Back to Journals » International Journal of General Medicine » Volume 15
Expression, Clinical Significance, Immune Infiltration, and Regulation Network of miR-3940-5p in Lung Adenocarcinoma Based on Bioinformatic Analysis and Experimental Validation
Authors Lin Z , Huang W, Xie Z, Yi Y, Li Z
Received 22 May 2022
Accepted for publication 20 July 2022
Published 6 August 2022 Volume 2022:15 Pages 6451—6464
DOI https://doi.org/10.2147/IJGM.S375761
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Scott Fraser
Zhichao Lin,* Wenhai Huang,* Zehua Xie,* Yongsheng Yi, Zumei Li
Department of Thoracic Surgery, Jiangmen Central Hospital, Affiliated Jiangmen Hospital of Sun Yat-sen University, Jiangmen, 529030, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhichao Lin, Department of Thoracic Surgery, Jiangmen Central Hospital, Affiliated Jiangmen Hospital of Sun Yat-sen University, No. 23 Haibang Street, Beijie, Jiangmen, 529030, People’s Republic of China, Tel +86 750-3989432, Email [email protected]
Background: Based on bioinformatics analysis and experimental validation, we investigated the expression, clinical significance, immune infiltration, and potential signaling pathways of miR-3940-5p in lung adenocarcinoma (LUAD).
Methods: 521 LUAD tissue samples and 46 normal lung tissue samples from The Cancer Genome Atlas (TCGA) database. We evaluated the relationship between clinical features and miR-3940-5p expression using Kruskal–Wallis, Wilcoxon sign-rank, and logistic regression, explored the relationship between miR-3940-5p expression and the prognosis of LUAD patients using Kaplan–Meier survival curve analysis. Several databases were used to identify miRNA targets. MiR-3940-5p target genes were analyzed based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. The significant role of miR-3940-5p in function was evaluated using immune infiltration analysis. LUAD cell lines were tested for miR-3940-5p expression using QRT-PCR.
Results: There was a significant association between high miR-3940-5p expression in LUAD and T stage (P=0.005), pathologic stage (P=0.047), race (White vs Asian & Black or African American) (P=0.041), residual tumor (P=0.043), and anatomic neoplasm subdivision2 (P=0.030). MiR-3940-5p expression predicted poor overall survival (HR: 1.35; 95% CI: 1.01– 1.81; P=0.045), disease-specific survival (HR: 1.53; 95% CI: 1.05– 2.23; P=0.026), and progression-free survival (HR: 1.35; 95% CI: 1.03– 1.77; P=0.032). BAP1, BBS1, CCR2, KCNE3, PEBP1, and RABL2A were all associated with poor OS in LUAD patients with low miR-3940-5p expression levels. According to GO and KEGG analyses, miR-3940-5p may play a role in LUAD development by regulating pathways such as measles, PI3K-Akt signaling pathway, and p53 signaling pathway. There was a correlation between the expression level of miR-3940-5p and immune infiltration. LUAD cell lines showed significantly higher levels of miR-3940-5p than Beas-2B cells.
Conclusion: A high expression of miR-3940-5p is significantly associated with a poor prognosis in patients with LUAD, suggesting that it could be used as a prognostic biomarker.
Keywords: microRNA-3940-5p, lung adenocarcinoma, clinical significance, immune infiltration, regulation network
Introduction
Worldwide, lung cancer is a serious threat to human health. Lung cancer is a highly heterogeneous disease, accounting for nearly a quarter of cancer-related deaths.1 Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer cases and is subdivided into adenocarcinoma of the lung and squamous lung cancer.2 The incidence of lung adenocarcinoma (LUAD) is increasing rapidly, and it is the most aggressive type of lung cancer.3 Despite recent advances in diagnosis and treatment, the long-term clinical prognosis and overall survival of patients with LUAD remains unsatisfactory.4 To guide the clinical diagnosis of LUAD, it is urgent to find validated tumor markers.
