Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 15
Gene Polymorphisms of m6A Erasers FTO and ALKBH1 Associated with Susceptibility to Gastric Cancer
Authors Li Y , Zhou D, Liu Q, Zhu W, Ye Z, He C
Received 2 February 2022
Accepted for publication 23 May 2022
Published 31 May 2022 Volume 2022:15 Pages 547—559
DOI https://doi.org/10.2147/PGPM.S360912
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Yue Li,* Dalei Zhou,* Qing Liu, Weijie Zhu, Zulu Ye, Caiyun He
Department of Molecular Diagnostics, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, 510060, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Caiyun He; Zulu Ye, Department of Molecular Diagnostics, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, No. 651, Dongfeng Road, Yuexiu District, Guangzhou, 510060, People’s Republic of China, Tel +86-18665593050 ; +86-15017590433, Fax +20-87340921, Email [email protected]; [email protected]
Purpose: Fat mass and obesity-associated protein (FTO) and AlkB homolog 1 (ALKBH1) are m6A demethylases that have been demonstrated to be associated with the overall survival of patients with gastric cancer (GC). This study investigates the influence of genetic variants of FTO and ALKBH1 on susceptibility to GC.
Patients and Methods: Potentially functional single nucleotide polymorphisms (SNPs) of FTO and ALKBH1 were genotyped in 419 patients with GC and 569 healthy controls by Kompetitive allele-specific PCR.
Results: The AG and AG/AA variants of FTO rs2287142 were significantly associated with a decreased GC risk (for AG/AA vs GG: adjusted OR = 0.73, p = 0.020). The GA and GA/GG variants of ALKBH1 rs1076496 were closely correlated with an increased risk of GC in people aged ≥ 55 years (for GA/GG vs AA: adjusted OR = 1.51, p = 0.041) but showed a decreasing tendency of risk of GC in people aged < 55 years (adjusted OR = 0.85, p = 0.444). FTO rs2287142 and ALKBH1 rs1076496 conformed to the principle of a dominant model. FTO haplotype rs1421091-rs1421092-rs2287142-rs9939609 CTAT was closely associated with a lower risk of total GC (adjusted OR = 0.62, p = 0.023), while CTGA was linked with an increased risk of intestinal GC (adjusted OR = 2.51, p = 0.005). ALKBH1 rs1048147-rs1076496-rs11159286 CAC haplotype was significantly associated with a decreased risk of GC in people aged ≥ 55 years (adjusted OR = 0.41, p = 0.008). The FTO rs2287142-rs9939609 AG/AA-TT combination was associated with a decreased risk of GC only in the presence of rs1421091 TC/TT (adjusted OR = 0.70, p = 0.047), demonstrating that these FTO SNPs might have a cooperative effect on susceptibility to GC.
Conclusion: FTO and ALKBH1 SNPs may have predictive value in evaluating susceptibility to GC with differing age or Lauren classification.
Keywords: FTO, ALKBH1, gastric cancer, susceptibility, Lauren classification
Introduction
Gastric cancer (GC) is one of the most common digestive malignancies worldwide.1,2 The carcinogenesis process of GC involves an evolution from an inflammatory to a precancerous stage, and finally to the carcinoma.3,4 Marked genetic heterogeneity may play a crucial role in this process, along with Helicobacter pylori infection, lifestyle, and other risk factors.5,6
N6-methyladenosine (m6A) is one of the most deeply studied RNA modifications in eukaryotic cells and is involved in multiple aspects of RNA metabolism, such as RNA decay, RNA nuclear export, mRNA translation, and mRNA stability.7 With increasing evidence demonstrating that m6A plays a key role in cancers, the “writers”, “erasers”, and “readers” that add, remove, or bind to m6A sites, respectively, have also garnered interest of the research community.8 In our previous study, we explored the associations of a series of proteins expressed by m6A-related genes with GC and found that a decreased level of fat mass and obesity-associated protein (FTO) was markedly associated with a shorter overall survival of patients with GC, and a lower expression of AlkB homolog 1 (ALKBH1) was correlated with larger tumor size (≥ 5 cm) and more advanced TNM stages of GC.9
FTO and ALKBH1 are primary m6A demethylases that are capable of reversing the methylation of RNA by oxidizing the N-methyl group of m6A sites to a hydroxymethyl group.10,11 As implied by the name, the FTO gene is located in the obesity susceptibility loci determined by a 2007 genome-wide association study.12,13 The most typical single nucleotide polymorphism (SNP) rs9939609, along with many other SNPs, such as rs8050136, rs1121980, and rs17817449, in the intron 1 region of FTO have been reported to affect the risk of obesity.14–16 The risk allele A of FTO rs9939609 was closely related to obesity and BMI across different populations such as Chinese, Brazilians, and Iranians.17–19 Moreover, several SNPs of FTO20,21 and one SNP located downstream of ALKBH122 have been demonstrated to be strongly related with the occurrence and development of cancers.
