Back to Journals » OncoTargets and Therapy » Volume 14
Epigenetic Mechanism of Enrichment of A549 Lung Cancer Stem Cells with 5-Fu
Authors Cao F, Shi M , Yu B, Cheng X, Li X, Jia X
Received 18 February 2021
Accepted for publication 14 May 2021
Published 18 June 2021 Volume 2021:14 Pages 3783—3794
DOI https://doi.org/10.2147/OTT.S233129
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Fangyuan Cao,1,2 Mumu Shi,1 Bo Yu,1 Xiangrong Cheng,3 Xin Li,4 Xinshan Jia5
1Department of Pathology, Science and Education Department of the Fifth People’s Hospital of Qinghai, Xining, Qinhai, 810000, People’s Republic of China; 2Department of Pediatrics, The Fifth Affiliated Hospital of Harbin Medical University, Daqing, Heilongjiang, 163319, People’s Republic of China; 3Department of Nuclear Medicine, The Fifth People’s Hospital of Qinghai, Xining, Qinhai, 810000, People’s Republic of China; 4Department of Physiology, College of Life Science and Biopharmaceutics of Shenyang Pharmaceutical University, Shenyang, Liaoning, 110001, People’s Republic of China; 5Department of Pathology, College of Basic Medical Sciences, China Medical University, Shenyang, Liaoning, 110001, People’s Republic of China
Correspondence: Mumu Shi
Department of Pathology, Science and Education Department of the Fifth People’s Hospital of Qinghai, Xining, Qinhai, 810000, People’s Republic of China
Email [email protected]
Bo Yu
Department of Pathology, Science and Education Department of the Fifth People’s Hospital of Qinghai, Xining, Qinhai, 810000, People’s Republic of China
Email [email protected]
Background: The influence of 5-fluorouracil (5-Fu) and cisplatin (CDDP) on the A549 and NCI-H226 cells was studied, and the epigenetic mechanism of enrichment of A549 lung cancer stem cells with 5-Fu was explored.
Materials and Methods: The cell proliferation of both A549 and NCI-H226 was detected by BrdU assay, and apoptosis condition was measured by flow cytometric analysis. The expressions of OCT3/4 and Nanog in cells treated with 5-Fu or CDDP were measured by immunofluorescence, Western blot and qPCR. qPCR was also performed to determine the relative expression of methyltransferase genes and miRNA. Sequencing after bisulfite treatment (BSP) was employed to detect the methylation of OCT3/4 promoter in A549 cells. And ChIP was conducted to detect the expression of H3K9Me3 and H3K9Ace.
Results: Both 5-Fu and CDDP result in the apoptosis of A549 and NCI-H226 cells and improve the expressions of has-miR-134 and has-miR-296. However, 5-Fu enhances the expression of OCT3/4 in A549 cells, and the change of methyltransferase genes and BSP results suggested some genetic differences between CDDP and 5-Fu treatment in A549 cells. ChIP assay showed that the expression of H3K9Me3 significantly decreased and H3K9Ace significantly increased in A549 cells.
Conclusion: The enrichment effect of CDDP on A549 and NCI-H226 carcinoma stem cells is inconsistent with the enrichment effect of 5-Fu. The enrichment of A549 lung cancer stem cells with 5-Fu might be related to the methylation of OCT3/4 promoter and the expression of H3K9Me3 and H3K9Ace.
Keywords: CDDP, 5-Fu, A549, epigenetic mechanism, methylation
Introduction
Lung carcinoma, one of the prominent causes of cancer-related deaths worldwide, has a high prevalence and high fatalities.1,2 Though surgical treatment, radiotherapy, chemotherapy and target therapy are the most useful lung cancer therapeutic methods, the prognosis of lung cancer is still unsatisfactory. About 70–80% of all lung cancer cases are non-small cell lung carcinoma (NSCLC), which stand for a heterogeneous group of tumors, mainly including adenocarcinoma (AD), squamous cell carcinoma (SCC) and large cell carcinoma.3 AD and SCC have a series of similarities and differences at the genetic level, clinical presentations, response to treatments and the livability of the patients.4,5 Several genes, such as KIAA0728 and KIAA0425, were differentially expressed in both lung adenocarcinomas and lung squamous cell carcinomas.6 Lung SCC and AD cause comparable deficiencies on hampering dendritic cell in vitro, switching monocyte differentiation from hampering dendritic cell to macrophage-like cells, but the majority of these changes occurred with diverse mediators.7 Therefore, it is necessary to explain the molecular mechanisms of the tumorigenesis and progression of lung carcinoma.
