Back to Journals » Journal of Inflammation Research » Volume 14
Exploration and Validation of a Novel Inflammatory Response-Associated Gene Signature to Predict Osteosarcoma Prognosis and Immune Infiltration
Authors Fu Y, He G, Liu Z, Wang J, Zhang Z, Bao Q, Wen J, Jin Z, Zhang W
Received 29 September 2021
Accepted for publication 1 December 2021
Published 9 December 2021 Volume 2021:14 Pages 6719—6734
DOI https://doi.org/10.2147/JIR.S340477
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Yucheng Fu, Guoyu He, Zhuochao Liu, Jun Wang, Zhusheng Zhang, Qiyuan Bao, Junxiang Wen, Zhijian Jin, Weibin Zhang
Department of Orthopedics, Shanghai Key Laboratory for Prevention and Treatment of Bone and Joint Diseases, Shanghai Institute of Traumatology and Orthopedics, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, 200025, People’s Republic of China
Correspondence: Weibin Zhang
Department of Orthopedics, Shanghai Key Laboratory for Prevention and Treatment of Bone and Joint Diseases, Shanghai Institute of Traumatology and Orthopedics, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, 197 Ruijin Er Road, Shanghai, 200025, People’s Republic of China
Tel +86-21-64370045, Ext 666983
Fax +86-21-54660217
Email [email protected]
Background: Inflammatory response took part in the progression of tumor and was regarded as the hallmark of cancer. However, the prognostic relationship between osteosarcoma and inflammatory response-associated genes (IRGs) was unclear. This research aimed to explore the correlations between osteosarcoma prognosis and IRG signature.
Methods: The inflammatory response-associated differentially expressed messenger RNAs (DEmRNAs) were screened out through Gene Expression Omnibus (GEO) and Molecular Signature Database (MSigDB) databases. Univariate and multivariate cox regression analyses were utilized to construct the IRG signature. The prognostic value of signature was investigated through Kaplan–Meier (KM) survival curve and nomogram. DEmRNAs among high and low inflammatory response-associated risks were identified and functional enrichment analyses were conducted. ESTIMATE, CIBERSORT and single-sample gene set enrichment analyses (ssGSEA) were implied to reveal the alterations in immune infiltration. All the above results were validated in Target database. The expression of IRGs was also validated in different cell lines by quantitative real-time PCR (qRT-PCR) and osteosarcoma patient samples by immunohistochemistry.
Results: The IRG signature that consisted of two genes (MYC, CLEC5A) was established. In training and validation datasets, patients with lower risk scores survived longer and the IRG signature was confirmed as the independent prognostic factor in osteosarcoma. The nomogram was constructed and the calibration curves demonstrated the reliability of this model. Functional analysis of risk score-associated DEmRNAs indicated that immune-related pathways and functions were significantly enriched. ssGSEA revealed that 14 immune cells and 11 immune functions were significantly dysregulated. The qRT-PCR results indicated IRGs were significantly differently expressed in osteosarcoma and osteoblast cell lines. The immunohistochemistry analyses of patients’ samples revealed the same result.
Conclusion: The novel osteosarcoma inflammatory response-associated prognostic signature was established and validated in this study. This model could serve as the biomarker and therapeutic target for osteosarcoma in the future.
Keywords: osteosarcoma, inflammatory response, metastasis, prognosis, immune
Introduction
Osteosarcoma was the predominant malignant primary bone tumor that mainly occurred in adolescents. It was the second leading cause of tumor-associated death in teenagers.1,2 In order to overcome the lethality of osteosarcoma, the radical treatment which included multi-drug neoadjuvant chemotherapy and aggressive surgical resection was applied in clinical. Since then, the 5-year survival rates of osteosarcoma patients were significantly improved.3 However, numerous patients still developed metastasis before or after treatment and their survival rates dramatically decreased to less than 20%.4 To improve the prognosis of these metastatic patients, novel therapeutic strategies such as immune checkpoint and molecular targeted inhibitors had been experimentally applied in clinical. Unfortunately, little progress had been achieved since the mechanism of metastasis was still elusive.
Inflammatory response, always accompanied with immune alterations in the tumor microenvironment, was involved in the development of tumor. In 1863, Virchow first found that “lymphoreticular infiltrate” was existed in the origin of tumor and this chronic inflammatory response might promote the progression of tumor.5 After that, the relationships between inflammation and cancer were widely investigated. To some extent, cancer was considered as the wound that did not heal and chronic inflammatory response would cause gene mutation and epigenetic alterations, which promoted the malignancy of cancer cells.6,7 Numerous studies also revealed that clinical blood inflammatory parameters were associated with the prognosis of cancers. In osteosarcoma, several elevated blood inflammatory-associated markers such as serum C-reactive protein (CRP) and erythrocyte sedimentation rate (ESR) were negatively related to patients’ prognosis.8,9
In addition, plenty of inflammatory response-associated genes were identified by microarray and next-generation sequencing technology in recent years.10 Several studies revealed that these genes could be combined as the signature to predict the prognosis of patients with malignant tumors and might be applied in clinical treatment.11,12 However, few studies focused on the relationship between osteosarcoma and inflammatory response.