To our knowledge, the influence of genetic polymorphisms of FTO and ALKBH1 on the risk of GC has not been previously investigated. In the present study, we selected and genotyped potentially functional SNPs of FTO and ALKBH1 in 419 Chinese patients with GC and 569 healthy controls, and the individual and interactive genetic effects of genetic polymorphisms of FTO and ALKBH1 on susceptibility to GC were comprehensively explored.
Materials and Methods
Participants
This study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center (Guangzhou, China) (approval NO. G2022-002-01). Written informed consent was provided by all participants at their first visit. All experiments were performed in accordance with the guidelines stated in the Declaration of Helsinki. In total, 988 subjects, which included 419 patients with GC and 569 healthy individuals, had been retrospectively recruited from the Sun Yat-sen University Cancer Center between July 2011 and June 2016. The diagnosis and Lauren classification of GC was confirmed by surgery, followed by pathological examination. The medical records were carefully reviewed to obtain demographic parameters. Healthy participants visiting for a physical check would be included if they had no history of precancerous lesions or other malignant or serous systemic diseases.
SNP Selection
Candidate SNPs were selected using tools from the SNPinfo Web Server (http://snpinfo.niehs.nih.gov/). Flanking sequences 5000 bp upstream and 1000 bp downstream of FTO and ALKBH1 genes were separately searched among the Han Chinese in Beijing, China and the SNPs annotated with functions such as nonsynonymous SNP, splicing regulation, stop codon, PolyPhen prediction, transcriptional factor binding site (TFBS), and microRNA binding site, and the nearby genes were screened. The SNPs with minor allele frequency < 0.05 were excluded. In addition, FTO rs9939609 was added to the candidate list because it has been reported to be linked with risk of cancer in a series of studies.21
Genotyping Assay
Genomic DNA was isolated from peripheral blood lymphocytes using the phenol-chloroform method and diluted to a working concentration of 50 ng/μL before genotyping. All sample DNAs were randomized on 384-well plates and blinded for the disease status. Genotyping was conducted using Kompetitive allele-specific PCR (KASP) by Gene Company (Shanghai, China) as described in previous publications.23,24 In brief, this procedure involved adding the SNP-specific KASP Assay mix and the universal KASP Master mix to DNA samples that were then subjected to a thermal cycling reaction (Table S1) followed by an end-point fluorescent read using a PHERAstar FSX Microplate Reader (BMG Labtech, Ortenberg, Germany). The KASP Assay mix contained three assay-specific non-labelled oligos, two allele specific forward primers, and one common reverse primer designed specific to each allele of the SNP (Table S2). For quality control, 10% of the total samples were repeatedly genotyped and the concordance rate reached 100%.
Determining the Optimal Genetic Models
With A defined as the variant allele and B as the common allele, odds ratio 1 (OR1) was determined for AA versus BB, OR2 for AB versus BB, and OR3 for AA versus AB. These three pairwise comparisons were evaluated to determine the best genetic models for candidate SNPs, as reported in a previous publication:25 (1) If OR1 = OR3 ≠ 1 and OR2 = 1, then a recessive model was considered and AA was compared with AB plus BB; (2) if OR1 = OR2 ≠ 1 and OR3 = 1, then a dominance model was used and AA plus AB was compared with BB; (3) if OR2 = 1/OR3 ≠ 1 and OR1 = 1, then a complete overdominance model was considered and AB was compared with AA plus BB; and (4) if OR1 > OR2 > 1 and OR1 > OR3 > 1 (or OR1 < OR2 < 1 and OR1 < OR3 < 1), then a codominance model was considered and AA was compared with AB and with BB.
