Huang,1 Wenjie Yang,3 Xueyan Dong,1 Qiong Chen,1 Min Yue,2,4,5 Daojun Yu1,3
1Department of Medical Laboratory, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, 310006, People’s Republic of China; 2Department of Veterinary Medicine, Institute of Preventive Veterinary Sciences, Zhejiang University College of Animal Sciences, Hangzhou, 310058, People’s Republic of China; 3Department of Medical Laboratory, The Fourth School of Clinical Medicine, Zhejiang Chinese Medical University, Hangzhou, 310053, People’s Republic of China; 4Hainan Institute, Zhejiang University, Sanya, 572025, People’s Republic of China; 5State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, National Medical Center for Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, 310003, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Min Yue; Daojun Yu, Email [email protected]; [email protected]
Purpose: Acinetobacter baumannii (A. baumannii or AB) is one of the most opportunistic, nosocomial pathogens threatening public healthcare across countries. A. baumannii has become a primary growing concern due to its exceptional ability to acquire antimicrobial resistance (AMR) to multiple antimicrobial agents which is increasingly reported and more prevalent every year. Therefore, there is an urgent need to evaluate the AMR knowledge of A. baumannii for effective clinical treatment of nosocomial infections. This study aimed to investigate the clinical distribution AMR phenotypes and genotypes, and genomic characteristics of A. baumannii isolates recovered from hospitalized patients of different clinical departments of a sentinel hospital to improve clinical practices.
Methods: A total of 123 clinical isolates were recovered from hospitalized patients of different clinical departments during 2019– 2021 to analyze AMR patterns, and further subjected to whole-genome sequencing (WGS) investigations. Multi-locus sequence typing (MLST), as well as the presence of antimicrobial-resistant genes (ARGs), virulence factor genes (VFGs) and insertion sequences (ISs) were also investigated from WGS data.
Results: The results highlighted that A. baumannii clinical isolates had shown a high AMR rate, particularly from the intensive care unit (ICU), towards routinely used antimicrobials, ie, β-lactams and fluoroquinolones. ST2 was the most prevalent ST in the clinical isolates, it was strongly associated to the resistance of cephalosporins and carbapenems, with blaOXA-23 and blaOXA-66 being the most frequent determinants; moreover, high carrier rate of VFGs was also observed such as all strains containing the ompA, adeF, pgaC, lpsB, and bfmR genes.
Conclusion: Acinetobacter baumannii clinical isolates are mostly ST2 with high rates of drug resistance and carrier of virulence factors. Therefore, it requires measurements to control its transmission and infection.
Keywords: Acinetobacter baumannii, antimicrobial resistance, genetic determinants, virulence factors, whole-genome sequencing
Introduction
Acinetobacter baumannii (A. baumannii or AB) is an opportunistic, gram-negative coccobacillus that widely exists in nature, has the strong adhesive ability and environmental adaptability, and can colonize the human respiratory system or skin/soft tissue surfaces.1,2 It has the potential to spread among hospitalized patients and is commonly associated with hospital-acquired nosocomial infections such as respiratory tract infections (RTIs), bacteremia, urinary tract infections (UTIs), secondary meningitis, and surgical site infections.3,4 Multi-drug resistant A. baumannii (MDR-AB) is considered the most prevalent pathogen in the intensive care unit (ICU) environment, and is currently recognized as an emerging cause of nosocomial epidemics worldwide. According to the Infectious Diseases Society of America (IDSA), A. baumannii is one of the six most deadly and alarming nosocomial priority pathogens. There are approximately 45,000 cases of A. baumannii clinical infections in the United States annually.5 The mortality rate of A. baumannii infections in the intensive care unit (ICU) is as high as 50%.6
MDR-AB is one of the most concerning bacteria in terms of control and treatment in both developed and developing countries, raising serious concerns for the MDR crisis.7–9 Recent studies have revealed the emergence of MDR bacterial infections in humans, birds, cattle and fish.10–12 Consequently, this has increased the necessity for routine antimicrobial susceptibility testing (AST) to find the recommended antibiotics and screen for the emergence of MDR strains. MDR pathogens resistant to various antimicrobials, including β-lactams, fluoroquinolones, tetracyclines, and aminoglycosides. Several virulence factor genes (VFGs) responsible for A. baumannii pathogenicity have been found, including outer membrane porins, pilus, lipopolysaccharides, capsular polysaccharides, phospholipases, protein secretion systems, and iron-chelating systems. Some clinical strains share genes that are involved in adhesion, invasion, and survival, and they can form biofilms.13,14 Importantly, A. baumannii forms biofilms on polystyrene and glass as well as on fungal filaments and epithelial cells. In addition, the adhesion and biofilm phenotypes of few clinical isolates were associated with the presence of broad-spectrum antibiotic resistance.15 Due to the high prevalence of infections and the shortage of effective antibiotics for treatment, A. baumannii is one of the most problematic pathogens causing nosocomial infections. To address this issue, it is important to identify antimicrobial resistance (AMR) and the VFGs determinants in clinical isolates.