In the present study, we identified inflammatory response-associated differentially expressed messenger RNA (DEmRNA) related to osteosarcoma metastasis in Gene Expression Omnibus (GEO) database. Univariate and multivariate cox regression analyses were conducted to screen out the prognosis-related genes and establish the prognostic signature. Next, patients were divided into low- and high-risk groups according to the median value of risk scores obtained previously. The Kaplan–Meier (KM) survival analysis was performed and the relationships between individual genes and clinical features were explored. In addition, the nomogram was established and the immune infiltrative features of signature were investigated. All the above results were validated in Therapeutically Applicable Research to Generate Effective Treatments (Target) database. The DEmRNAs in signature were also validated in different cell lines by quantitative real-time PCR (qRT-PCR) and in osteosarcoma patients’ samples by immunohistochemistry (IHC). The flow chart of this study is shown in Figure 1. We believed that our research constructed a reliable gene signature to reveal the relationship between inflammatory response and osteosarcoma patients’ prognosis. We hoped that this signature could act as the novel prognostic biomarker and would be applied in clinical practice in the future.
Materials and Methods
Screening of Datasets
The datasets of osteosarcoma patients were searched from Target (https://ocg.cancer.gov/programs/target) and GEO (http://www.ncbi.nlm.nih.gov/geo) databases. The inclusion criteria for dataset were shown as follows: (1) the patients were diagnosed as osteosarcoma by pathology; (2) the datasets should contain complete prognostic information; (3) the number of patients in datasets should be more than 50. Finally, one GEO dataset (GSE21257, platform GPL10295, Illumina human-6 v2.0 expression beadchip Illumina, Inc., San Diego, CA, United States) was screened out as the training group and another dataset in Target database was selected as the validation group. The clinical characteristics of patients in two datasets are shown in Supplementary Table 1. The inflammatory response-associated genes were obtained from the hallmark gene sets in the Molecular Signature Database (MSigDB, https://www.gsea-msigdb.org/gsea/msigdb/).
Identification of Inflammatory Response-Associated DEmRNAs
The mRNA expression file and patients’ clinical information were downloaded from each dataset. Patients in GSE21257 were separated into metastatic and non-metastatic groups and the DEmRNAs among these groups were screened out through Linear Models for Microarray Analysis (limma) package in R software. The cutoff values of screening were |fold change (FC)| >1.5 and P < 0.05. The characteristic distribution of DEmRNAs was shown by “pheatmap” package in R software. Inflammatory response-associated DEmRNAs were identified through overlapping DEmRNAs and inflammatory response-associated genes.
Construction and Validation of the Inflammatory Response-Associated Prognostic Signature
Inflammatory response-associated DEmRNAs related to osteosarcoma prognosis were identified by univariate cox regression analysis. The multivariate cox regression analysis was utilized to construct the inflammatory response-associated gene signature. The least numbers of DEmRNAs that represented the signature were screened through akaike information criterion (AIC).13 The risk score of signature was calculated by the following formulation:
Risk score = ∑i Coefficient mRNAi * Expression mRNAi
Then the patients in GSE21257 were divided into high- and low-risk groups according to the median value of risk score. The relationships between risk score and patients’ survival were conducted by KM survival curve in R software. The reliability of the signature was evaluated by time-related receiver operating characteristic (ROC) curves.
After that, the univariate and multivariate cox regression analyses were applied to evaluate the prognostic value of this gene signature. The correlations between individual mRNAs in signature and clinical features were explored through the follow-up information. The KM survival analysis of mRNAs in signature was also conducted. The above features of signature were validated in Target database and the KM survival analysis of individual mRNAs was examined in R2 database (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi) once more. P < 0.05 was considered as statistically significant.
Establishment of the Nomogram
Recently, the nomogram that contained patients’ clinical information was constructed to stratify the different risk patients in various studies.14 The nomogram of the osteosarcoma patients in the present study was established through “rms” package in R software. The predictive value of clinical factors such as gender, age and metastasis were calculated by multivariate cox regression analysis. The reliability of the nomogram was evaluated by Harrell’s concordance index (C-index). The nomogram was also established and validated in Target database.
Functional Analyses of DEmRNAs Related to Inflammatory Response-Associated Prognostic Signature
The DEmRNAs related to this gene signature were screened out through high- and low-risk groups in Target and GSE21257 datasets. The overlapped DEmRNAs among these two datasets were considered as the risk score-related DEmRNAs. The heatmaps of DEmRNAs in two datasets were drawn by the “pheatmap” package in R software. The functional analyses of these DEmRNAs were performed by “clusterProfiler” and “enrichplot” packages in R software. P- and q-values less than 0.05 were defined as statistically significant.