Cancer stem cells (CSCs) can initiate and sustain the growth of tumor. The hypothesis of CSCs comes up with a better understanding of tumor initiation, recurrence and metastasis. Based on the theory of cancer stem cells, it can be inferred that lung cancer stem cells with self-renewal and infinite proliferation ability are the root of lung cancer, and the malignant phenotypic characteristics of lung cancer, such as recurrence, invasion and metastasis, drug resistance and radiation resistance, are all bound up with lung cancer stem cells.8
OCT4 (also called OCT3) is an important regulator of stem cell properties and is related to the regulation of symmetric division.9 OCT4 is also a key regulator of cancer cells; in particular it can participate in regulating various states and functions of CSCs.10,11 CSCs have stronger cloning and drug resistance in human colon cancer cells overexpressing OCT4.11 4T1 cells overexpressing OCT4 have higher expression level of stem cell marker genes, such as Sca-1, CD133 and ALDH1.10 These findings suggest that high expression of OCT4 may promote the function of CSCs. More CSC-like cells could undergo apoptosis through OCT4/Tcl1/Akt1 pathway after OCT4 was knocked down in MCF-7 cells, and this suggested that OCT4 has the ability to inhibit apoptosis in cancer cells.12 OCT4 and Nanog could induce cancer stem cell-like properties, enhance EMT and promote the migration ability of cancer cells, suggesting that there was a correlation between the worse prognosis of lung adenocarcinoma patients and the high levels of Oct4/Nanog/Slug.13 These findings show that OCT3/4 plays a key role in the maintenance of dryness, self-renewal and metastasis of cancer cells and CSCs.
Cisplatin (CDDP) is the most widely used chemotherapy drug. The chemotherapeutic agent 5-fluorouracil (5-Fu) is a member of the pyrimidine anti-metabolite drug family which suppresses the growth of proliferative cells, eventually leading to cell death. By targeting thymidylate synthase and by the direct incorporation of 5-Fu metabolites into DNA and RNA, 5-Fu initiates apoptosis.14 Around half of lung cancer patients have primary drug resistance.15 Current anticancer drugs are resistant to resting cancer stem cells, thus it is particularly important to elucidate the biological characteristics of cancer stem cells and develop cell regulation technology.
The expression of eukaryotic genes is a complex process which is dependent on the cooperative action of genetic and epigenetic regulation. Epigenetic regulation can be divided into two broad types: one is regulation of gene selective transcription expression such as DNA methylation, gene imprinting, histone modification, chromatin remodeling; the other is post-transcriptional regulation of genes such as non-coding RNA, microRNA, antisense RNA, etc. Previous studies have found that 5-Fu enriches OCT3/4-positive lung adenocarcinoma SPC cell lines16 and lung adenocarcinoma A549 cell lines.13 However, whether 5-Fu can enrich lung squamous carcinoma cells, whether other chemotherapeutic drugs can enrich lung adenocarcinoma cells or lung squamous carcinoma cells, and the mechanism of epigenetic regulation of the enrichment of lung cancer stem cells by chemotherapeutic drugs has not been reported. The lung adenocarcinoma cell lines A549 and NCI-H226 were treated with 5-Fu and CDDP, respectively. Then, the effect was examined, which would provide new insight to the development of novel therapeutic strategies in the management of lung cancer.