MiRNAs are ubiquitous small non coding RNAs, ranging from 19–24 nucleotides in length.5 There is an association between aberrant miRNA expression and the diagnosis and prognosis of cancer.6 MiRNAs regulate the growth and proliferation of immune and stromal cells in the tumor microenvironment (TME).7 Serum hsa-miRNA-3940-5p is an independent prognostic factor in colorectal cancer (CRC).8 The miRNA index (miR.index), constructed from nine miRNAs (hsa-miR-3940 and eight other miRNAs), is an independent predictor of survival in LUAD patients.9 There is no clear understanding of miR-3940-5p’s role in LUAD.
In this study, we explored the expression, clinical significance, immune infiltration, and regulation network of miR-3940-5p in lung adenocarcinoma based on bioinformatic analysis and experimental validation. It provides patients with LUAD with a reliable prognostic and diagnostic marker.
Materials and Methods
Patients
521 LUAD tissue samples and 46 normal lung tissue samples from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The clinical information included T stage, N stage, M stage, pathologic stage, primary therapy outcome (CR vs PD&SD&PR), gender, race, age, residual tumor, anatomic neoplasm subdivision (right vs left), anatomic neoplasm subdivision2 (peripheral lung vs central lung), number_pack_years_smoked (≥40 vs <40), and smoker.
Clinical Information
The analysis was performed according to the reference.10–12 Molecule: hsa-miR-3940-5p [MIMAT0019229]. In The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/), LUAD (lung adenocarcinoma) project, miRNAseq data and clinical data have been collected. The RPM (Reads per Million mapped reads) format of miRNAseq data is grouped according to molecular expression.
Differential Expression of miR-3940-5p
Unpaired Samples
In accordance with the reference, we conducted the analysis.10–12 Molecule: hsa-miR-3940-5p. In the TCGA LUAD project, miRNAseq data were collected from the level 3 BCGSC miRNA profiling. For inter-sample expression comparison, log2 transformation of miRNAseq data in RPM format is used. Data filtering keeps paired samples.
Paired Samples
In accordance with the reference, we conducted the analysis.13,14 Molecule: hsa-miR-3940-5p. In the TCGA LUAD project, miRNAseq data were collected from the level 3 BCGSC miRNA profiling. For inter-sample expression comparison, log2 transformation of miRNAseq data in RPM format is used. There is no data filtering.
ROC Curves
In accordance with the reference, we conducted the analysis.10–12 Molecule: hsa-miR-3940-5p. Clinical variables included Normal vs Tumor. In the TCGA LUAD project, miRNAseq data were collected from the level 3 BCGSC miRNA profiling.
The Correlation Between miR-3940-5p Expression and Clinical Information
In accordance with the reference, we conducted the analysis.10–12 Molecule: hsa-miR-3940-5p. Subgroup: median. Clinical variables: T stage, pathological stage, residual tumor, and anatomic neoplasm subdivision2. In the TCGA LUAD project, miRNAseq data were collected from the level 3 BCGSC miRNA profiling.
Patient Prognosis Based on miR-3940-5p Expression in LUAD
In accordance with the reference, we conducted the analysis.10–12 Molecule: hsa-miR-3940-5p. Prognosis type included overall survival (OS), disease specific survival (DSS), and progression-free survival (PFS). The time from randomization to progression or death is called the PFS. In the TCGA LUAD project, miRNAseq data were collected from the level 3 BCGSC miRNA profiling.
Prediction of miR-3940-5p Targets
TargetScan, miRanda, TarBase, miRTarBase, miR2Disease, miRecords, and miRWalk were used to predict the targets of miR-3940-5p.15–18 The website (http://bioinfo.life.hust.edu.cn/miR_path/download.html) was used to download the LUAD mRNA average expression data. LUAD downregulated genes and common genes predicted by databases were analyzed using Venn diagrams.