Assessment of Tumor Immune Infiltration Status Related to the Inflammatory Response
The infiltrative levels of immune and stromal cells in osteosarcoma samples were evaluated through immune and stromal scores by ESTIMATE software. The relationships between these scores and inflammatory response-associated risks were conducted by Spearman correlate analysis.15 Next, the compositions of 22 immune cells in osteosarcoma were explored through the CIBERSORT deconvolution algorithm.16,17 In brief, the mRNA signature matrix of 22 immune cells was downloaded from the CIBERSORT platform (https://cibersortx.stanford.edu). Then, it was matched with the mRNAs from GSE21257 and Target datasets to generate the infiltrative proportions of immune cells in osteosarcoma.
After that, the immune infiltrative differences between low- and high-risk patients were analyzed by single-sample gene set enrichment analysis (ssGSEA). The intersected differences with the same alternative trends in GSE21257 and Target datasets were regarded as the inflammatory response-associated immune changes in osteosarcoma. The immune infiltrative features between lowly and highly expressed genes in signature were also investigated.
Cell Culture and Reagents
Human osteosarcoma cell lines 143B, MNNG/HOS, SAOS2, U2OS, MG63 and human osteoblast cell line hFOB1.19 were obtained from ATCC (Manassas, VA, USA). Osteosarcoma cell line WELL5 was a kind gift from Dr. Wu Zhang of Shanghai Institute of Hematology.18 All osteosarcoma cell lines were cultured in high glucose DMEM (Gibco, 11965092) supplemented with 10% fetal bovine serum (FBS, Gibco, 10099–141) and 1% penicillin-streptomycin (Gibco, 15070063) at 37°C in 5% CO2. The hFOB1.19 cell was cultured in DMEM/F12 medium (Gibco, 11320033) containing 10% FBS and 0.3mg/mL G418 (Gibco, 10131035) at 34°C in 5% CO2.
Quantitative Real-Time PCR Analysis
The mRNA expressions of MYC and CLEC5A in osteosarcoma and osteoblast cell lines were quantified by quantitative real-time PCR (qRT-PCR) analyses. In brief, whole cellular RNA was extracted by Trizol reagent (Invitrogen, 15596018) and the cDNA templates were obtained by reverse transcription. The amplification was conducted through Roche LightCycler System. The whole procedure was performed according to the protocol of TB Green Premix Ex Taq II kit (Takara RR820A, Japan). The primer sequences of genes were: MYC: 5ʹ- CGACGAGACCTTCATCAAAAAC-3ʹ (forward) and 5ʹ-CTTCTCTGAGACGAGCTTGG-3ʹ (reverse); CLEC5A: 5ʹ-GAAAAGGATCCACATTGGCAAT-3ʹ (forward) and 5ʹ-GTCGCACAGTTGAAATTCTGA-3ʹ (reverse); GAPDH: 5ʹ-GCACCGTCAAGGCTGAGAAC-3ʹ (forward) and 5ʹ-TGGTGAAGACGCCAGTGGA-3ʹ (reverse). The expression of genes was calculated by formula 2−ΔΔCT. The expressive difference between osteosarcoma and osteoblast cell lines or high- and low-metastatic osteosarcoma cell lines were calculated by Student’s t test. P value less than 0.05 was considered as statistically significant.
Immunohistochemistry Analysis
Ten patients who were diagnosed as osteosarcoma by pathology in Ruijin Hospital were selected for IHC assay. Among these patients, five developed metastasis in 3 years and the others lived without metastasis. Each osteosarcoma sample was fixed with 10% formalin, embedded in paraffin and sliced as 4 μm thickness. The sections were deparaffinized and rehydrated by xylene and ethanol. After heat-induced antigen retrieval, endogenous peroxidase activity blockage and serum sealing, the samples were incubated with rabbit anti-CLEC5A (Biosis, bs-2663R, 1:200) and anti-MYC (Affinity, AF0358, 1:100) antibodies at 4°C overnight. Then, slices were covered with horseradish peroxidase-coupled goat anti-rabbit secondary antibody (CST, 7074, 1:200) at room temperature for 50 minutes and stained with diaminobenzidine. The nuclei were counterstained by hematoxylin. At last, the samples were dehydrated and mounted. All slices were visualized by the microscope (Nikon DS-U3).
Results
Identification of Inflammatory Response-Associated DEmRNAs Related to Osteosarcoma Metastasis
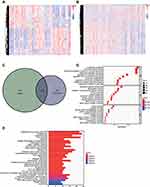
The expressions of mRNAs in GSE21257 that included 34 metastatic and 19 non-metastatic osteosarcoma patients were downloaded from GEO database. Two hundred and forty-seven up-regulated and 217 down-regulated DEmRNAs related to osteosarcoma metastasis were identified (Figure 2A). Two hundred inflammatory response-associated genes were obtained from hallmark gene sets in MSigDB and then overlapped with previously obtained DEmRNAs (Figure 2B). In the end, 24 inflammatory response-associated DEmRNAs were screened out (Table 1).