The progress in whole genome sequencing (WGS) technology has promoted research on bacterial typing, whole genome characteristics, drug resistance mechanisms and pathogenesis of A. baumannii.16,17 The rapid development of this technology, accompanied by a significant reduction in price, has enabled WGS to be widely adopted, with over 3500 A. baumannii genomes available by early April 2019.18 However, A. baumannii genome information has distorted geographical distribution, with 69% (2481/3575) of all sequenced strains across the five countries (the United States, the People’s Republic of China, Thailand, Australia, and Pakistan). The analysis of the genome sequences revealed that ST2, ST1, ST79, and ST25 account for more than 71% of all the genomes that have been sequenced to date, with ST2 being the most widespread type.18,19
Nevertheless, our previous investigations have demonstrated the routine use of WGS technology for studying molecular epidemiology, AMR pattern, MLST types, VFGs and the correlation between VFGs and AMR profiles.20–23 Therefore, WGS technology was used to investigate the genomic features of 123 A. baumannii clinical isolates recovered from 2019 to 2021 from the specimens of hospitalized patients from different clinical departments of the First People’s Hospital in Hangzhou, Zhejiang province, China. In addition, we also statistically analyzed the clinical distribution, AMR phenotypes and genotypes, and the relationship between clinical diagnosis and antimicrobial susceptibility testing (AST) of isolates. Therefore, this comprehensive and integrated research study could improve our understanding regarding AMR pattern in clinical A. baumannii isolates and provide essential knowledge to clinicians for clinical therapy and management as well as pinpointing the nosocomial infections.24,25
Materials and Methods
Bacterial Isolation and Identification of Acinetobacter baumannii
A total of 123 isolates were recovered from specimens from different clinical departments of the First People’s Hospital in Hangzhou, Zhejiang Province, China, during the three years (2019–2021). All clinical specimens from patients were inoculated onto blood agar plate and incubated at 37°C for 18–24 hrs. The dominant growing strains were transferred to another blood agar plates for pure culture for 18–24 hrs, followed by Matrix Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) (Bruker Microflex LT/SH, Billerica, Massachusetts, USA) and antimicrobial susceptibility testing (AST). The identified A. baumannii strains were stored at −80°C using glycerol with a final concentration of 20% (v/v). The isolates were resuscitated when used and re-identified by mass spectrometry. Each patient was matched to one specimen, and each specimen was matched to one strain of A. baumannii. The clinical data of 123 patients were statistically analyzed, and the distribution of ages, genders, clinical diagnoses, and treatments was determined.
Antimicrobial Susceptibility Testing
In this study, the phenotypic antimicrobial resistance (AMR) of A. baumannii isolates was evaluated by automated identification/susceptibility testing system bioMérieux (VITEK2 Compact, Durham, USA) and Kirby-Bauer disc diffusion method as recommended by CLSI guidelines (CLSI, 2019). The minimum inhibitory concentrations (MICs) and zone size/diameters for all isolates were determined against twelve antibiotics belonging to six antimicrobial classes according to the recommendations of the Clinical Laboratory Standards Institute guidelines (CLSI, 2019).26 Escherichia coli ATCC25922 and Pseudomonas aeruginosa ATCC27853 were used as quality control strains to validate antimicrobial susceptibility testing (AST) as recommended by the CLSI. Isolates were nonsusceptible to ≥1 agent in ≥3 antimicrobial classes were considered multi-drug resistant (MDR).27,28 The tested antimicrobials and their detection concentration are mentioned in Table S1.
DNA Isolation, Whole Genome Sequencing (WGS), and Genome Assembly
Genomic DNA was extracted from isolates using the Vazyme Bacterial DNA Kit (Vazyme Biotech, Nanjing, China) according to the manufacturer’s recommended protocol. The extracted DNA was quantified using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s instructions. Whole-genome sequencing (WGS) was performed by next-generation sequencing (NGS) using the Illumina NovaSeq platform (San Diego, California, USA) and the NovaSeq 6000 SP kit (300 cycles). The quality of the whole-genome library was checked by quality control. The sequence data were trimmed with Trimmomatic v0.39. De novo assemblies of our 123 A. baumannii genomes were generated using SPAdes v3.12.0.29
Bioinformatics Analysis
Sequence analyses were performed using several bioinformatics tools. The sequence types (STs), antimicrobial-resistance genes (ARGs), and plasmid types were identified using assemblies of the samples on the Galaxy platform,30,31 combined with mlstv2.16.1 (https://github.com/tseemann/mlst2016) and ABRicate v1.0.1 (https://github.com/tseemann/abricate),32 inclu-ding ResFinder v3.2 (https://cge.cbs.dtu.dk/services/ResFinder/) with a 90% similarity cutoff for ARGs.33–35 The plasmid replicons were identified using PlasmidFinder database (https://cge.cbs.dtu.dk/services/PlasmidFinder/) with a similarity cutoff of 95%.36 The virulence factor genes (VFGs) were identified using virulence factors database (VFDB).37 The presence of insertion sequence (IS) elements was predicted using ABRicate v1.0.1 with the default parameter against the IS database from ISfinder (https://www-is.biotoul.fr/).38 In addition, single nucleotide polymorphisms (SNPs) alignment was matched using package Snippy v4.4.4, and phylogenetic trees were constructed by IQ-tree v1.6.12 and TVM+F+R3 models. Interactive Tree of Life (iTOL) v6 software (https://itol.embl.de/)39 was to visualize and edit the phylogenetic tree.