Materials and Methods
Cell Culture
Two human lung cancer cell lines including A549 (adenocarcinoma) and NCI-H226 (squamous cell carcinoma) were purchased from ATCC. A549 cells and NCI-H226 cells were cultured in F-12K, RPMI-1640 medium containing 10% fetal bovine serum (FBS) (Hyclone, Logan, UT), respectively. They were maintained at 37°C in a 5% CO2 incubator, and the media were changed every two days.
IC50 of A549 and NCI-H226 Cells Were Detected with MTT Method
Exponentially growing cells of A549 and NCI-H226 were seeded into 96-well plates in corresponding medium. Cells were allowed to attach for 24 hours before treatment. Then, both cell lines were treated with fresh medium containing cisplatin (Sigma Aldrich, Germany) at different concentrations (0, 0.0016, 0.008, 0.04, 0.2, 1, 5, 25, 125, 625 μg/mL), and the plates were incubated for 48 hours. Then, 5 mg/mL MTT (Sigma Aldrich, Germany) was added into each well, and the plates were incubated at 37°C in an incubator for 4 h (5% CO2). Media containing MTT were removed and 150 µL dimethyl sulfoxide (DMSO) was added in each well to dissolve the formazan crystals. Absorbance of the wells was read at 490 nm (A490) after 10 minutes of incubation at room temperature (RT) with microplate reader (PerkinElmer/envision). For each experiment, three independent experiments at least were done in triplicate.
Preparation of the Samples
Exponentially growing cells of A549 and NCI-H226 were seeded into 24/96-well plates in corresponding medium, respectively. After overnight cultivation, A549 cells were treated with 100 μg/mL 5-Fu (Sigma-Aldrich, Germany) and IC50 concentration of CDDP (8 μg/mL) for 48 hours (the concentration based on the above MTT experiment), named as CDDP group and 5-Fu group, respectively. NCI-H226 cells were also treated with 5-Fu (100 μg/mL) and IC50 concentration of CDDP (4 μg/mL) for 48 hours (the concentration based on the above MTT experiment), respectively. Normal cultured cells served as blank group.
Cell Proliferation Activity
The cell proliferation was measured using a BrdU (5-bromo-2ʹ-deoxyuridine) cell proliferation assay kit (Biovision, USA) according to the manufacturers’ instructions. Briefly, BrdU (the final concentration was 1×) was added into cells after drug treatment, and incubated for 4 hours. Subsequently, 100 μL cell immobilization/denaturation solution was added for 30 minutes at RT after cell culture supernatant was discarded. Then 100 μL BrdU detection antibodies was added after cell immobilization/denaturation solution was removed, and plates were slightly shaken at RT for 1 hour. Each well was washed three times after BrdU was removed; HRP labeled sheep diluted anti-mice (100 μL/well) was added and incubated at 37°C for 1 hour, then washed again. After incubating with 100 μL TMB substrate solution for 5–15 minutes, 50 μL substrate termination solution was added, and absorbance value was measured at 450 nm using a 96-well plate reader.
Cell Apoptosis Assay
To determine the extent of apoptosis and necrosis, Annexin V/FITC staining (Annexin V/FITC Apoptosis Detection Kit, Vazyme Biotech Co., Ltd, Nanjing, China) was used. Briefly, the cells were harvested and washed with PBS three times after treatment with CDDP and 5-Fu for 48 hours. The cells were suspended in 1× Annexin binding buffer. After the addition of Annexin V-FITC, the cells were stained in the dark for 15 minutes at RT. Then, PI dye was added and incubated for 15 minutes of total incubation time at RT in the dark. Flow cytometry was done within 1 hour using a flow cytometer (Beckman Coulter, USA).
Immunofluorescence
Fixed samples were washed with PBS to remove paraformaldehyde, and then rabbit polyclonal anti–OCT4 antibody (1:1000) (Abcam, USA) in PBST (PBS+0.1% Triton-X) supplemented with 10% normal goat serum was added to each well and incubated overnight at 4°C. After rabbit polyclonal anti-OCT4 antibody was removed and samples were washed with PBS, goat anti-rabbit Alexa-Fluor 568 (1:500) in PBS-Triton supplemented with 10% NGS were added to each well and incubated for 45 min at RT in dark. DAPI (1:500) was also added to goat anti-rabbit Alexa-Fluor 568 solution to counterstain cell nuclei. About 10–15 characteristic pictures/well were taken by a fluorescent microscope (Olympus IX-71) equipped with a Canon EOS digital camera (Canon, Tokyo, Japan). Then, cells were counted after merging images by Adobe Photoshop CS4, and the results were displayed as a percentage of total cells counted.