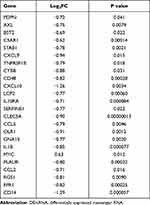 |
Table 1 The Inflammatory Response-Associated DEmRNAs in GSE21257 |
Construction and Validation of the Inflammatory Response-Associated Prognostic Signature in GEO and Target Databases
Based on the 24 DEmRNAs acquired previously, univariate and multivariate cox regression analyses were utilized to establish the inflammatory response-associated prognostic signature. In univariate cox regression analysis, three genes were associated with osteosarcoma prognosis (Figure 3A). Then, two of these three DEmRNAs were screened out to construct the inflammatory response-associated prognostic signature according to the multivariate cox regression result (Table 2). The risk score formula was established through mRNAs’ regression coefficients multiplied by expression levels:
 |
Table 2 The Multivariate Cox Regression Analysis of GSE21257 |
Risk score = (0.41*MYC) + (−0.96*CLEC5A)
Based on the median value of risk score, 26 patients were assigned to high-risk group and the rest 27 were regarded as low-risk. At the same time, patients in high-risk group showed worse mortality rates and shorter survival times than low-risk patients (Figure 3B). The KM survival curve also indicated poorer survival rates in high-risk patients (Figure 3C). Finally, the area under the time-dependent ROC curves (AUC) were 0.65 at 1-year, 0.80 at 3- and 5-year, which demonstrated the robustness of this inflammatory response-associated prognostic signature (Figure 3D).
The reliability of the inflammatory response-associated gene signature was validated in Target database. Ninety-five patients with integrated follow-up information were included in the test. Based on the median value of risk scores, 48 patients were separated into low-risk group and the rest were considered as high risk (Figure 3E). The risk plot (Figure 3E) and KM survival analysis (Figure 3F) showed that low-risk patients had better prognoses. In addition, the time-dependent ROC curve revealed the signature was robust (Figure 3G).
Exploration of the Relationship Between Clinical Features and Inflammatory Response-Associated Prognostic Signature
The prognostic value of gene signature was investigated through univariate and multivariate cox regression analyses. Figure 4A and B showed that the risk score was the independent prognostic factor in osteosarcoma. The same result was validated in Target database (Supplementary Figure 1A and 1B).
After that, the correlations between clinical features and genes in signature were investigated. In GSE21257, CLEC5A was associated with patients’ metastasis status as well as inflammatory response-associated risk and MYC was related to gender, metastasis status and inflammatory response-associated risk (Figure 4C–J). In Target database, CLEC5A and MYC were only correlated to risk scores (Supplementary Figure 1C–J). The KM survival analyses indicated the expression of CLEC5A positively related to the survival rates of osteosarcoma patients in GSE21257, Target and online R2 databases (Supplementary Figure 2A, C, E). Instead, the higher expression of MYC related to the worse prognosis of osteosarcoma patients (Supplementary Figure 2B, D, F).
Establishment of the Nomogram
The nomogram was a powerful tool that integrated clinical variables to evaluate the prognosis of patients. In this research, the nomogram was constructed in GSE21257 and Target databases to predict the 3- and 5-year survival rates of osteosarcoma patients (Figure 5A and Supplementary Figure 3A). The 3-year and 5-year calibration curves were close to the standard curve in two datasets (Figure 5B, C and Supplementary Figure 3B, C). The C-index was 0.99 in GSE21257 and 0.77 in Target dataset, which indicated the high accuracy of the predictive model.
Functional Analysis of DEmRNAs Related to Inflammatory Response-Associated Prognostic Signature
The DEmRNAs among high- and low-risk patients were explored in GSE21257 and Target databases. A total of 736 DEmRNAs were screened out in GSE21257 and 1619 DEmRNAs were identified in Target database (Figure 6A and B). Then, 216 DEmRNAs overlapped in two databases were selected for further analysis (Figure 6C).
The biological functions of these DEmRNAs were investigated through GO functional and KEGG pathway analyses (Figure 6D and E). A number of inflammatory response-associated biological processes such as neutrophil activation, regulation of leukocyte activation and antigen processing and presentation were significantly enriched. Furthermore, some immune-related functions (ie, regulation of immune effector process, Th17 cell differentiation) were also enriched, which implied the underlying relationships between inflammatory response and immune alterations in osteosarcoma.
Evaluation of the Correlation Between Inflammatory Response and Immune Infiltration
The relationship between tumor microenvironment and inflammatory response-associated risks was evaluated through ESTIMATE software. The results showed that the risk score was negatively related to the immune and stromal scores in GSE21257 and Target datasets (Figure 7A, B and Supplementary Figure 4A, B). Then the immune infiltration of 22 immune cells was investigated by CIBERSORT algorithm. The statistically significant abundance ratios of immune cells in osteosarcoma were shown in Figure 7C and Supplementary Figure 4C. Then the specific relationships between immune functions and inflammatory response-associated risks were evaluated through ssGSEA analysis. The outcome revealed that the vast majority of immune cells (14) and functions (11) were down-regulated in high-risk group in GSE21257 (Figure 7D and E). At the same time, eight different immune cells and twelve immune functions were dysregulated in Target dataset (Supplementary Figure 4D, E). The overlapped items in both datasets that included eight immune cells and eleven immune functions were considered as the alterations of inflammatory response-associated immune infiltration (Figure 7F). In addition, the expression of MYC was negatively related to 10 immune cells and 10 immune functions in GSE21257, while CLEC5A showed the positive correlations (Supplementary Figure 5A–D). The same dysregulated trends were observed in Target dataset (Supplementary Figure 5E–H).