Statistical and Data Projection Analysis
Clinical data were analyzed using SPSS software Version 19.0 (IBM SPSS Inc., Chicago, USA), and the figures of STs, AMR profiles and VFGs were generated by GraphPad Prism 8 (San Diego, CA). The distribution of various STs and genetic relatedness of A. baumannii clinical isolates are illustrated in the minimum spanning tree (MST) using PHYLOViZ software 2.0 for providing scalable data integration and visualization according to the goeBURST algorithm.40 Pearson correlations (r) among AMR genotype, phenotype, VFGs and STs of A. baumannii clinical isolates were determined using R software. Binary data (0/1) were imported into R software (version 4.1.0, R Foundation for Statistical Computing, Austria), and correlations were determined using the “cor” function and visualized using the “ggcorplot” function. Correlation significance (P<0.05) was also determined by the “cor. mtest” function. The correlation is strong if r ≥ 0.7, moderate if the r value is between 0.4 and 0.69, and weak if r < 0.4.41
Results
Distribution of Acinetobacter baumannii
The distribution of clinical A. baumannii isolates in different clinical departments recovered or isolated from various clinical specimens is shown in the Table 1. The prevalence of A. baumannii isolates was high in intensive care units (ICU)(62.60%, 77/123), followed by the internal medicine department. The respiratory system was the most common body site for A. baumannii infections (86.18%, 106/123), and all specimens isolated from blood (7.32%, 9/123) were from the ICU department.
 |
Table 1 Distribution of Acinetobacter baumannii Based on Mode of Acquisition of Infection and in Various Clinical Specimens |
The A. baumannii isolates were obtained from 123 clinically infected patients, of which 82 cases (64.1%) were males, and 41 cases (33.3%) were females. Hence, a gender ratio (M/F) was 2:1. Centralized trend indicators showed that the median age of 82 males was 71 years (interquartile range: 14–96 years) and the median age of 41 females was 76 years (interquartile range: 27–97 years), and the distribution of age showed that 1/4 of the population was younger than 62 years, 1/2 was younger than 73 years, and 1/4 was older than 81 years. In addition, 70% of the patients were between 62 and 89 years, predominantly middle-aged and older adults.
Among the 96 patients with clear clinical diagnosis and treatment information (27 patients with Incomplete treatment information) were counted, the clinical diagnosis was dominated by pulmonary infections (88.54%, 85/96) followed by bloodstream infections (9.37%, 9/96). In terms of clinical treatment, enzyme-inhibitor complexes were the predominant agents, in which penicillin complexes and cephalosporin complexes account for 35.42% (34/96), 23.96% (23/96), respectively (Table 2).
 |
Table 2 Correlation Between the Clinical Diagnosis and Antibiotics Used in the Infected Patients |
Antimicrobial Susceptibility Testing
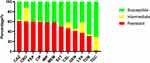
The antimicrobial resistance profiles of all 123 clinical A. baumannii isolates against twelve antibiotics belonging to six antimicrobial classes are shown in Figure 1 and Table S2. The results showed a high resistance rate towards β-lactams (cephalosporins), with ceftazidime (60.97%, 75/123), ceftriaxone (60.16%, 74/123), and cefepime (60.16%, 74/123) followed by carbapenems (60.16%, 74/123) and quinolones, ciprofloxacin (60.16%, 74/123) and. The lowest resistance rate was observed against tigecycline (1.62%, 2/123). In addition, 75 (60.97%) of the 123 isolates were multi-drug resistance (MDR), 17.07% (21/123) were resistant to four antimicrobial classes, 22.76% (28/123) were resistant to five antimicrobial classes, and 20.33% (25/123) were resistant to all six antimicrobial classes.
Prediction of Antimicrobial Resistance Genes (ARGs)
The genome sequences of 123 clinical A. baumannii isolates were analyzed for ARGs. A total of 40 different ARGs encoding resistance towards six classes of antimicrobial drugs were found in A. baumannii genomes (Figure 2). The most prevalent genes were blaADC-25 (100%,123/123), encoding resistance to cephalosporins, followed by blaOXA-23 and blaOXA-66 (60.16%, 74/123), harbored resistance to carbapenems; sul2 (47.97%, 59/123) encoding resistance to sulfonamides; tet(B) (48.78%, 60/123) encoding resistance to tetracyclines and aph (3”)-Ib (47.97%,59/123), aph (6)-Id (47.97%,59/123) harbored resistance to aminoglycosides. Furthermore, the trend of ARGs in our hospital over the last three years revealed a significant increase in the proportion of aminoglycosides, and macrolide ARGs, such as armA from 21.05% to 49.02% and mph(E) from 19.30% to 31.37%. However, the ARGs for β-lactam and sulfonamides showed a decreasing trend over time. Compared with 2019, blaTEM-1D decreased from 45.61% to 36.36% in 2021, and sul decreased by about 10% (Figure S1).