Western Blot
The protein of the cells treated was extracted, and BCA Kit (Sigma-Aldrich, Germany) was used to measure the protein concentrations. Proteins were separated by 12% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Danvers, MA, USA), and 5% nonfat milk was used to block PVDF membranes for 1 hour at 37°C. The dilution degrees of anti-OCT4 antibody (Abcam, USA), anti-Nanog antibody (Abcam, USA) and GAPDH (Abcam, USA) were 1:1000, 1:1000 and 1:2000, respectively. Three antibodies were hybridized overnight at 4°C and were combined with the HRP‑linked secondary antibody (BOSTER, Wuhan, China) for 2 hours at RT after PVDF membranes were washed. Finally, enhanced chemiluminescence (ECL) (General Electric Healthcare, Aurora, OH, USA) was used to visualize the bands of the proteins.
qRT-PCR Was Performed to Determine the Relative Expression of Methyltransferase Genes and miRNA
Total RNA was isolated using MiniBEST Universal RNA Extraction Kit (TaKaRa, Japan) and regarded as a template. The random primer Oligo (dT) was reverse-transcribed into cDNA with Primescript RT reagent kit with gDNA Eraser (perfect real time) (TaKaRa, Japan), and cDNA served as the template for qPCR. BioRad’s SYBR Green kit was used to carry out qPCR experiment. The GAPDH gene was regarded as the reference. qPCR was performed using a CFX-96 Real-Time System (Bio-Rad), at 95°C for 10 minutes, then 95°C for 10 seconds, 60°C for 15 seconds and 72°C for 20 seconds for 40 cycles. The sequences of the primers used for detecting genes are shown in Supplemental Table 1.
MiniBEST Universal RNA Extraction Kit (TaKaRa, Japan) was used to extract miRNA from the cell samples, and the reverse primers corresponding to each miRNA were used to reverse-transcribe with Primescript RT reagent kit with gDNA Eraser (TaKaRa, Japan); and the product served as the templates of qPCR. U6 was considered as the reference. qPCR was carried out according to the above method. The universal reverse primer of miRNA detected is 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ. The reverse primers and forward primers of miRNA measured are displayed in Supplemental Table 2. All experiments were repeated three times in each group; 2−ΔΔCt was used in the quantification of mRNA and miRNA.
Detection of OCT3/4 Promoter Methylation by Sequencing After Bisulfite Treatment
The genome of the samples was extracted according to the instructions of the blood/cell/tissue DNA extraction kit (Tiangen), and the concentration and purity were measured by ultraviolet spectrophotometer. DNA was treated with bisulfite using bisulfite DNA methylation kit (QIAGEN). The forward and reverse primers of OCT3/4 promoter methylation were GGGTAATAAAGTGAGATTTTGTTTTAAAAA(5ʹ-3ʹ) and ATCCCACCCACTAACCTTAACCTCTAAC(5ʹ-3ʹ), respectively.17 The 50 μL PCR system included 25 μL 2×TransTaq DNA Polymerase High Fidelity (TransGen, China), 5 μL DNA, 0.5 μL forward primer and 0.5 μL reverse primer, as well as 19 μL water. PCR was performed at 95°C for 10 minutes, then 95°C for 30 seconds, 60°C for 30 seconds and 72°C for 30 seconds, for 35 cycles, and finally 72°C extended for 7 min; 1.5% of agarose gel electrophoresis was used to detect PCR products. The recovered and purified PCR fragments were linked to T-carrier, and the positive clones were sequenced.