Expression of miR-3940-5p Target Genes Correlating with Prognosis
In accordance with the reference, we conducted the analysis.19 Molecules: BAP1[ENSG00000163930], BBS1[ENSG00000174483], CCR2[ENSG00000121807], KCNE3[ENSG00000175538], PEBP1[ENSG00000089220], and RABL2A[ENSG00000144134]. The prognosis type was OS. In the TCGA LUAD project, RNAseq data and clinical data are provided in level 3 HTSeq-FPKM format.
GO and KEGG Analysis of miR-3940-5p
GO and KEGG analyses were conducted on miR-3940-5p targets using the DAVID database,20,21 focusing with BP (biological process), MF (molecular function), CC (cellular component), and pathway analysis.
Immune Infiltration Analysis by ssGSEA
In accordance with the reference, we conducted the analysis.14,19,22,23 Target molecule: hsa-miR-3940-5p.
QRT-PCR
Cells originally purchased from the Chinese Academy of Sciences included BEAS-2B, A549, and PC9. At 37 °C, 5% CO2 was introduced to Beas-2B, A549, and PC9 cells in high-glucose DMEM medium (SH30022.01B; HyClone, Beijing, China) containing 10% fetal bovine serum (11,011–8611; Sijiqing Biotechnology, Hangzhou, China) and 1% penicillin-streptomycin (3810–74-0; Sigma, USA). QRT-PCR was used to identify miR-3940-5p levels in BEAS-2B, A549, and PC9 cell lines. In accordance with the reference, the specific steps were performed.24 Following are the primer sequences used: U6, forward: CTCGCTTCGGCAGCACA, reverse: AACGCTTCACGAATTTGCGT; miR-3940-5p, forward: 5′-TAAAAGTGGGTTGGGGCGG-3′, reverse: 5′-GTGCAGGGTCCGAGGT-3′.
Statistical Analysis
According to the reference, a statistical analysis was conducted.19
Results
Clinical Characteristics
This study included 521 patients, as shown in Table 1. The T stage was made up of 173 T1 (33.4%), 277 T2 (53.5%), 49 T3 (9.5%), and 19 T4 (3.7%). The N stage consisted of 336 N0 (66.3%), 95 N1 (18.7%), 74 N2 (14.6%), and 2 N3 (0.4%). The M stage consisted of 351 M0 (93.9%), and 23 M1 (6.1%). The pathological stage consisted of 283 Stage I (55.1%), 123 Stage II (23.9%), 84 Stage III (16.3%), and 24 Stage IV (4.7%). The primary therapy outcome consisted of 68 PD (15.7%), 35 SD (8.1%), 6 PR (1.4%), and 324 CR (74.8%). The gender consisted of 278 females (53.4%) and 243 males (46.6%). The race consisted of 8 Asian (1.8%), 53 Black or African American (11.6%), and 394 White (86.6%). The age consisted of 240 patients (≤65, 47.8%), and 262 patients (>65, 52.2%). The residual tumor consisted of 342 R0 (95.3%), 13 R1 (3.6%), and 4 R2 (1.1%). The anatomic neoplasm subdivision consisted of 202 left (39.9%) and 304 right (60.1%). The anatomic neoplasm subdivision2 consisted of 62 central lungs (33.3%) and 124 peripheral lungs (66.7%). The number pack years smoked consisted of 176 patients (<40, 49.7%) and 178 patients (≥40, 50.3%). The smokers consisted of 76 no (15%) and 431 yes (85%). There was a range of 59 to 72 years of age, with a median age of 66.5 years.