Multi-Locus Sequence Typing (MLST)
The MLST results of 123 clinical A. baumannii isolates revealed 36 STs according to the Pasteur scheme (Figure 3a). Among them, ST2 was the predominant ST, 74/123 (60.16%), followed by ST40, 8/123 (6.50%). The MLST of isolates showed that ST2 was the most common ST from respiratory tract origin, 64/106 (60.38%), followed by ST40, 5.66% (6/106). All the clinical isolates from blood were ST2 while all isolates from bile belonged to ST40. The MLST of isolates from urine were identified as ST10, ST25, ST57, ST218, and ST2265. According to the years of isolations, 16 STs were isolated in 2019, 8STs were recovered in 2020, and in 2021 a total of 19 STs were isolated (Figure S2). The minimum spanning tree (MST) showed phylogenetic relationships of the sequence types (STs) using allele profiles of 123 clinical A. baumannii isolates. According to the MST, ST2 was primarily from ICU patients (82.89%,63/76), whereas ST40 was distributed across ICU patients (25%, 2/8), patients in internal medicine (37.5%, 3/8), and patients in surgery (37.5%, 3/8). Compared to the ICU (12 STs) and surgery departments (4STs), internal medicine (28 STs) has a more significant number of STs (Figure 3b).
Prediction of Virulence Factor Genes (VFGs) in Acinetobacter baumannii Genomes
The results of VFGs were predicted based on the virulence factor database (VFDB) presented in Table S3. 123 clinical isolates of A. baumannii contained 7 major categories of VFGs, including adherence, biofilm formation, enzyme, immune evasion, iron uptake, regulation, and serum resistance. A total of 47 different VFGs belonging to 12 subcategories were found. All isolates possessed genes encoding OmpA (outer membrane protein A), AdeFGH efflux pump, Phospholipase C, Phospholipase D, Acinetobactin, LPS (lipopolysaccharide), BfmRS (regulation of biofilm formation), and PbpG (penicillin-binding protein). 82 (66.67%), 86 (69.92%) to 97 (78.86%), 120 (97.56%) to 123 (100%), and 102 (82.93%) to 104 (84.55%) isolates possessed virulence factors including biofilm-associated proteins (bap), Csu fimbriae, poly-N-acetylglucosamine (PANG) and quorum sensing, respectively. In addition, 56.76% (42/74) of ST2 isolates harbored all 7 major classes of VFGs.
Differences in Antimicrobial Resistance Phenotypes, Genotypes, STs and VFGs in Clinical Departments
Among the 123 strains of A. baumannii, the resistance rate of ICU isolates was higher than that of the other two departments, especially ciprofloxacin, ceftriaxone, cefepime and carbapenems. In addition, ICU isolates also have the highest carrying rate of ARG, such as aminoglycosides (aph (3”)-Ib, aph (6)-Id), most cephalosporins (blaOXA-23, blaOXA-66and blaTEM-1D), sulfonamides (sul2), and tetracyclines (tetB). The ICU isolates were mainly ST2, while the ST40 strain mainly come from surgery. The isolates from three departments all carried a large number of VFGs. The abaI/R carrying rate of ICU isolates was high, but the csu pili of the isolates from internal medicine and surgery departments was higher than that of ICU isolates. There were significant differences in AMR phenotypes (P<0.0001), resistance genotypes (P=0.0002), and VFGs (P<0.0001) between isolates from different clinical departments (Figure 4).
Correlation Analysis Between AMR Profiles, STs and VFGs
The correlation analysis showed that all observed correlation results were statistically significant (P<0.05) (Figure S3a–d). ARGs and phenotypic antibiotic resistance correlation analysis showed that carbapenems were most strongly correlated with β-lactamase resistance genes blaOXA-23, blaOXA-66, blaTEM-1D, aminoglycoside genes aph (3”)-Ib, aph (6)-Id and tetracycline gene tet(B) (Figure S3a). STs and phenotypic antibiotic resistance correlation analysis demonstrated a strong correlation between ST2 strains and resistance to cephalosporins, carbapenems, quinolones, and aminoglycosides (Figure S3b). STs and ARGs correlation analysis illustrated that ST2 was strongly correlated with β-lactamase resistance genes blaOXA-23 and blaOXA-66, the aminoglycoside genes aph (3”)-Ib, aph (3’)-Ia, aph (6)-Id, the tetracycline gene tet(B), and the sulfonamide gene sul2 (Figure S3c). STs and VFGs correlation analysis displayed a strong correlation between ST2 and bap, and some negative correlation with virulence factor Csu fimbriae; ST723 and pgaA showed absolute negative correlation (Figure S3d).
Prediction of Insertion Sequence (ISs)
The results of insertion sequences of 123 strains of A. baumannii showed that 124 kinds of insertion sequences (ISs) were identified. We found that the most common ISs were ISAba22 (86.18%,106/123) and ISAba26 (81.30%, 100/123). ISAba50, ISAba64, ISAba46 and ISAba16 were detected in 85 (69.10%), 83 (67.48%), 80 (65.04%) and 78 (63.41%) strains, respectively (Table S4). ISAba1 was detected in all CRAB isolates.
Genetic Relationship and Phylogenomic Analysis of the Isolates
The maximum likelihood (ML) phylogenetic tree was constructed from the best model TVM+F+R3 of IQ tree v1.6.12 using 4744 SNPs, identified by core genome matching in Snippy v3.1. The phylogenetic tree (Figure 5) showed that the upper branch was dominated by strains from internal medicine “(blue)” and surgery “(green)”, with a large number of strains from the ICU “(orange)” in the lower branch. Phylogenomic analysis showed that most of the non-ST2 strains did not carry resistance genes such as aph (3”)-Ib and aph (6)-Id, and most strains showed the absence of chloramphenicol resistance genes, and respiratory isolates comprising of ST 40 were very similar to the bile isolates.