Statistical Analysis
The association of each SNP with the risk of GC was explored by multivariate logistic regression, with or without adjusting for sex and age, and was recorded as OR with 95% confidence interval (CI). To explore the modifying effects of sex and age on the influence of candidate SNPs on the risk of GC, we used the Breslow–Day test, which is commonly used to test for homogeneity by comparing the differences between the ORs from each stratum. The cooperative effect of SNP-A on SNP-B was explored by comparing the associations between SNP-B and GC by a multivariate logistic regression analysis adjusted for sex and age when all data was grouped according to the two different genotypes of SNP-A. All statistical analyses were performed using the SPSS 17.0 software (SPSS, Chicago, IL, USA), excluding the calculations of D′ and r2 values for linkage disequilibrium (LD) and the haplotype analysis, which were performed using an online tool (https://www.snpstats.net/start.htm?). Each haplotype with a minimum frequency of 0.03 in the control group was explored using the common haplotype as the reference. All p values were two-sided and statistical significance was set at p < 0.05.
Results
Individual Genetic Effects of Candidate SNPs on the Risk of GC
Four SNPs of FTO and three SNPs of ALKBH1 predicted to have potential function were chosen and genotyped (Table 1). The clinicopathological features of the studied population are presented in Table 2. We investigated the existence of individual associations between the seven SNPs and the total risk of GC, which were evaluated by crude ORs and adjusted ORs while controlling by sex and age. We then explored the potential influence of modification factors on the candidate SNPs. Regardless of performing a crude analysis or adjusted analysis, FTO rs2287142 AG (adjusted OR = 0.73, 95% CI: 0.55–0.96, p = 0.025) and AG/AA (adjusted OR = 0.73, 95% CI: 0.56–0.95, p = 0.020) genotypes were both significantly associated with a lower risk of GC compared with that of the GG genotype, while the other SNPs did not show any individual effects on susceptibility to GC (Table 3).
 |
Table 1 Basic Information of SNPs Explored in the Present Study |
 |
Table 2 The Clinicopathological Characteristics of Studied Population |
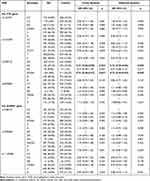 |
Table 3 The Individual Genetic Effect of Candidate SNPs on the Risk of GC |
Considering the discrepant genetic background for GC with differing Lauren classification, we further grouped patients with GC into intestinal, diffuse, and mixed-type subgroups. Due to a very limited sample size in each subgroup (intestinal type, 128; diffuse type, 151; and mixed type, 116), this part of the analysis was regarded as a preliminary exploration. After gender- and age-adjusted analysis, we observed borderline associations between ALKBH1 rs1048147 and the risk of mixed-type GC (AC/AA vs CC: OR = 1.54, 95% CI: 1.02–2.33, p = 0.042), and between FTO rs1421092 and the risk of intestinal type GC (TT vs TC/CC: OR = 1.59, 95% CI: 1.02–2.48, p = 0.042). No statistical association was found between these variants and diffuse type GC.
Influence of Candidate SNPs Modified by Sex and Age on the Risk of GC
There were no differences between ORs in males and females for all the investigated SNPs (all Breslow–Day test p values > 0.05, Table 4), which indicated no modifying effect of sex on these SNPs. The FTO rs2287142 AG and AG/AA genotypes were still statistically related to a low risk of GC under the modifying effects of sex and age (Tables 4 and 5).