Validation of the Inflammatory Response-Associated Genes in Cell Lines and Osteosarcoma Samples
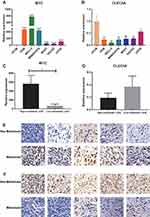
The expressive levels of inflammatory response-associated genes in human osteosarcoma and osteoclast cell lines were detected through qRT-PCR analyses. The results showed that MYC was up-regulated and CLEC5A was down-regulated in all osteosarcoma cell lines (Figure 8A and B). Then, osteosarcoma cell lines were divided into high- (143B, WELL5, MNNG/HOS) and low-metastatic (U20S, SAOS2, MG63) groups according to their metastasis ability. The MYC was up-regulated in high-metastatic osteosarcoma cell lines and CLEC5A showed the opposite trend but with no statistically significant (Figure 8C and D). At the same time, the expressions of MYC and CLEC5A in metastasis and non-metastasis patients were validated by IHC. The result revealed that MYC was down-regulated in non-metastasis patients (Figure 8E) while CLEC5A showed the opposite trends (Figure 8F).
Discussion
Osteosarcoma, one of the most common bone malignant tumors, often developed lung metastasis and the 5-year survival rates of these patients were extremely low.2 In clinical studies, the diagnosis of osteosarcoma metastasis mainly relied on the radiology tests such as X-ray, computerized tomography (CT) and positron emission tomography-computed tomography (PET-CT). Unfortunately, the sensitivity and specificity of these methods were not high enough and the patients with metastatic possibilities always lost the best chance to receive early treatment. Thus, the novel reliable evaluation of early-stage metastasis was urgently needed.
In 1889, Paget first described the mechanism of tumor metastasis which was widely known as the “seed and soil” theory.19 After that, various theories related to metastasis were discovered, such as epithelial-mesenchymal transition (EMT), angiogenesis, gene mutation and so on.20 Among these hallmarks, the inflammatory response was extensively investigated. Numerous inflammatory biomarkers, such as neutrophils, lymphocytes, platelets and so on, had been implied as the prognostic predictor for different cancers.21–23 In osteosarcoma, some researchers revealed that lymphocyte-to-monocyte ratio (LMR) was positively related to overall and event-free survival rates of patients, while the neutrophil-lymphocyte ratio (NLR) and platelet-to-lymphocyte ratio (PLR) showed the opposite trends.24–26 Several studies also implied that nonsteroidal anti-inflammatory drugs (NSAIDs) could inhibit the progression of osteosarcoma.27,28 In addition, the relationship between inflammatory response-associated genes and osteosarcoma had also been widely studied. For instance, Jiang et al found that the inflammatory gene polymorphisms were closely related to the occurrence and progression of osteosarcoma.29 Recent researches also indicated that several inflammatory response-associated genes could combine as the prognostic signature in different cancers.11,12 Unfortunately, none of this signature had been established in osteosarcoma.
In the present study, we first analyzed the relationship between osteosarcoma and inflammatory response-associated genes. We screened out 464 DEmRNAs related to osteosarcoma metastasis in GSE21257 dataset. By overlapping with MSigDB database, 24 of these DEmRNAs were identified as the inflammatory response-associated genes. Then, two of these genes were selected to construct the inflammatory response-associated gene signature through univariate and multivariate cox regression analyses. Next, patients in GSE21257 were separated into high- and low-risk groups according to the median value of risk scores. The KM survival curve indicated that the risk score was negatively correlated with patients’ survival rates and the AUC values of ROC curves revealed that this signature was reliable. Most importantly, the same results were obtained in Target database. Besides that, we verified that the risk score was the independent prognostic factor for osteosarcoma patients.
The nomogram was a convincible tool that incorporated clinical features to estimate the patients’ prognosis. It could stratify the patients with unfavorable prognoses and guide clinical treatments. In this research, the nomogram was established in both datasets and the calibration curves, as well as C indexes, indicating the robustness of this nomogram. To sum up, we believed that this inflammatory response-associated gene signature was a reliable prognostic predictor for osteosarcoma and it could be easier to apply in clinics because only two genes were involved in it.
In the last decades, numerous studies demonstrated that the tumor infiltrative immune cells and immune-related genes played vital roles in the development and progression of tumors.30 In osteosarcoma, tumor cells could also recruit different immune infiltrating cells and establish the specific local immune microenvironment, which finally promoted tumor growth, drug resistance and metastasis.31 For instance, Cao et al found that the expression of BMPR2 was negatively related to the infiltration of CD8+ T cells, monocytes, and M2 macrophages, which resulted in the poor prognosis of patients.32 In addition, Deng et al revealed that after neoadjuvant chemotherapy, more immune cells such as CD3+ T cells, CD8+ T cells and Ki67+/CD8+ T cells were infiltrated in tumor microenvironment and this might benefit for immunotherapy.33 However, no studies focused the relationship between inflammatory response and immune infiltration in osteosarcoma.