Discussion
In this study, 75 of 123 patients with Acinetobacter baumannii had MDR-AB infections. 70% of the patients were between the ages of 62 and 89, with the majority being middle-aged or elderly individuals. Similar findings were observed in a previous study where A.baumannii infections were more prevalent in elder patients.42 We found that most of the isolates recovered from the ICU department, and the infection site was predominantly identified in the respiratory tract, which may be due to their low immunity, and the use of ventilators for assisted ventilation and/or invasive surgery, which made a higher risk of nosocomial infection,43,44 indicating that older patients in ICU were highly prone to A. baumannii infections. Our current findings were consistent with the global data.45 In addition, our study found significant differences between isolates from ICU and other clinical departments in terms of AMR phenotypes, genetic determinants and virulence factor genes (VFGs). Many nosocomial infections were treatable with medicines; however, the increase in AMR rate has resulted in favoring the use of wide-spectrum antimicrobials in clinical practices, which in turn could lead to antimicrobial selection effects, conversely, this made it challenging to treat AB-infected patients.46,47
The results of phenotypic AMR profiling showed that the resistance rate of MDR-AB to cephalosporins and carbapenems was higher than aminoglycosides, fluoroquinolones, tigecycline, and cefoperazone-sulbactam sodium.48 The analysis of clinical diagnosis and antibiotics used in the infected patients revealed that the most used antimicrobial drug in clinical practice was the enzyme-inhibitor complex (cefoperazone-sulbactam sodium or piperacillin-sulbactam), followed by the combined application of two or more antimicrobial drugs. For the anti-infective treatment of MDR-AB infection, enzyme-inhibitor complexes are preferred, and the combination of antimicrobial drugs could control the development of the disease as soon as possible.49–51
The results of ARGs analysis showed that a variety of resistance genes were identified in clinical A. baumannii isolates from hospitalized patients in different clinical departments. All isolates contained blaADC-25, which is generally considered to be an important genetic determinant of cephalosporin resistance in A. baumannii.52 Moreover, β-lactamase resistance gene blaOXA-23 and blaOXA-51-like variant (blaOXA-66) were more prevalent in the 123 genomes and associated with resistance to all β-lactam based compounds.53 A few isolates carried blaTEM-1D, which encodes a class A β-lactamase. In Carbapenem-resistant Acinetobacter baumannii (CRAB) from different regions of the world, it was found that the isolates carrying blaTEM-1D were sensitive to carbapenems,54 cephalosporins and enzyme-inhibitor complex, but showed resistance to other β-lactams, aminoglycosides and quinolones. In addition to β-lactam resistance genes, these isolates contained genes that likely conferred resistance to aminoglycoside, tetracycline, chloramphenicol, sulfonamide, and macrolide. In addition, the three-year trend of ARGs showed that the proportion of aminoglycosides, macrolides, and tetracycline ARGs increased over time. The increase in bacterial ARGs and phenotypic resistance indicated that hospitals had to take strict measures to control the use of relevant antimicrobial drugs.
ST2 was the most dominant ST, which belonged to “international clone 2 (IC2)”.55 The resistance rate of ST2 isolates to antibacterial drugs and carrying rate of ARGs was relatively far higher than other STs, especially the resistance rate to β- lactam drugs. These results were consistent with the previous findings.18 The correlation analysis between STs and antibiotic resistance phenotypes and ARGs showed that ST2 isolate showed a strong correlation with the resistance genes blaOXA-23 and blaOXA-66, cephalosporins and carbapenems. In this study, ST2 was mainly identified from ICU isolates, while ST40 was distributed in ICU, internal medicine and surgery department, and the number of STs from internal medicine was much higher than the other two clinical departments. Notably, all isolates from blood specimens belonged to ST2. Although STs were complicated, the major STs did not change over three years. ST2 was the most prevalent and mainly from ICU, indicating that the isolates recovered from ICU were genetically homogenous and had the ability for horizontal transmission of resistance genes.
Nevertheless, A. baumannii was once thought to be a low-virulent pathogen, but in recent years, researchers have revealed the presence of highly virulent A. baumannii.56,57 In this study, we found many virulence factor genes, including adherence, biofilm formation, enzyme, immune evasion, iron uptake, regulation, and serum resistance. These results were consistent with the previous study.13 In our study, all the isolates had BfmRS indicating that these isolates were motile. It was well known that A. baumannii exhibited loss of BfmRS58 or AbaI/R quorum sensing system59 leading to a significant reduction in motility. More than half of ST2 isolates contained all seven major classes of VFGs in this study, implying that ST2 isolates carried more VFGs and were more viable in the host environment. This might be the reason that ST2 had become a prevalent clone worldwide.