Chromatin Immunoprecipitation (ChIP)
The cell samples were submerged in 1% formaldehyde and vacuumed for 10 minutes at 37°C to cross-link the genomic DNA and proteins. The cross-linking reaction was stopped by adding glycine to a final concentration of 0.125 M for 5 minutes at RT. After removing culture medium, the samples were placed for 5 minutes at RT. Subsequently, 5–10 mL ice bath precooling PBS including 1 mM PMSF was used to wash cells for 2–3 times. Then, 1 mL ice bath precooling PBS including 1 mM PMSF was added to the plates, and cells were scrapped and centrifuged. The cells were divided into two parts: one of them was used to optimize the conditions of ultrasound (ultrasound conditions: 3 seconds on, 3 seconds off, 20% power, 2, 4, 6, 8, 10, 12 minutes processing time) to breaking most genomic DNA into 200–1000 bp sizes, and the agarose gel electrophoresis was used to observed the shear effect of ultrasonic treatment on genomic DNA.
Ultrasound samples were centrifuged, supernatant was taken and a suitable amount of ChIP dilution buffer containing 1 mM PMSF was added. Two tubes of 100 µL supernatant were taken from the same sample for input detection. The samples were processed according to the instructions of the ChIP assay kit (Abcam, ab500, USA). One tube was treated with acetyl-histone H3-K9 (ABClonal, China), trimethyl-histone H3-K9 (ABClonal, China) and incubated overnight at 4°C, and the other tube was used as control without one primary antibody. For assays, the isolated DNA was considered as a template for ChIP-qPCR amplification. The forward and reverse primers of OCT3/4 gene for ChIP-qPCR were ATCGGGATGGGAATGCCTAG (5ʹ-3ʹ) and CTCCCACACCTCCATGTTCT (5ʹ-3ʹ), respectively.
Statistical Analysis
Data analysis was carried out by SPSS 26.0 software using one-way ANOVA followed by Tukey’s post-hoc test. All data were presented as mean ± SEM, and P<0.05 was considered as the significance level between groups. Significance is indicated by * P<0.05, ** P<0.01 and *** P<0.001 vs the blank group.
Results
IC50 of A549 and NCI-H226 Cells and Cell Proliferation Assay
When different concentrations of CDDP acted on A549 and NCI-H226 cells, the cell proliferation ability decreased with the increasing concentration (Supplemental Figure 1). Fifty percent of cell cytotoxicity/proliferation inhibition caused by the concentration of CDDP was regarded as IC50. These values for A549 and NCI-H226 at 48 hours were found to be 7.337 μg/mL and 3.268 μg/mL, respectively. There was a remarkable difference between cells treated and untreated with CDDP in all experiments by one-way ANOVA analysis (P<0.05).
The results of cell proliferation assay by BrdU incorporation (Figure 1) showed that the proliferation activity of A549 and NCI-H226 cell lines significantly decreased under the action of CDDP and 5-Fu, respectively (P<0.05), compared to the blank group. These concentrations of CDDP and 5-Fu caused inhibition on A549 and NCI-H226 cell lines compared to untreated cell.
 |
Figure 1 Cell proliferation assay by BrdU incorporation. Note: **P<0.01 and ***P<0.001 vs Blank group. |
Cell Apoptosis After 5-Fu and CDDP Treatment by Flow Cytometry
Flow cytometry showed that apoptosis of A549 and NCI-H226 cell lines increased remarkably with the treatment of 5-Fu and CDDP, and the effect of CDDP was more obvious. Statistical analysis suggested that NCI-H226 cells were more susceptible to apoptosis induced by CDDP, and A549 cells were more susceptible to apoptosis induced by 5-Fu than NCI-H226, especially early apoptosis (Figure 2A-B).