In this study, we screened out the overlapped DEmRNAs related to inflammatory response-associated risks in GSE21257 and Target datasets. The functional analyses of these genes indicated that they were involved in the dysregulated immune functions such as regulation of immune effector process, Th17 cell differentiation and so on. According to the above results, we speculated that the inflammatory response might transform the tumor microenvironment, especially for the immune system. We then used Estimate algorithm to evaluate the correlation between inflammatory response-associated risks and stromal or immune infiltration. Both stromal and immune scores were negatively related to risk scores, which indicated higher purity of osteosarcoma cells existed in high-risk groups. These results implied that high-risk osteosarcoma was poorly immunogenic infiltrated, which was widely known as immunological cold tumors and thus was not sensitive to immunogenic therapy.34 To further investigate the specific dysregulation of immune functions, ssGSEA analysis was conducted. The results showed that high-risk patients related to decreased infiltrative immune cells and functions. These conclusions were validated in various studies. For example, Muraro et al found that osteosarcoma cells could inhibit the maturation and function of dendritic cells (DCs) and these effects would be revised by adding inflammatory-associated cytokines (rhIL-12) or compounds (Indometacin).35 The follicular helper T (Tfh) cells were related to the prognosis of patients in several tumors.36 In osteosarcoma, Gao et al revealed that impaired functions of Tfhs lead to lower IL-21 secretion in CD4+ T cells, which might result in the immunosuppressive microenvironment.37 In a word, the inflammatory response was closely related to the osteosarcoma immune infiltrative alteration and the down-regulated immune functions might contribute the tumor development.
What is more, the expression of individual mRNAs in the signature was investigated. The results showed that both of them related to the survival rates of patients in GSE21257, Target and R2 online datasets. For in vitro validation, the qRT-PCR analyses revealed that the expression of MYC was up-regulated in all osteosarcoma cell lines while CLEC5A showed the opposite trend. Compared to low-metastatic cell lines, MYC was over-expressed in high-metastatic cell lines but CLEC5A exhibited the contrary tendency. For the IHC study, the same expressive trends of two genes were observed between metastasis and non-metastasis osteosarcoma patients’ samples. Besides that, the ssGSEA analysis revealed that in both datasets, CLEC5A was positively related to the immune cells’ infiltration and immune functions while MYC showed the opposite trends. These results indicated that the mRNAs in our inflammatory response-associated signature played vital roles in the dysregulation of immune system, which finally promoted tumor progression.
MYC, one of the most common oncogene, was mutated in more than 10% of osteosarcoma patients.38 High expression of MYC promoted proliferation, migration and spheroid formation of osteosarcoma cells and it was also upregulated in metastatic samples.39,40 In addition, Kortlever et al revealed that MYC promoted the inflammatory, angiogenic and immunosuppressive tumor microenvironment by elevating IL-23 and CCL9, which accelerated pulmonary tumor progression. C-type lectin domain family 5 member A (CLEC5A) was one of the C-type lectin receptors that involved in inflammatory diseases.41 It regulated the release of pro-inflammatory cytokines from macrophages in infection and was involved in the process of collagen-induced arthritis.42 It played vital roles in the innate immune system by regulating the differentiation and activation of macrophages as well as neutrophils.43 Lu et al found that the expression of CLEC5A in osteosarcoma was higher than adjacent normal tissues and it could prevent the metastasis of osteosarcoma.44 However, the specific mechanisms of CLEC5A in osteosarcoma progression were still unknown and further research should be carried out.
For all we know, this research first revealed the relationship between inflammatory-associated gene signature and osteosarcoma prognosis. However, several defects still existed. First, the sample size of datasets was relatively small. This might owe to the extremely low incidence of osteosarcoma and the large cohort researches in osteosarcoma were limited. Second, the correlations between immune infiltration and gene signature might be variable because osteosarcoma was high heterogeneity. Most importantly, our research was a retrospective study and the result should be verified in other prospective studies in the future.
Conclusion
In summary, a novel inflammatory response-associated gene signature that related to osteosarcoma prognosis was constructed and validated in our study. Besides that, we provided a new perspective on the correlation between immune infiltration and inflammatory response. Meanwhile, this signature could be regarded as the prognostic biomarker for osteosarcoma and acted as the therapeutic target in clinical studies. Further experimental and clinical studies should be conducted to illuminate the precise mechanisms of inflammatory response in osteosarcoma.
Abbreviations
IRGs, inflammatory response-associated genes; DEmRNAs, differentially expressed messenger RNAs; GEO, Gene Expression Omnibus; MSigDB, Molecular Signature Database; qRT-PCR, quantitative real-time PCR; ssGSEA, single-sample gene set enrichment analyses; KM, Kaplan–Meier; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; Target, Therapeutically Applicable Research to Generate Effective Treatments; Limma, Linear Models for Microarray Analysis; AIC, Akaike information criterion; ROC, receiver operating characteristic; C-index, concordance index; CT, computerized tomography; PET-CT, positron emission tomography-computed tomography; EMT, epithelial-mesenchymal transition; LMR, lymphocyte-to-monocyte ratio; PLR, platelet-to-lymphocyte ratio; NLR, neutrophil-lymphocyte ratio; NSAIDs, nonsteroidal anti-inflammatory drugs; DCs, dendritic cells; Tfh, follicular helper T; CLEC5A, C-type lectin domain family 5 member A; IHC, immunohistochemistry.