 |
Table 1 Relationships Between miR-3940-5p Expression and Clinical Characteristics |
Clinical Characteristics of LUAD are Negatively Correlated with MiR-3940-5p Expression
As shown in Figure 1A, LUAD tissues (0.631 ± 0.028, n=521) expressed significantly higher miR-3940-5p levels than normal tissues (0.393 ± 0.128, n=46) (P<0.001). As shown in Figure 1B, LUAD tissues (0.586±0.085, n=46) expressed significantly higher miR-3940-5p levels than matched normal tissues (0.393 ± 0.128, n=46) (P=0.02). As shown in Figure 1C, the AUC of miR-3940-5p was 0.709. As shown in Table 1, miR-3940-5p expression was associated with T stage (P< 0.001), primary therapy outcome (P=0.016), anatomic neoplasm subdivision2 (P=0.043), and median age (P=0.036; low miR-3940-5p expression 67 (60, 74) vs high miR-3940-5p expression 65 (58, 72)). As shown in Figure 2 and Table 2 (dichotomous logistic model), miR-3940-5p expression was significantly related to T stage (P=0.005), pathologic stage (P=0.047), race (P=0.041), residual tumor (P=0.043), and anatomic neoplasm subdivision2 (P=0.030). Overall, the findings indicated a link between high miR-3940-5p expression and a poor prognosis for LUAD patients.
 |
Table 2 MiR-3940-5p Expression Associated with Clinical Characteristics (Logistic Regression) |
 |
Figure 2 Association with miR-3940-5p and clinical characteristics in LUAD. (A) T Stage, (B) Pathologic stage, (C) Residual tumor, (D) Anatomic neoplasm subdivision 2. **P<0.01; ***P<0.001. |
Survival of LUAD Patients is Influenced by miR-3940-5p
As shown in Figure 3, miR-3940-5p expression was negatively associated with poor OS (HR: 1.35; 95% CI: 1.01–1.81; P=0.045), DSS (HR: 1.53; 95% CI: 1.05–2.23; P=0.026) and PFS (HR: 1.35; 95% CI: 1.03–1.77; P=0.032) in patients with LUAD. According to the above data, miR-3940-5P levels are associated with poor OS and high levels are associated with poor prognosis.
 |
Figure 3 Relationship between miR-3940-5p expression and prognosis. (A) Overall survival, OS, (B) Disease specific survival, DSS, (C) Progress free interval, PFS. |
Relationship Between miR-3940-5p Target Genes and Survival of LUAD Patients
As shown in Figure 4, there were 414 target genes of miR-3940-5p, 3459 LUAD down regulated genes, and 74 common genes. The low expression of BAP1 predicted was associated with poor OS (HR: 0.72; 95% CI: 0.54–2.23; P=0.96), the low expression of BBS1 predicted was associated with poor OS (HR: 0.72; 95% CI: 0.54–0.96; P=0.027), the low expression of CCR2 predicted was associated with poor OS (HR: 0.56; 95% CI: 0.42–0.76; P<0.001), the low expression of KCNE3 predicted was associated with poor OS (HR: 0.72; 95% CI: 0.54–0.96; P=0.024), the low expression of PEBP1 predicted was associated with poor OS (HR: 0.65; 95% CI: 0.49–0.87; P=0.004), and the low expression of RABL2A predicted was associated with poor OS (HR: 0.74; 95% CI: 0.55–0.99; P=0.04) (Figure 5).
 |
Figure 4 Venn diagram of common genes between target genes of miR-3940-5p and LUAD down regulated genes. |
 |
Figure 5 Relationship between expression of miR-3940-5p target genes and prognosis. (A) BAP1, (B) BBS1, (C) CCR2, (D) KCNE3, (E) PEBP1, (F) RABL2A. Overall survival, OS. |
GO and KEGG Analysis of miR-3940-5p Target Genes in LUAD Patients
The common genes are involved in biological processes, including actomyosin structure organization, brown fat cell differentiation, magnesium ion homeostasis, response to stimulus, mammary gland epithelial cell proliferation, positive regulation of protein import into nucleus, translocation, hemidesmosome assembly, and lateral ventricle development; cellular components, including ruffle, cell junction, dendrite, growth cone, cytosol, actin filament, cell-cell adherens junction, and synaptic vesicle; molecular functions, including phosphatase activity, protein binding, actin filament binding, protein kinase binding, phosphatidylinositol transporter activity, and actin binding (Figure 6). The common genes have been found to be associated with measles, PI3K-Akt signaling pathway, and p53 pathways (Figure 7).