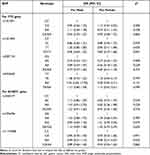 |
Table 4 The Modification Effect of Sex on the Associations of Candidate SNPs with GC Risk |
 |
Table 5 The Modification Effect of Age on the Associations of Candidate SNPs with GC Risk |
Interestingly, the associations of the ALKBH1 rs1076496 GA genotype with risk of GC were significantly different between young (< 55 years) and elderly people (≥ 55 years) (Breslow–Day test p value = 0.048), and the GA/GG genotype of the same SNP showed a borderline difference (Breslow–Day test p value = 0.051) (Table 5). We measured the layer-specific ORs in each age group using logistic regression. Compared with the AA genotype, GA and GA/GG genotypes were both associated with a significantly increased risk of GC in the people aged ≥ 55 years (OR = 1.60, 95% CI: 1.06–2.42, p = 0.026; OR = 1.51, 95% CI: 1.01–2.23, p = 0.041, respectively) but exhibited a decreased tendency of risk of GC in the people aged < 55 years (OR = 0.87, 95% CI: 0.56–1.35, p = 0.528; OR = 0.85, 95% CI: 0.56–1.29, p = 0.444, respectively). These results may suggest a modifying effect of age on ALKBH1 rs1076496.
Determining the Optimal Genetic Model for Candidate SNPs
The genetic model represents the best-fitting manner in which an SNP influences the biology of a disease in population-based molecular association studies; therefore, determining the optimal genetic model for each candidate SNP by means of an appropriate method is crucial. For rs2287142, OR1, OR2, and OR3 were 0.77 (p = 0.238), 0.74 (p = 0.003), and 1.03 (p = 0.893), respectively, which was concordant with the principle represented by “OR1 = OR2 ≠ 1 and OR3 = 1.” This data, therefore, fits well with a dominant model. For ALKBH1 rs1076496 in people aged ≥ 55 years, OR1, OR2, and OR3 were 1.28 (p = 0.350), 1.60 (p = 0.026), and 0.80 (p = 0.340), respectively, which was also compatible with a dominant model (Table 6).
 |
Table 6 The Optimal Genetic Model for Candidate SNPs |
Haplotype Analysis
Figure 1 shows the statistical results of D’ and r2 used for evaluating LD between SNPs in FTO and ALKBH1 genes. As Lauren classification is an important prognostic factor of GC, we also explored the influence of haplotypes on the risk conferred by each Lauren subtype of GC. When compared with the most common haplotype CTGT, the FTO rs1421091-rs1421092-rs2287142-rs9939609 CTAT haplotype showed a significant association with a lower total risk of GC (adjusted OR = 0.62, 95% CI: 0.41–0.93, p = 0.023), and the FTO CTGA haplotype was closely associated with an increased risk of intestinal GC (adjusted OR = 2.51, 95% CI: 1.32–4.76, p = 0.005) (Table 7).
 |
Table 7 The Influence of FTO Haplotypes on the Risk of Total GC and GC of Each Lauren’s Classification |
 |
Figure 1 The D’ and r2 for linkage disequilibrium evaluation of FTO SNPs and ALKBH1 SNPs. |
Haplotypes of ALKBH1 were also explored in different age groups as ALKBH1 rs1076496 AA exhibited a connection with a lower risk of GC in the people aged ≥ 55 years. Consistent with these results, the ALKBH1 rs1048147-rs1076496-rs11159286 CAC haplotype was significantly associated with a decreased risk of GC in people aged ≥ 55 years (adjusted OR = 0.41, 95% CI: 0.21–0.79, p = 0.008) (Table 8). ALKBH1 haplotypes had no significant effect on the risk of total GC (Table 8) or GC defined by any Lauren classification (Table S3).
 |
Table 8 The Associations of ALKBH1 Haplotypes with the Risk of Total GC and GC of Different Age Group |
Cooperative Effect of SNPs Within FTO Haplotypes
A comparison of the FTO haplotypes CTAT and CTGA, which had opposite effects on the risk of GC, revealed that the variants at rs1421091 and rs1421092 were both CT, while the variants at rs2287142 and rs9939609 were different, suggesting that the variant combination at rs2287142 and rs9939609 was the crucial factor. In the subsequent analysis, we compared the risk of GC conferred by rs2287142 and rs9939609 genotype combinations with the remaining genotypes pooled as the reference and found that people simultaneously carrying rs2287142 AG/AA and rs9939609 TT had a markedly decreased risk of GC (adjusted OR = 0.71, 95% CI: 0.54–0.92, p = 0.011) (Table 9). The analysis of SNP combinations showed evidence of cooperative effects between the rs2287142 and rs9939609 sites on the FTO gene.