Data Sharing Statement
The datasets used in this study are available at Target (https://ocg.cancer.gov/programs/target) and GEO (http://www.ncbi.nlm.nih.gov/geo) databases.
Ethics Approval and Informed Consent
The study was conducted according to the guidelines of the Helsinki declaration and was approved by Ruijin Hospital Ethics Committee (Registration number: 2020409). All patients signed the written informed consent to participate in this study.
Consent for Publication
All authors agreed to publish this research.
Acknowledgments
We appreciated researchers and patients participated in GEO and Target datasets.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC, NO. 81702661,82072957).
Disclosure
All authors have no conflicts of interest to declare.
References
1. Beck O, Paret C, Russo A, et al. Safety and activity of the combination of ceritinib and dasatinib in osteosarcoma. Cancers. 2020;12(4):793. doi:10.3390/cancers12040793
2. Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21 Suppl 7:vii320–325. doi:10.1093/annonc/mdq276
3. Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47(1):283–292. doi:10.1016/j.ocl.2015.08.022
4. Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14(11):722–735. doi:10.1038/nrc3838
5. Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–545. doi:10.1016/S0140-6736(00)04046-0
6. Sipos F, Firneisz G, Műzes G. Therapeutic aspects of c-MYC signaling in inflammatory and cancerous colonic diseases. World j Gastroenterol. 2016;22(35):7938–7950. doi:10.3748/wjg.v22.i35.7938
7. Dvorak HF, Underhill LH, Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–1659. doi:10.1056/NEJM198612253152606
8. Jettoo P, Tan G, Gerrand CH, Rankin KS. Role of routine blood tests for predicting clinical outcomes in osteosarcoma patients. J Orthop Surg Res. 2019;27(2):2309499019838293. doi:10.1177/2309499019838293
9. Li X, Tian F, Wang F, Li Y. Serum C-reactive protein and overall survival of patients with osteosarcoma. Tumor Biol. 2015;36(7):5663–5666. doi:10.1007/s13277-015-3240-6
10. Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15(11):e493–503. doi:10.1016/S1470-2045(14)70263-3
11. Lin Z, Xu Q, Miao D, Yu F. An inflammatory response-related gene signature can impact the immune status and predict the prognosis of hepatocellular carcinoma. Front Oncol. 2021;11:644416. doi:10.3389/fonc.2021.644416
12. Zhang C, Zhang Z, Zhang G, et al. Clinical significance and inflammatory landscapes of a novel recurrence-associated immune signature in early-stage lung adenocarcinoma. Cancer Lett. 2020;479:31–41. doi:10.1016/j.canlet.2020.03.016
13. Vrieze SI. Model selection and psychological theory: a discussion of the differences between the Akaike information criterion (AIC) and the Bayesian information criterion (BIC). Psychol Methods. 2012;17(2):228–243. doi:10.1037/a0027127
14. Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364–1370. doi:10.1200/JCO.2007.12.9791
15. Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi:10.1038/ncomms3612
16. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
17. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259.
18. Zhang W, Zhao JM, Lin J, et al. Adaptive fibrogenic reprogramming of osteosarcoma stem cells promotes metastatic growth. Cell Rep. 2018;24(5):1266–1277.e1265. doi:10.1016/j.celrep.2018.06.103
19. Fokas E, Engenhart-Cabillic R, Daniilidis K, Rose F, An HX. Metastasis: the seed and soil theory gains identity. Cancer Metastasis Rev. 2007;26(3–4):705–715. doi:10.1007/s10555-007-9088-5
20. Han Y, Guo W, Ren T, et al. Tumor-associated macrophages promote lung metastasis and induce epithelial-mesenchymal transition in osteosarcoma by activating the COX-2/STAT3 axis. Cancer Lett. 2019;440–441:116–125. doi:10.1016/j.canlet.2018.10.011
21. Dai Y, Liu M, Lei L, Lu S. Prognostic significance of preoperative prognostic nutritional index in ovarian cancer: a systematic review and meta-analysis. Medicine. 2020;99(38):e21840. doi:10.1097/MD.0000000000021840
22. Templeton AJ, Ace O, McNamara MG, et al. Prognostic role of platelet to lymphocyte ratio in solid tumors: a systematic review and meta-analysis. Cancer Epidemiol, Biomarkers Prevent. 2014;23(7):1204–1212. doi:10.1158/1055-9965.EPI-14-0146