 |
Figure 6 GO analysis of miR-3940-5p target genes. |
 |
Figure 7 KEGG analysis of miR-3940-5p target genes. |
The Correlation Between miR-3940-5p Expression and Immune Infiltration
As shown in Figures 8 and 9, and Table 3, analysis of the relationship between miR-3940-5p and immune infiltration based on ssGESA with Spearman r showed that there was a positive correlation between miR-3940-5p expression and the expression of NK CD56dim cells (P<0.001) and Th2 cells (P<0.001), and a negative correlation with the expression of DC (P=0.003), Eosinophils (P<0.001), iDC (P<0.001), Macrophages (P=0.021), Mast cells (P<0.001), NK CD56dim cells (P<0.001), NK cells (P<0.001), pDC (P=0.002), Tcm (P=0.043), Tem (P=0.045), TFH (P<0.001), and Th2 cells (P<0.001).
 |
Table 3 MiR-3940-5p Expression Associated with Immune Cells (Spearman Method) |
 |
Figure 8 The expression level of miR-3940-5p was related to the immune infiltration in the tumor microenvironment (group comparison chart). Ns, P>0.05; *P<0.05, **P<0.01; ***P<0.001. |
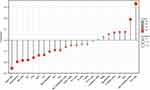 |
Figure 9 The expression level of miR-3940-5p was related to the immune infiltration in the tumor microenvironment (lollipop chart). |
Validating the Expression of miR-3940-5p in Cell Lines
As shown in Figure 10, A549 expressed significantly more miR-3940-5p than Beas-2B (2.608±0.253 vs 0.874±0.233, P<0.001), while PC9 expressed significantly more miR-3940-5p than Beas-2B (1.823±0.055 vs 0.874±0.233, P<0.001). LUAD cell lines showed significantly higher levels of miR-3940-5p in contrast to normal Beas-2B cells from the human bronchial epithelium.
 |
Figure 10 Expression of miR-3940-5p in A549, PC9, and Beas-2B. *P<0.05. |
Discussion
MiRNAs have potentially important implications for regulating the development of malignancies, and their expression profile correlates well with specific clinical cancer features.25,26 Patients with LUAD with low miR-99a expression have a poor prognosis.27 LUAD can be diagnosed using miR-210-3p as a biomarker.28 There is an essential role for miR-375 in the progression of LUAD.29 NSCLC is influenced by miR-139-5p, which acts as a tumor suppressor.30 This emphasizes the importance of developing novel biomarkers for LUAD.
Levels of miR-3940-5p in NSCLC tumor tissues were lower than those in matched tumor-adjacent tissues.31 The results of the study in 90 NSCLC patients with tumor, tumor-adjacent, and normal lung parenchymal tissues, and 17 embryonic lung cDNAs showed a significant downregulation of miR-3940-5p expression in tumor and embryonic lung tissues.32 However, miR-3940-5p was highly expressed in LUAD in this study. The difference in findings may be related to the size of the sample. There was a significant association between miR-3940-5p expression and T stage (P=0.005), pathologic stage (P=0.047), race (P=0.041), residual tumor (P=0.043), and anatomic neoplasm subdivision2 (P=0.030). In patients with advanced T stage or clinical stage III–IV, LUAD expressed more miR-3940-5p than adjacent normal lung tissue, suggesting that miR-3940-5p may be involved in tumorigenesis and promote proliferation. There may be a high expression of miR-3940-5p in tumors with positive R1 and R2 resection margins. MiR-3940-5p expression was associated with OS (HR: 1.35; 95% CI: 1.01–1.81; P=0.045), DSS (HR: 1.53; 95% CI: 1.05–2.23; P=0.026), and PFI (HR: 1.35; 95% CI: 1.03–1.77; P=0.032). Poor OS was associated with low expression of miR-3940-5p target genes, including BAP1, BBS1, CCR2, KCNE3, PEBP1, and RABL2A. BAP1 promotes breast cancer cell proliferation and metastasis through deubiquitination of KLF5.33 High BBS1 gene expression facilitates survival of malignant pleural mesothelioma.34 Crosstalk between macrophages and cancer cells via CCR2 and CX3CR1 is the underlying mechanism driving lung cancer.35 RIPK4 overexpression promotes pancreatic cancer cell migration and invasion through PEBP1 degradation-induced activation of the RAF1/MEK/ERK pathway.36 The RABL2A-CCDC34 axis plays an important role in mediating p38/MAPK and JNK/MAPK signaling, leading to acquired sorafenib resistance in hepatocellular carcinoma.37 The specific mechanisms by which miR3940-5p mediates LUAD development through the regulation of BAP1, BBS1, CCR2, KCNE3, PEBP1, and RABL2A need to be further investigated.