We predicted the insertion sequence (IS) of 123 A. baumannii strains, and among the 124 ISs identified in our study, we found ISAba22 to be the most prevalent IS, followed by ISAba26, ISAba50, ISAb64a, and ISAba46. In addition to functioning as a promoter for the expression of many ARGs, IS usually provided a copy-and-paste mechanism of transposition.60 Based on our findings, ISAba1 was identified in all blaOXA-23 positive CRAB isolates. Many previous reports suggested that ISAba1 was usually located upstream of blaOXA-23-like genes and that it provided a powerful promoter that drive the expression of blaOXA23-like genes.61
Our phylogenetic analysis showed that the distribution of isolates from respiratory specimens was relatively concentrated, indicating that A. baumannii can be easily transmitted through the respiratory tract route. The isolates from the respiratory tract were similar to those from bile, indicating that bile-derived A. baumannii might have come from respiratory infection. Moreover, the blood isolates were closely linked with respiratory isolates, which suggested that the bloodstream infection of the clinical A. baumannii isolates was more likely to be from a respiratory infection.13,62 The phylogenetic distribution of our blood-infected isolates was dispersed, suggesting a distant relationship and perhaps not a single route of entry into the bloodstream from the respiratory tract or other sites. Treating patients in different clinical departments could be another factor that causes the risk of cross-sectoral transmission of A. baumannii. Hence, it was necessary to enhance hygienic and sanitizing measures as well as detection capabilities for A. baumannii. Additional recording of the traceability of individual patient could help to limit the persistence and dissemination of A. baumannii in and between clinical departments.
Conclusion
The present study concluded that 123 clinical isolates of A. baumannii showed a high rate of antimicrobial resistance pattern. Especially, isolates from the ICU department were more resistant to antimicrobial drugs and might be more virulent. Most of the A. baumannii were found in ICU respiratory specimens, which indicated that it is challenging to treat ICU-acquired pneumonia. This was most common in older people, who speeded long periods of time on artificial ventilation; ST2 was found to be the prevalent sequence type in our hospital. A. baumannii isolates revealed high rate of virulence factor carriage, which indicated the pathogenicity of various prevalent clones.
Data Sharing Statement
The datasets generated for this study can be found in the NCBI Bioproject with the accession number PRJNA873189.
Ethics Approval and Informed Consent
This study was approved by the Ethics Committee of Hangzhou First People’ Hospital [Approval numbers 2019-020-01]. Waiving of informed consent was given due to the retrospective, non-interventional study design. All patient’s data were collected anonymously and ensured about the confidentiality of their information.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant number 81930111), the Natural Science Foundation of Zhejiang Province (grant number LZ22H190002), the Health & Medical Sci-Tech Project of Hangzhou Municipal Health Commission (Z2021005), the Science and Technology Project of Hangzhou Municipal (grant number 202204A001), and the Health & Medical Sci-Tech Project of Health Commission of Zhejiang Province (grant number 2020KY206, 2022PY016).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Martin-Aspas A, Guerrero-Sanchez FM, Garcia-Colchero F, Rodriguez-Roca S, Giron-Gonzalez JA. Differential characteristics of Acinetobacter baumannii colonization and infection: risk factors, clinical picture, and mortality. Infect Drug Resist. 2018;11:861–872. doi:10.2147/IDR.S163944
2. Morris FC, Dexter C, Kostoulias X, Uddin MI, Peleg AY. The mechanisms of disease caused by Acinetobacter baumannii. Front Microbiol. 2019;10:1601. doi:10.3389/fmicb.2019.01601
3. Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–582. doi:10.1128/cmr.00058-07
4. Al-Kadmy IMS, Ali ANM, Salman IMA, Khazaal SS. Molecular characterization of Acinetobacter baumannii isolated from Iraqi hospital environment. New Microbes New Infect. 2018;21:51–57. doi:10.1016/j.nmni.2017.10.010
5. Spellberg B, Rex JH. The value of single-pathogen antibacterial agents. Nat Rev Drug Discov. 2013;12(12):963. doi:10.1038/nrd3957-c1
6. Bianco A, Quirino A, Giordano M, et al. Control of carbapenem-resistant Acinetobacter baumannii outbreak in an intensive care unit of a teaching hospital in Southern Italy. BMC Infect Dis. 2016;16(1):747. doi:10.1186/s12879-016-2036-7
7. Zarrilli R, Casillo R, Di Popolo A, et al. Molecular epidemiology of a clonal outbreak of multidrug-resistant Acinetobacter baumannii in a university hospital in Italy. Clin Microbiol Infect. 2007;13(5):481–489. doi:10.1111/j.1469-0691.2006.01675.x
8. Yadav SK, Bhujel R, Hamal P, Mishra SK, Sharma S, Sherchand JB. Burden of multidrug-resistant Acinetobacter baumannii infection in hospitalized patients in a tertiary care hospital of Nepal. Infect Drug Resist. 2020;13:725–732. doi:10.2147/idr.S239514