 |
Table 9 The Cooperation Effect of FTO rs2287142 and rs9939609 on GC Risk |
Two other FTO haplotypes, ACAT and CCAT, that also contained the AT variant of rs2287142-rs9939609, were not closely related with the risk of GC (Table 7). Unlike the GC-associated haplotype CTAT, the alleles at rs1421092 for these two haplotypes were C but not T. It was further observed that the AG/AA-TT combination of rs2287142 and rs9939609 was significantly associated with a decreased risk of GC only in the rs1421092 TC/TT group (adjusted OR = 0.70, 95% CI: 0.49–0.99, p = 0.047) but not in the rs1421092 CC group (adjusted OR = 0.77, 95% CI: 0.30–2.00, p = 0.592). These results led us to distinguishing three loci on FTO that might share biological mechanisms in GC.
Discussion
In this study, we genotyped seven potentially functional SNPs of FTO and ALKBH1 using the DNA samples collected from 419 patients with GC and 569 healthy controls. According to our results, the FTO rs2287142 AG and AG/AA variants were stably associated with a reduced risk of GC, regardless of sex and age. This SNP was predicted to be an exonic splicing enhancer/silencer, a type of splicing regulatory cis-element that can recruit trans-acting factors and determine splicing sites, thus possibly generating functionally different isoforms.26 Therefore, we speculated that FTO rs2287142 might affect susceptibility to GC by modulating FTO splicing, which might produce transcripts that differ in function or quantity.
SNPs, together with certain features of a particular population, are known to affect the risk of cancer. For instance, rs298982 GA/AA of METTL14 encoding an m6A methyltransferase exerts a protective effect against acute lymphoblastic leukemia only in children aged < 10 years and in males.27 Similarly, we found that the GA and GA/GG genotypes of ALKBH1 rs1076496 were closely associated with a higher risk of GC in people aged ≥ 55 years, but showed a decreasing tendency of risk of GC in people aged < 55 years. The rs9939609 AA showed a significant p value for the Breslow–Day test, possibly due to the small sample size of this group (Table 5), which was confirmed by subsequent analysis that revealed a negative result. In addition to sex and age, the Lauren classification is another common stratification factor that has been widely accepted as an independent prognostic indicator of GC.28 The FTO CTGA haplotype was linked with an increased risk of intestinal GC, but not with that of mixed or diffuse GC, suggesting that this haplotype might only exert a genetic effect on certain histological subtypes of GC.
Haplotypes tend to have enhanced power of predicting disease-related genes compared to that of single SNPs.23 CTAT carriers of the FTO haplotype rs1421091-rs1421092-rs2287142-rs9939609 had an OR showing a significantly lower susceptibility to GC (adjusted OR = 0.62), which was lower than the OR of carriers with the single SNP rs2287142 (adjusted OR = 0.73). The ALKBH1 CAC haplotype was associated with a lower OR for the risk of GC (adjusted OR = 0.41) than that of the single rs1076496 variant AA (adjusted OR = 0.56) in people aged ≥ 55 years. Within the FTO haplotype, rs1421092 TC/TT presentation was the prerequisite for the AG/AA-TT combination of rs2287142 and rs9939609 to influence the risk of GC. This emphasized the essential role of SNP-SNP interaction analysis, without which the cooperative effect of rs1421092, rs2287142 and rs9939609 would have not been detected.