There are studies on the regulatory pathway of miR-3940-5p in some solid tumors. LncRNA-SNHG14/miRNA-3940-5p/NAP12 mRNA may be potential for CRC diagnosis and prognosis.8 miR-3940-5p negatively regulates CUL7 and activates NF-κB to promote gliomagenesis in human gliomas.38 miR-3940-5p targets CCND1 and USP28 to inhibit proliferation and induce apoptosis in NSCLC cells.31 In this study, miR-3940-5p target genes participate in several pathways, including measles, PI3K-Akt signaling pathway, and p53 signaling pathway.
It is important to understand the immune infiltration in LUAD to develop immunotherapy for it. The present study found miR-3940-5p expression to be associated with infiltration of NK CD56dim cells, Th2 cells, DC, Eosinophils, iDC, Macrophages, Mast cells, NK CD56dim cells, NK cells, pDC, Tcm, Tem, TFH, and Th2 cells. In other words, miR-3940-5p promotes the function of NK CD56dim cells and Th2 cells, and inhibits the function of DC, Eosinophils, iDC, Macrophages, Mast cells, NK CD56dim cells, NK cells, pDC, Tcm, Tem, TFH, and Th2 cells.
Its limitations lie in its reliance on database data and bioinformatics analysis as well as some experimental validation. More biological experiments will be carried out to further explore the function of miR-3940-5p in LUAD. Circulating tumor cells (CTCs) are an independent prognostic indicator of PFS and OS.39 In future studies, the role of miR-3940-5p can be compared with the role of circulating tumor cells and other prognostic factors in LUAD.
Conclusion
MiR-3940-5p showed high expression in LUAD tissues and cell lines. High expression of MiR-3940-5p was associated with poor prognosis in LUAD patients. Poor OS in LUAD was associated with low expression of miR-3940-5p target genes, including BAP1, BBS1, CCR2, KCNE3, PEBP1, and RABL2A. Several pathways may play a role in the development of LUAD, including measles, PI3K-Akt signaling pathway, and p53 signaling pathway. Immune infiltration was associated with miR-3940-5p expression. MiR-3940-5p may provide insight into LUAD patient prognosis.
Data Sharing Statement
The data used to support the findings of this study are included within the article.
Ethics Approval
The Medical Ethics Committee of Jiangmen Central Hospital reviewed this study, and the ethics requirement was waived (Approval No: [2021]01).
Acknowledgments
The authors thank TCGA for providing the data. For technology support, the authors would like to thank MyGene Diagnostics Co., Ltd., especially Dr. Dongbing Li who did an excellent job.
Funding
This work was supported by grants from the Medical Scientific Research Foundation of Guangdong Province of China (A2016419 and B2018239) and The Bethune Charitable Foundation (HZB-20181119-29).