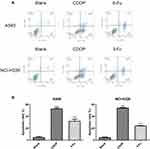 |
Figure 2 The result of Annexin V/PI experiments on both A549 and NCI-226 cell lines. (A) The cells were measured by flow cytometric analysis. (B) The apoptotic rate. ***P<0.001 vs Blank group |
5-Fu Induces the Expression of OCT3/4 in A549 Cells
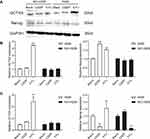
To analyze the effects of 5-Fu and CDDP exposure on OCT3/4 expression, A549 and NCI-H226 were exposed to 5-Fu (100 μg/mL) and CDDP as mentioned earlier and then plated in differentiation culture medium for 48 h. Immunofluorescence analysis showed that the expression of OCT3/4-positive cells increased in A549 cells and was significantly increased with 5-Fu treatment (Figure 3A, P<0.05). The expression of OCT3/4-positive cells in NCI-H226 increased with 5-Fu and CDDP treatment, but there was no statistical significance (Figure 3B, P>0.05).
The expression of OCT3/4 and Nanog was analyzed in A549 and NCI-H266 cells after 5-Fu and CDDP treatment by Western blotting and qPCR (Figure 4). Interestingly, there were fewer Nanog- and OCT3/4-positive cells in A549 blank group compared to the 5-Fu-treated group (Figure 4A and B). These results suggested that 5-Fu promoted the expression of OCT3/4 and Nanog during repair of the lung adenocarcinoma.
The levels of OCT3/4 and Nanog mRNA were examined using RT-PCR (Figure 4C). These data revealed higher OCT3/4 and Nanog expression in the 5-Fu-treated group compared to the blank group for A549 cells. The expression of Nanog mRNA increased after CDDP treatment in the NCL-H226 cell line, but decreased after 5-Fu treatment, and there was no significant change in OCT3/4 expression before and after treatment. Thus, the expression of OCT3/4 and Nanog was enhanced by 5-Fu in A549 cells.
DNA Methyltransferase Activity
Compared with the blank group, DNMT1 activity significantly decreased after 5-Fu or CDDP treatment at 48 h for A549 and NCI-H226 cells (Figure 5A and B). DNMT3a, DNMT3b and MBD2 activity was similar to those of normal cells after 5-Fu treatment (48 hours). Thus, DNMT3a in NCI-H226 cells significantly increased, but DNMT3a significantly decreased after 5-Fu/CDDP treatment. Thus, significant differences were observed in DNA methyltransferase activity between normal cells and 5-Fu/CDDP treated cells.
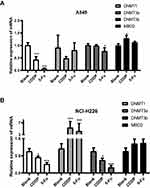 |
Figure 5 DNA methyltransferase activity. (A and B) The expressions of DNMT1, DNMT3a, DNMT3b and MBD2 in A549 cells and NCI-H226 cells, respectively. *P<0.05 and ***P<0.001 vs Blank group. |
Enhanced Expression of has-miR-134 and has-miR-296 in A549 Cells
To further understand the mechanism of 5-Fu treatment in A549 and NCI-H226 cells, we examined the level of has-miR-134 and has-miR-296. Figure 6A demonstrates that these miRNAs were significantly increased with CDDP and 5-Fu treatment in A549 cells. These miRNAs also had significantly increased levels of NCI-H226 after treatment with 5-Fu compared to blank group as shown in Figure 6B, but there was no significant difference between CDDP group and blank group.
The Methylation of OCT3/4 Promoter in A549 Cells
Based on the results of immunofluorescence, sequencing after bisulfite treatment (BSP) was used to detect the methylation of OCT3/4 promoter in human lung adenocarcinoma A549 cells. As shown in Figure 7, OCT3/4 promoter demethylation occurred at site 153 under the action of 5-Fu, and OCT3/4 promoter demethylation occurred at sites 194 bp and 201 bp under the action of CDDP. Because the amplification efficiency of methylated PCR is affected by the methylation sites and the degree of methylation, the blue and white spot screening of TA clone showed that the positive clone spots obtained in CDDP group and 5-Fu group were significantly decreased. Cloning and sequencing comparison showed that the methylation sites of all positive clone spots were significantly different in the drug group compared with the control group. The methylation sites were found at 200 bp in the blank group, while methylation sites were found at 151 bp and 193 bp in the 5-Fu group. There are some genetic differences between CDDP and 5-Fu treatment in A549 cells from the above results.
 |
Figure 7 The methylation of OCT3/4 promoter in A549 cells. |
The Expression of H3K9Me3 and H3K9Ace by ChIP
ChIP assay showed that the expression of H3K9Me3 significantly decreased and H3K9Ace increased in the A549 cell line as shown in Figure 8A and B. There was a similar trend in NCI-H226 cells, but the level of H3K9Ace was not significantly different between CDDP group and blank group.