23. Gu L, Li H, Chen L, et al. Prognostic role of lymphocyte to monocyte ratio for patients with cancer: evidence from a systematic review and meta-analysis. Oncotarget. 2016;7(22):31926–31942. doi:10.18632/oncotarget.7876
24. Liu T, Fang XC, Ding Z, Sun ZG, Sun LM, Wang YL. Pre-operative lymphocyte-to-monocyte ratio as a predictor of overall survival in patients suffering from osteosarcoma. FEBS Open Bio. 2015;5:682–687. doi:10.1016/j.fob.2015.08.002
25. Liu B, Huang Y, Sun Y, et al. Prognostic value of inflammation-based scores in patients with osteosarcoma. Sci Rep. 2016;6:39862. doi:10.1038/srep39862
26. Xia WK, Liu ZL, Shen D, Lin QF, Su J, Mao WD. Prognostic performance of pre-treatment NLR and PLR in patients suffering from osteosarcoma. World J Surg Oncol. 2016;14:127. doi:10.1186/s12957-016-0889-2
27. Zuckerman LM, Frames WL, Elsissy JG, et al. The effect of non-steroidal anti-inflammatory drugs on osteosarcoma cells. Eur Rev Med Pharmacol Sci. 2019;23(6):2681–2690. doi:10.26355/eurrev_201903_17416
28. Liu B, Yan S, Qu L, Zhu J. Celecoxib enhances anticancer effect of cisplatin and induces anoikis in osteosarcoma via PI3K/Akt pathway. Cancer Cell Int. 2017;17:1. doi:10.1186/s12935-016-0378-2
29. Jiang Y, Wang X, Cheng Y, et al. Associations between inflammatory gene polymorphisms (TNF-α 308G/A, TNF-α 238G/A, TNF-β 252A/G, TGF-β1 29T/C, IL-6 174G/C and IL-10 1082A/G) and susceptibility to osteosarcoma: a meta-analysis and literature review. Oncotarget. 2017;8(57):97571–97583. doi:10.18632/oncotarget.18813
30. Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32(19–20):1267–1284. doi:10.1101/gad.314617.118
31. Heymann MF, Lézot F, Heymann D. The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell Immunol. 2019;343:103711. doi:10.1016/j.cellimm.2017.10.011
32. Cao H, Quan S, Zhang L, Chen Y, Jiao G. BMPR2 expression level is correlated with low immune infiltration and predicts metastasis and poor survival in osteosarcoma. Oncol Lett. 2021;21(5):391. doi:10.3892/ol.2021.12652
33. Deng C, Xu Y, Fu J, et al. Reprogramming the tumor immunologic microenvironment using neoadjuvant chemotherapy in osteosarcoma. Cancer Sci. 2020;111(6):1899–1909. doi:10.1111/cas.14398
34. Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi:10.1126/science.aaa8172
35. Muraro M, Mereuta OM, Saglio F, et al. Interactions between osteosarcoma cell lines and dendritic cells immune function: an in vitro study. Cell Immunol. 2008;253(1–2):71–80. doi:10.1016/j.cellimm.2008.05.002
36. Gu-Trantien C, Loi S, Garaud S, et al. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest. 2013;123(7):2873–2892. doi:10.1172/JCI67428
37. Gao W, Zhou J, Ji B. Evidence of interleukin 21 reduction in osteosarcoma patients due to PD-1/PD-L1-mediated suppression of follicular helper T cell functionality. DNA Cell Biol. 2017;36(9):794–800. doi:10.1089/dna.2017.3669
38. Ueda T, Healey JH, Huvos AG, Ladanyi M. Amplification of the MYC gene in osteosarcoma secondary to paget’s disease of bone. Sarcoma. 1997;1(3–4):131–134. doi:10.1080/13577149778209
39. Chen D, Zhao Z, Huang Z, et al. Super enhancer inhibitors suppress MYC driven transcriptional amplification and tumor progression in osteosarcoma. Bone Res. 2018;6:11. doi:10.1038/s41413-018-0009-8
40. Han G, Wang Y, Bi W. C-Myc overexpression promotes osteosarcoma cell invasion via activation of MEK-ERK pathway. Oncol Res. 2012;20(4):149–156. doi:10.3727/096504012X13522227232237
41. Xiong W, Wang H, Lu L, et al. The macrophage C-type lectin receptor CLEC5A (MDL-1) expression is associated with early plaque progression and promotes macrophage survival. J Transl Med. 2017;15(1):234. doi:10.1186/s12967-017-1336-z
42. Chen X, Eksioglu EA, Carter JD, et al. Inactivation of DAP12 in PMN inhibits TREM1-mediated activation in rheumatoid arthritis. PLoS One. 2015;10(2):e0115116. doi:10.1371/journal.pone.0115116
43. Huang YL, Chen ST, Liu RS, et al. CLEC5A is critical for dengue virus-induced osteoclast activation and bone homeostasis. J Mol Med. 2016;94(9):1025–1037. doi:10.1007/s00109-016-1409-0
44. Lu J, Chen W, Liu H, Yang H, Liu T. Transcription factor CEBPB inhibits the proliferation of osteosarcoma by regulating downstream target gene CLEC5A. J Clin Lab Anal. 2019;33(9):e22985. doi:10.1002/jcla.22985
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.