Disclosure
The authors declare that they have no conflicts of interest relevant to this study.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
2. Zhang S, Liu J, Yuan T, Liu H, Wan C, Le Y. Circular RNA 0001313 knockdown suppresses non-small cell lung cancer cell proliferation and invasion via the microRNA-452/HMGB3/ERK/MAPK axis. Int J Gen Med. 2020;13:1495–1507. doi:10.2147/ijgm.s272996
3. Matsuda T, Machii R. Morphological distribution of lung cancer from cancer incidence in five continents Vol. X. Jpn J Clin Oncol. 2015;45(4):404. doi:10.1093/jjco/hyv041
4. Wang J, Jia Y, Zhao S, et al. BIN1 reverses PD-L1-mediated immune escape by inactivating the c-MYC and EGFR/MAPK signaling pathways in non-small cell lung cancer. Oncogene. 2017;36(45):6235–6243. doi:10.1038/onc.2017.217
5. Zhang T, Li W, Gu M, et al. Clinical significance of miR-183-3p and miR-182-5p in NSCLC and their correlation. Cancer Manag Res. 2021;13:3539–3550. doi:10.2147/CMAR.S305179
6. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi:10.1038/nrc3932
7. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235–246. doi:10.1158/2159-8290.cd-15-0893
8. Matboli M, Shafei AE, Ali MA, et al. Role of extracellular LncRNA-SNHG14/miRNA-3940-5p/NAP12 mRNA in colorectal cancer. Arch Physiol Biochem. 2019:1–7. doi:10.1080/13813455.2019.1650070
9. Xin G, Cao X, Zhao W, et al. MicroRNA expression profile and TNM staging system predict survival in patients with lung adenocarcinoma. Math Biosci Eng. 2020;17(6):8074–8083. doi:10.3934/mbe.2020409
10. Chen B, Jin X, Wang H, Zhou Q, Li G, Lu X. Expression, clinical significance, and prospective pathway signaling of miR-501-3p in ovarian cancer based on database and informatics analysis. Int J Gen Med. 2021;14:5193–5201. doi:10.2147/ijgm.s327673
11. Lao Y, Li T, Xie X, Chen K, Li M, Huang L. MiR-195-3p is a novel prognostic biomarker associated with immune infiltrates of lung adenocarcinoma. Int J Gen Med. 2022;15:191–203. doi:10.2147/ijgm.s350340
12. Lu X, Jing L, Liu S, Wang H, Chen B. miR-149-3p is a potential prognosis biomarker and correlated with immune infiltrates in uterine corpus endometrial carcinoma. Int J Endocrinol. 2022;2022:5006123. doi:10.1155/2022/5006123
13. Chen T, Zhu C, Wang X, Pan Y. LncRNA ELF3-AS1 is a prognostic biomarker and correlated with immune infiltrates in hepatocellular carcinoma. Can J Gastroenterol Hepatol. 2021;2021:8323487. doi:10.1155/2021/8323487
14. Lin Z, Huang W, Yi Y, et al. LncRNA ADAMTS9-AS2 is a prognostic biomarker and correlated with immune infiltrates in lung adenocarcinoma. Int J Gen Med. 2021;14:8541–8555. doi:10.2147/ijgm.s340683
15. Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi:10.7554/eLife.05005
16. Chiang HR, Schoenfeld LW, Ruby JG, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24(10):992–1009. doi:10.1101/gad.1884710
17. Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(1):18. doi:10.1186/s13059-019-1629-z
18. Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi:10.1038/ng1536
19. Lu X, Li G, Liu S, Wang H, Zhang Z, Chen B. Bioinformatics analysis of KIF1A expression and gene regulation network in ovarian carcinoma. Int J Gen Med. 2021;14:3707–3717. doi:10.2147/ijgm.s323591
20. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
21. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi:10.1093/nar/gkn923
22. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
23. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi:10.1016/j.immuni.2013.10.003