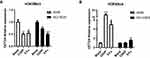 |
Figure 8 The expression of H3K9Me3 and H3K9Ace by ChIP. (A and B) The expression of H3K9Me3 and H3K9Ace in A549 and NCI-H226 cells, respectively. *P<0.05, **P<0.01 and ***P<0.001 vs Blank group. |
Discussion
OCT3/4 and Nanog are important transcription factors, which are involved in regulating the self-renewal of embryonic stem cells and maintaining their totipotency, especially OCT3/4, and OCT3/4 is also a key gene for inducing multifunctional stem cells in vitro. Both in vivo and in vitro chemoradiotherapy experiments have shown that the resistance of lung cancer was associated with OCT3/4 gene expression.18,19 In this study, the expression and methylation status of the OCT3/4 and Nanog genes in A549 and NCI-H226 cells with 5-Fu/CDDP treatment were analyzed. Our results indicated that 5-Fu can enrich A549 lung adenocarcinoma cells, and OCT3/4 can be used as cancer stem cell markers in A549. The number of OCT3/4-positive cells remarkably increased in A549 cells treated with 5-Fu; this suggests that OCT3/4-expressing lung cancer stem cells were selectively activated in response to 5-Fu. For A549 cells, the expression of Nanog decreased after CDDP treatment, and that of NCI-H226 cells after 5-Fu treatment. This may be due to insufficient sample size. This problem will be avoided in future experiments.
DNA methylation mainly occurs at the CpG dinucleotide position and is mainly catalyzed by two important types of DNA methyltransferase (Dnmt). The DNMT family, especially Dnmt1, Dnmt3a and Dnmt3b, has been found to play an important role in the initial methylation of DNA and the maintenance of methylation. Abnormal activity of DNMT family is involved in the occurrence and development of tumors and is an important factor leading to methylation of tumor suppressor genes.20 Dnmt1 is a maintenance enzyme that maintains the semi-methylation of CpG dinucleotide in the newly cloned DNA of DNA in the methylation state.21 Therefore, its function is very important for the maintenance of methylation in the genomes of expanding cells.22 Dnmt3a and Dnmt3b are very important for de novo methylation in vivo and for establishing a new methylation state.23 Both of them cooperate to methylate the promoter regions of the OCT3/4 and Nanog genes during the differentiation of embryonic carcinoma cells and ES cells, and the removal of them can lead to insufficient methylation and disordered OCT3/4 expression.24,25 Here, we demonstrated that in A549 and NCI-H226 lung cells the activity of the maintenance methyltransferase DNMT1 and DNMT3b decreased after 5-Fu/CDDP treatment, but DNMT3a in NCI-H226 cells significantly increased. DNMT3a was increased in the NCI-H226 cell line, which may be related to the interference of other factors in the NCI-H226 cell line. We will continue to find relevant literature and conduct research on this issue. The difference of DNA in the methylation state in A549 and NCI-H226 might contribute to the expression of OCT3/4. The methylation sites were also different in A549 cells treated with CDDP and 5-Fu by BSP. This suggested that the mechanism of CDDP and 5-Fu were different on the different lung cells, and the mechanisms of CDDP and 5-Fu on the same lung cell were also different.