9. Ibrahim S, Al-Saryi N, Al-Kadmy IMS, Aziz SN. Multidrug-resistant Acinetobacter baumannii as an emerging concern in hospitals. Mol Biol Rep. 2021;48(10):6987–6998. doi:10.1007/s11033-021-06690-6
10. Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5(12):939–951. doi:10.1038/nrmicro1789
11. Lopinska A, Indykiewicz P, Skiebe E, et al. Low occurrence of Acinetobacter baumannii in gulls and songbirds. Pol J Microbiol. 2020;69:1–6. doi:10.33073/pjm-2020-011
12. Mateo-Estrada V, Vali L, Hamouda A, Evans BA, Castillo-Ramirez S. Acinetobacter baumannii sampled from cattle and pigs represent novel clones. Microbiol Spectr. 2022;10(4):e0128922. doi:10.1128/spectrum.01289-22
13. Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91–102. doi:10.1038/nrmicro.2017.148
14. Ayoub Moubareck C, Hammoudi Halat D. Insights into Acinetobacter baumannii: a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics. 2020;9(3):119. doi:10.3390/antibiotics9030119
15. Gaddy JA, Actis LA. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 2009;4(3):273–278. doi:10.2217/fmb.09.5
16. Wareth G, Brandt C, Sprague LD, Neubauer H, Pletz MW. WGS based analysis of acquired antimicrobial resistance in human and non-human Acinetobacter baumannii isolates from a German perspective. BMC Microbiol. 2021;21(1):210. doi:10.1186/s12866-021-02270-7
17. Roberts LW, Forde BM, Hurst T, et al. Genomic surveillance, characterization and intervention of a polymicrobial multidrug-resistant outbreak in critical care. Microb Genom. 2021;7(3). doi:10.1099/mgen.0.000530
18. Hamidian M, Nigro SJ. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom. 2019;5(10). doi:10.1099/mgen.0.000306
19. Cherubini S, Perilli M, Segatore B, et al. Whole-genome sequencing of ST2 A. baumannii causing bloodstream infections in COVID-19 patients. Antibiotics. 2022;11(7):955. doi:10.3390/antibiotics11070955
20. Li Y, Kang X, Ed-Dra A, et al. Genome-based assessment of antimicrobial resistance and virulence potential of isolates of non-pullorum/gallinarum salmonella serovars recovered from dead poultry in China. Microbiol Spectr. 2022;10(4):e0096522. doi:10.1128/spectrum.00965-22
21. Pan H, Jia C, Paudyal N, et al. Comprehensive assessment of subtyping methods for improved surveillance of foodborne salmonella. Microbiol Spectr. 2022;10(5):e0247922. doi:10.1128/spectrum.02479-22
22. Zhou W, Lin R, Zhou Z, et al. Antimicrobial resistance and genomic characterization of Escherichia coli from pigs and chickens in Zhejiang, China. Front Microbiol. 2022;13:1018682. doi:10.3389/fmicb.2022.1018682
23. Tang B, Ni J, Lin J, et al. Genomic characterization of multidrug-resistance gene CFR in Escherichia coli recovered from food animals in Eastern China. Front Microbiol. 2022;13:999778. doi:10.3389/fmicb.2022.999778
24. Xu X, Chen Y, Pan H, et al. Genomic characterization of Salmonella Uzaramo for human invasive infection. Microb Genom. 2020;6(7). doi:10.1099/mgen.0.000401
25. Elbediwi M, Pan H, Biswas S, Li Y, Yue M. Emerging colistin resistance in Salmonella enterica serovar Newport isolates from human infections. Emerg Microbes Infect. 2020;9(1):535–538. doi:10.1080/22221751.2020.1733439
26. Clinical, Institute LS. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2019.
27. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
28. Van An N, Hoang LH, Le HHL, et al. Distribution and antibiotic resistance characteristics of bacteria isolated from blood culture in a teaching hospital in Vietnam during 2014–2021. Infect Drug Resist. 2023;16:1677–1692. doi:10.2147/IDR.S402278
29. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi:10.1089/cmb.2012.0021
30. Afgan E, Baker D, van den Beek M, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44(W1):W3–W10. doi:10.1093/nar/gkw343
31. Biswas S, Elbediwi M, Gu G, Yue M. Genomic characterization of new variant of hydrogen sulfide (H(2)S)-producing Escherichia coli with multidrug resistance properties carrying the mcr-1 gene in China†. Antibiotics. 2020;9(2):80. doi:10.3390/antibiotics9020080
32. Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi:10.1093/jac/dks261
33. Liu Q, Chen W, Elbediwi M, et al. Characterization of salmonella resistome and plasmidome in pork production system in Jiangsu, China. Front Vet Sci. 2020;7:617. doi:10.3389/fvets.2020.00617
34. Wu B, Ed-Dra A, Pan H, Dong C, Jia C, Yue M. Genomic investigation of salmonella isolates recovered from a pig slaughtering process in Hangzhou, China. Front Microbiol. 2021;12:704636. doi:10.3389/fmicb.2021.704636
35. Liu Y, Jiang J, Ed-Dra A, et al. Prevalence and genomic investigation of Salmonella isolates recovered from animal food-chain in Xinjiang, China. Food Res Int. 2021;142:110198. doi:10.1016/j.foodres.2021.110198
36. Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi:10.1128/aac.02412-14
37. Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47(D1):D687–d692. doi:10.1093/nar/gky1080
38. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(Databaseissue):D32–6. doi:10.1093/nar/gkj014
39. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–5. doi:10.1093/nar/gkw290
40. Nascimento M, Sousa A, Ramirez M, Francisco AP, Carrico JA, Vaz C. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33(1):128–129. doi:10.1093/bioinformatics/btw582
41. Schober P, Boer C, Schwarte LA. Correlation coefficients: appropriate use and interpretation. Anesth Analg. 2018;126(5):1763–1768. doi:10.1213/ANE.0000000000002864
42. Girija ASS, Priyadharsini JV. Prevalence of Acb and non-Acb complex in elderly population with urinary tract infection (UTI). Acta Clin Belg. 2021;76(2):106–112. doi:10.1080/17843286.2019.1669274
43. Uwingabiye J, Lemnouer A, Baidoo S, et al. Intensive care unit-acquired Acinetobacter baumannii infections in a Moroccan teaching hospital: epidemiology, risk factors and outcome. Germs. 2017;7(4):193–205. doi:10.18683/germs.2017.1126
44. Ferrer M, Torres A. Epidemiology of ICU-acquired pneumonia. Curr Opin Crit Care. 2018;24(5):325–331. doi:10.1097/MCC.0000000000000536
45. Zheng W, Yuan S, Li L. Analysis of hospital departmental distribution and antibiotic susceptibility of Acinetobacter isolated from sputum samples. Am J Infect Control. 2013;41(8):e73–6. doi:10.1016/j.ajic.2012.11.004
46. Alrahmany D, Omar AF, Harb G, El Nekidy WS, Ghazi IM. Acinetobacter baumannii infections in hospitalized patients, treatment outcomes. Antibiotics. 2021;10(6):630. doi:10.3390/antibiotics10060630
47. Granata G, Taglietti F, Schiavone F, Petrosillo N. Durlobactam in the treatment of multidrug-resistant Acinetobacter baumannii infections: a systematic review. J Clin Med. 2022;11(12):3258. doi:10.3390/jcm11123258
48. Finegold SM. In vitro efficacy of beta-lactam/beta-lactamase inhibitor combinations against bacteria involved in mixed infections. Int J Antimicrob Agents. 1999;12(Suppl 1):S9-14;discussion S26–7. doi:10.1016/s0924-8579(99)00086-2
49. Smolyakov R, Borer A, Riesenberg K, et al. Nosocomial multi-drug resistant Acinetobacter baumannii bloodstream infection: risk factors and outcome with ampicillin-sulbactam treatment. J Hosp Infect. 2003;54(1):32–38. doi:10.1016/s0195-6701(03)00046-x
50. Lee CR, Lee JH, Park M, et al. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017;7:55. doi:10.3389/fcimb.2017.00055
51. Sakoulas G, Rose W, Berti A, et al. Classical beta-lactamase inhibitors potentiate the activity of daptomycin against methicillin-resistant Staphylococcus aureus and colistin against Acinetobacter baumannii. Antimicrob Agents Chemother. 2017;61(2). doi:10.1128/AAC.01745-16
52. Wareth G, Linde J, Nguyen NH, et al. WGS-based analysis of carbapenem-resistant Acinetobacter baumannii in Vietnam and molecular characterization of antimicrobial determinants and MLST in Southeast Asia. Antibiotics. 2021;10(5):563. doi:10.3390/antibiotics10050563
53. Heritier C, Poirel L, Fournier PE, Claverie JM, Raoult D, Nordmann P. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49(10):4174–4179. doi:10.1128/AAC.49.10.4174-4179.2005
54. Chopjitt P, Kerdsin A, Takeuchi D, et al. Whole genome analysis of extensively drug-resistant Acinetobacter baumannii clinical isolates in Thailand. Infect Disord Drug Targets. 2021;21(5):e270421188042. doi:10.2174/1871526520999201116201911
55. Levy-Blitchtein S, Roca I, Plasencia-Rebata S, et al. Emergence and spread of carbapenem-resistant Acinetobacter baumannii international clones II and III in Lima, Peru. Emerg Microbes Infect. 2018;7(1):119. doi:10.1038/s41426-018-0127-9
56. Li J, Yu T, Luo Y, et al. Characterization of carbapenem-resistant hypervirulent Acinetobacter baumannii strains isolated from hospitalized patients in the mid-south region of China. BMC Microbiol. 2020;20(1):281. doi:10.1186/s12866-020-01957-7
57. Allen JL, Tomlinson BR, Casella LG, Shaw LN. Regulatory networks important for survival of Acinetobacter baumannii within the host. Curr Opin Microbiol. 2020;55:74–80. doi:10.1016/j.mib.2020.03.001
58. Clemmer KM, Bonomo RA, Rather PN. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology. 2011;157(Pt 9):2534–2544. doi:10.1099/mic.0.049791-0
59. Tang J, Chen Y, Wang X, Ding Y, Sun X, Ni Z. Contribution of the AbaI/AbaR quorum sensing system to resistance and virulence of Acinetobacter baumannii clinical strains. Infect Drug Resist. 2020;13:4273–4281. doi:10.2147/IDR.S276970
60. Yoon EJ, Kim JO, Yang JW, et al. The blaOXA-23-associated transposons in the genome of Acinetobacter spp. represent an epidemiological situation of the species encountering carbapenems. J Antimicrob Chemother. 2017;72(10):2708–2714. doi:10.1093/jac/dkx205
61. Khurshid M, Rasool MH, Ashfaq UA, et al. Dissemination of bla(OXA-23)-harbouring carbapenem-resistant Acinetobacter baumannii clones in Pakistan. J Glob Antimicrob Resist. 2020;21:357–362. doi:10.1016/j.jgar.2020.01.001
62. Yu K, Zeng W, Xu Y, et al. Bloodstream infections caused by ST2 Acinetobacter baumannii: risk factors, antibiotic regimens, and virulence over 6 years period in China. Antimicrob Resist Infect Control. 2021;10(1):16. doi:10.1186/s13756-020-00876-6
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.