24. Lu X, Li G, Liu S, Wang H, Chen B. MiR-585-3p suppresses tumor proliferation and migration by directly targeting CAPN9 in high grade serous ovarian cancer. J Ovarian Res. 2021;14(1):90. doi:10.1186/s13048-021-00841-w
25. Wang S, Zhao X, Wang J, et al. Upregulation of microRNA-203 is associated with advanced tumor progression and poor prognosis in epithelial ovarian cancer. Med Oncol. 2013;30(3):681. doi:10.1007/s12032-013-0681-x
26. Võsa U, Vooder T, Kolde R, et al. Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer. 2011;50(10):812–822. doi:10.1002/gcc.20902
27. Gu W, Fang S, Gao L, Tan Y, Yang Z. Clinic significance of microRNA-99a expression in human lung adenocarcinoma. J Surg Oncol. 2013;108(4):248–255. doi:10.1002/jso.23381
28. Świtlik WZ, Karbownik MS, Suwalski M, Kozak J, Szemraj J. Serum miR-210-3p as a potential noninvasive biomarker of lung adenocarcinoma: a preliminary study. Genet Test Mol Biomarkers. 2019;23(5):353–358. doi:10.1089/gtmb.2018.0275
29. Gan T-Q, Chen W-J, Qin H, et al. Clinical value and prospective pathway signaling of MicroRNA-375 in lung adenocarcinoma: a study based on the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and bioinformatics analysis. Med Sci Monit. 2017;23:2453–2464. doi:10.12659/msm.901460
30. Yong-Hao Y, Xian-Guo W, Ming X, Jin-Ping Z. Expression and clinical significance of miR-139-5p in non-small cell lung cancer. J Int Med Res. 2019;47(2):867–874. doi:10.1177/0300060518815379
31. Ren K, Li Y, Lu H, Li Z, Han X. miR-3940-5p functions as a tumor suppressor in non-small cell lung cancer cells by targeting Cyclin D1 and ubiquitin specific Peptidase-28. Transl Oncol. 2017;10(1):80–89. doi:10.1016/j.tranon.2016.11.004
32. Sun Y, Su B, Zhang P, et al. Expression of miR-150 and miR-3940-5p is reduced in non-small cell lung carcinoma and correlates with clinicopathological features. Oncol Rep. 2013;29(2):704–712. doi:10.3892/or.2012.2152
33. Qin J, Zhou Z, Chen W, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471. doi:10.1038/ncomms9471
34. Vavougios GD, Solenov EI, Hatzoglou C, et al. Computational genomic analysis of PARK7 interactome reveals high BBS1 gene expression as a prognostic factor favoring survival in malignant pleural mesothelioma. Am J Physiol Lung Cell Mol Physiol. 2015;309(7):L677–L686. doi:10.1152/ajplung.00051.2015
35. Schmall A, Al-Tamari HM, Herold S, et al. Macrophage and cancer cell cross-talk via CCR2 and CX3CR1 is a fundamental mechanism driving lung cancer. Am J Respir Crit Care Med. 2015;191(4):437–447. doi:10.1164/rccm.201406-1137OC
36. Qi ZH, Xu HX, Zhang SR, et al. RIPK4/PEBP1 axis promotes pancreatic cancer cell migration and invasion by activating RAF1/MEK/ERK signaling. Int J Oncol. 2018;52(4):1105–1116. doi:10.3892/ijo.2018.4269
37. Zhou M, Chen X, Bai H, et al. RABL2A-CCDC34 axis promotes sorafenib resistance in hepatocellular carcinoma. DNA Cell Biol. 2021;40(11):1418–1427. doi:10.1089/dna.2021.0473
38. Xu J, Zhang Z, Qian M, et al. Cullin-7 (CUL7) is overexpressed in glioma cells and promotes tumorigenesis via NF-κB activation. J Exp Clin Cancer Res. 2020;39(1):59. doi:10.1186/s13046-020-01553-7
39. Rossi E, Aieta M, Tartarone A, et al. A fully automated assay to detect the expression of pan-cytokeratins and of EML4-ALK fusion protein in circulating tumour cells (CTCs) predicts outcome of non-small cell lung cancer (NSCLC) patients. Transl Lung Cancer Res. 2021;10(1):80–92. doi:10.21037/tlcr-20-855
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