FTO, also known as ALKBH9, belongs to the non-heme Fe II/α-ketoglutarate-dependent dioxygenase AlkB family that also contains ALKBH1–ALKBH8.11 FTO and ALKBH1 have been established as important regulators of malignant phenotype and the therapeutic response of cancer cells.29 Decreased FTO mRNA levels have been demonstrated to be associated with a poor prognosis in renal cell carcinoma,30 and the silencing of FTO is considered to be associated with selective reduction of the in vitro and in vivo survival of von Hippel–Lindau-deficient renal carcinoma cells.31 The cytotoxicity induced by cisplatin in bladder cancer cells is known to be reverted by co-treatment with the FTO selective inhibitor MA2, an ethyl ester derivative of meclofenamic acid.32 FTO is also known to be related to tumor immune infiltration observed in various cancers.33 With immunotherapy attracting extensive attention for treating multiple cancers, including advanced GC,34 genetic variants may potentially serve as novel biomarkers for predicting immunological response. ALKBH1 has been reported to be up-regulated in lung cancer. Silencing and overexpression of ALKBH1 could, respectively, suppress and promote the invasion and migration of lung cancer cells.35 As the change in expression of FTO or ALKBH1 is relevant to cancer development, it could be speculated that SNPs localized at the key regulatory sites of FTO and ALKBH1 might make a difference in cancer development, possibly by influencing the expression levels of the host genes.
FTO polymorphisms are known to affect risk and prognosis of cancers. FTO rs16953002 and rs12596638 have been reported to markedly influence melanoma susceptibility.36 FTO rs7202116 has a statistically significant association with a shorter overall survival of patients with hepatocellular carcinoma treated with transhepatic arterial chemotherapy and embolization.37 The ALKBH1 SNP has not be extensively studied. The SNP rs3850370, which is 360 kb downstream of ALKBH1, has been reported to modulate the survival of non-small cell lung cancer across Chinese and Caucasian populations.22 Apart from FTO rs9939609, all the other SNPs researched in this study had not been previously explored in terms of their association with any disease. FTO rs9939609 variant A is a protective factor for lung cancer.37 Our data showed the FTO haplotype rs1421091-rs1421092-rs2287142-rs9939609 CTGA to be linked to an increased susceptibility to intestinal GC, illustrating that the same allele of a SNP can play different roles in different cancer types.
This study, however, has some limitations to be acknowledged. The sample size could be increased to obtain more reliable results. Additionally, we could not analyze the relationship between the SNP genotypes and the expression levels of FTO and ALKBH1 due to the unavailability of sufficient GC tissues. Moreover, the possibility of SNPs affecting the alternative splicing sites remained unexplored in this study. These limitations and the results warrant deeper research to help further understand the mechanisms behind the association of SNPs with GC in the future.
Conclusion
In summary, FTO rs2287142, individually or within a haplotype, notably decreased the risk of GC, whereas ALKBH1 rs1076496 was closely associated with an increased risk of GC in the people aged ≥ 55 years. Our data shows that FTO and ALKBH1 SNPs may have predictive value in evaluating susceptibility to GC with differing age or Lauren classification. Large-scale multicenter studies and functional experiments are needed in future to further validate these results.
Abbreviations
ALKBH1, alkB homolog 1; CI, confidence interval; ESE/ESS, exonic splicing enhancer/silencer; FTO, fat mass and obesity-associated protein; GC, gastric cancer; LD, linkage disequilibrium; m6A, N6-methyladenosine; OR, odds ratio; SNP, single nucleotide polymorphism; TFBS, transcriptional factor binding site.
Data Sharing Statement
The datasets used or analyzed in the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
This study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center (Guangzhou, China) (approval NO. G2022-002-01). Written informed consent was provided by all participants at their first visit. All experiments were performed in accordance with the guidelines stated in the Declaration of Helsinki.
Consent for Publication
The purpose of this study was well informed to the all participants and written informed consent was obtained from the all participants for publication of this report.
Acknowledgments
We are grateful to the individuals for their participation in this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was funded by the National Natural Science Foundation of China (grant number 82002561); and the Guangdong Basic and Applied Basic Research Foundation (grant number 2020A1515010098).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020. doi:10.3322/caac.21590
2. Libânio D, Rodrigues JR, Bento MJ, et al. Gastric cancer incidence and mortality trends 2007–2016 in three European countries. Endoscopy. 2021. doi:10.1055/a-1673-1118
3. Shah SC, Piazuelo MB, Kuipers EJ, Li D. AGA clinical practice update on the diagnosis and management of atrophic gastritis: expert review. Gastroenterology. 2021;161(4):1325–1332.e7. doi:10.1053/j.gastro.2021.06.078
4. Waddingham W, Nieuwenburg SAV, Carlson S, et al. Recent advances in the detection and management of early gastric cancer and its precursors. Frontline Gastroenterol. 2021;12(4):322–331. doi:10.1136/flgastro-2018-101089
5. Lam SY, Mommersteeg MC, Yu B, et al. Toll-like receptor 1 locus re-examined in a genome-wide association study update on anti-Helicobacter pylori IgG titers. Gastroenterology. 2022;162:1705–1715. doi:10.1053/j.gastro.2022.01.011
6. Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 2020;21(11):4012. doi:10.3390/ijms21114012
7. Batista PJ, Molinie B, Wang J, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2015;15(6):707–719. DOI:10.1016/j.stem.2014.09.019.m
8. Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9(2):124. doi:10.1038/s41419-017-0129-x
9. Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H. Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Dig Dis Sci. 2019;64(6):1503–1513. doi:10.1007/s10620-018-5452-2
10. Alemu E, He C, Klungland A. ALKBHs-facilitated RNA modifications and de-modifications. DNA Repair. 2016;44(0027):87–91. doi:10.1016/j.dnarep.2016.05.026
11. Wu G, Yan Y, Cai Y, et al. ALKBH1-8 and FTO: potential therapeutic targets and prognostic biomarkers in lung adenocarcinoma pathogenesis. Front Cell Dev Biol. 2021;9:1–12. doi:10.3389/fcell.2021.633927
12. Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–726. doi:10.1038/ng2048
13. Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi:10.1126/science.1141634
14. Haupt A, Thamer C, Machann J, et al. Impact of variation in the FTO gene on whole body fat distribution, ectopic fat, and weight loss. Obesity. 2008;16(8):1969–1972. doi:10.1038/oby.2008.283
15. Cauchi S, Stutzmann F, Cavalcanti-Proença C, et al. Combined effects of MC4R and FTO common genetic variants on obesity in European general populations. J Mol Med. 2009;87(5):537–546. doi:10.1007/s00109-009-0451-6
16. Wen W, Cho Y-S, Zheng W, et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet. 2012;44(3):307–311. doi:10.1038/ng.1087
17. Jiang Y, Mei H, Lin Q, et al. Interaction effects of FTO rs9939609 polymorphism and lifestyle factors on obesity indices in early adolescence. Obes Res Clin Pract. 2019;13(4):352–357. doi:10.1016/j.orcp.2019.06.004
18. da Fonseca ACP, Abreu GM, Zembrzuski VM, et al. The association of the fat mass and obesity-associated gene (FTO) rs9939609 polymorphism and the severe obesity in a Brazilian population. Diabetes Metab Syndr Obes. 2019;12:667–684. doi:10.2147/DMSO.S199542
19. Mehrdad M, Fardaei M, Fararouei M, Eftekhari MH. The association between FTO rs9939609 gene polymorphism and anthropometric indices in adults. J Physiol Anthropol. 2020;39(1):14. doi:10.1186/s40101-020-00224-y
20. Kaklamani V, Yi N, Sadim M, et al. The role of the fat mass and obesity associated gene (FTO) in breast cancer risk. BMC Med Genet. 2011;12:52. doi:10.1186/1471-2350-12-52
21. Hernández-Caballero ME, Sierra-Ramírez JA. Single nucleotide polymorphisms of the FTO gene and cancer risk: an overview. Mol Biol Rep. 2015;42(3):699–704. doi:10.1007/s11033-014-3817-y
22. Hu L, Wu C, Zhao X, et al. Genome-wide association study of prognosis in advanced non-small cell lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2012;18(19):5507–5514. doi:10.1158/1078-0432.CCR-12-1202
23. Chen LZ, He CY, Su X, et al. SPP1 rs4754 and its epistatic interactions with SPARC polymorphisms in gastric cancer susceptibility. Gene. 2018;640:43–50. doi:10.1016/j.gene.2017.09.053
24. Li Y, He HC, Zhou DL, et al. Associations between lncRNA-related polymorphisms and hepatocellular carcinoma risk: a two-stage case–control study. J Gastroenterol Hepatol. 2021;36(1):233–239. doi:10.1111/jgh.15118
25. Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24(9):1291–1306. doi:10.1002/sim.2010
26. Wang F, Fu X, Chen P, et al. SPSB1-mediated HnRNP A1 ubiquitylation regulates alternative splicing and cell migration in EGF signaling. Cell Res. 2017;27(4):540–558. doi:10.1038/cr.2017.7
27. Luo A, Yang L, Liu X, Yang X. Genetic variants in METTL14 are associated with the risk of acute lymphoblastic leukemia in Southern Chinese children: a five-center case-control study. Cancer Manag Res. 2021;Volume 13:9189–9200. doi:10.2147/CMAR.S335925
28. Li Y, Xue XW, Luo YF, Wu HW, Chen J, Zhou WX. Clinicopathologic features of gastric adenocarcinoma based on the revised Lauren’s classification. Zhonghua bing li xue za zhi. 2018;47(7):486–491. doi:10.3760/cma.j.issn.0529-5807.2018.07.002
29. Lan N, Lu Y, Zhang Y, et al. FTO – a common genetic basis for obesity and cancer. Front Genet. 2020;11:1–12. doi:10.3389/fgene.2020.559138
30. Guimarães-Teixeira C, Barros-Silva D, Lobo J, et al. Deregulation of N6-methyladenosine RNA modification and its erasers FTO/ALKBH5 among the main renal cell tumor subtypes. J Pers Med. 2021;11(10). doi:10.3390/jpm11100996
31. Xiao Y, Thakkar KN, Zhao H, et al. The m(6)A RNA demethylase FTO is a HIF-independent synthetic lethal partner with the VHL tumor suppressor. Proc Natl Acad Sci U S A. 2020;117(35):21441–21449. doi:10.1073/pnas.2000516117
32. Wen L, Pan X, Yu Y, Yang B. Down-regulation of FTO promotes proliferation and migration, and protects bladder cancer cells from cisplatin-induced cytotoxicity. BMC Urol. 2020;20(1):39. doi:10.1186/s12894-020-00612-7
33. Zhao C, Liu Y, Ju S, Wang X. Pan-cancer analysis of the n6-methyladenosine eraser FTO as a potential prognostic and immunological biomarker. Int J Gen Med. 2021;14:7411–7422. doi:10.2147/IJGM.S331752
34. Kodach LL, Peppelenbosch MP. Targeting the myeloid-derived suppressor cell compartment for inducing responsiveness to immune checkpoint blockade is best limited to specific subtypes of gastric cancers. Gastroenterology. 2021;161(2):727. doi:10.1053/j.gastro.2021.03.047
35. Li H, Zhang Y, Guo Y, et al. ALKBH1 promotes lung cancer by regulating m6A RNA demethylation. Biochem Pharmacol. 2021;189:114284. doi:10.1016/j.bcp.2020.114284
36. Iles MM, Law MH, Stacey SN, et al. A variant in FTO shows association with melanoma risk not due to BMI. Nat Genet. 2013;45(4):428–432, 432e1. doi:10.1038/ng.2571
37. Liu J, Wang D, Zhou J, et al. N6-methyladenosine reader YTHDC2 and eraser FTO may determine hepatocellular carcinoma prognoses after transarterial chemoembolization. Arch Toxicol. 2021;95(5):1621–1629. doi:10.1007/s00204-021-03021-3
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
The Effects of Resistin Gene Polymorphism on Pain Thresholds and Postoperative Sufentanil Consumption in Gastric Cancer Patients
Li J, Li S, Yu L, Wei J, Li S, Tan H
Journal of Pain Research 2022, 15:1995-2004
Published Date: 17 July 2022