Histone modifications which were related to transcriptional activation such as H3K9Ace were largely absent in OCT3/4-negative cells, while markers linked with transcriptional repression were present, for example H3K9Me.26 This state was in agreement with maintenance of a heterochromatin state. The absence of the OCT3/4 transcription factor in these cells showed that the cells were differentiated. However, H3K9Ace was present at high levels in OCT3/4-positive cells, while H3K9Me3 was largely absent.27 This state was in agreement with a euchromatin state. The presence of the OCT3/4 transcription factor indicated that the cells were undifferentiated. In this study, the results showed that OCT3/4-positive cells increased remarkably in A549 cells treated with 5-Fu by immunofluorescence and Western blot experiments. And the expression of H3K9Me3 decreased significantly and the expression of H3K9Ace increased significantly in A549 cells treated with 5-Fu. According to the results of previous studies, it shows that A549 cells treated with 5-Fu were in a euchromatin state and were undifferentiated. This suggests that 5-Fu can enrich A549 lung cancer stem cells by participating in regulating the expression of OCT3/4.
CDDP and 5-Fu can suppress the growth of cancer cells. In this study, the proliferation of both A549 and NCI-H226 cells was suppressed by 5-Fu/CDDP. The apoptosis of both cells increasedremarkably after 5-Fu/CDDP treatment, whereas the effect of CDDP was more obvious. NCI-H226 cells were more susceptible to apoptosis induced by CDDP, while A549 cells were more susceptible to apoptosis induced by 5-Fu than NCI-H226, especially early apoptosis.
MicroRNAs (miRNAs), which are a class of small non-coding RNA molecules, are proposed to play key roles in the development of tumor progression and invasion. A number of publications have reported that dysregulation of miRNAs plays a key role in many types of cancers.28,29 Tumor proliferation, apoptosis, senescence, cell identity, stem cell maintenance and metastasis are related to miRNA expression.30,31 MiR-134 played a pivotal role on NSCLC through inhibiting cell proliferation, migration and invasion, and promoting apoptosis by targeting oncogenic CCND1.32 MiR-134 suppresses epithelial-to-mesenchymal transition by targeting Forkhead Box M1 in A549 cells.33 MiR-296-5p suppresses NSCLC viability by taking aim at Polo-like kinase 1 through combining to the putative binding sites in its 3ʹ-untranslated region (3ʹ-UTR).34 MiR-134, an expression inhibitor of pluripotency markers including OCT4, Sox2 and Nanog, was decreased both in BPA-treated mESC and mEB.35 MiR-296, miR-470 and miR-134 were all significantly upregulated relative to their expression at day 0 in undifferentiated mouse ESCs, and were also predicted computationally by rna22 to target the CDSs of Nanog, OCT4 and Sox2 in various combinations.36 These studies suggest that has-miR-134 and has-miR-296 help to suppress the development of tumor. In our study, the levels of has-miR-134 and has-miR-296 were significantly improved after CDDP or 5-Fu treatment, indicating that CDDP and 5-Fu enhance the expression of has-miR-134 and has-miR-296 to inhibit the development of lung cancer. This result is consistent with the result of Annexin V/PI and BrdU incorporation experiments, where both CDDP and 5-Fu promoted lung cell apoptosis and inhibited cell proliferation.
The enrichment effect of CDDP on lung adenocarcinoma stem cells and lung squamous cell carcinoma stem cells is inconsistent with the enrichment effect of 5-Fu on lung adenocarcinoma stem cells. Nanog cannot be a marker of lung cancer stem cells. 5-Fu and CDDP cause epigenetic modification of lung adenocarcinoma and lung squamous cell carcinoma cells which provides new ideas for the identification of lung cancer stem cells and new clues and intervention targets for lung cancer treatment and drug resistance mechanism.
Data Sharing Statement
All data generated or analyzed during this study are included in this article.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81370100 (Xinshan Jia) and No. 30170407 (Xinshan Jia).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
We confirm that all authors have no conflict of interest. All authors have seen and approved the manuscript.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19.
3. Shao Y, Liang B, Long F, Jiang SJ. Diagnostic MicroRNA biomarker discovery for non-small-cell lung cancer adenocarcinoma by integrative bioinformatics analysis. Biomed Res Int. 2017;2017:2563085. doi:10.1155/2017/2563085
4. Niklinski J, Niklinska W, Laudanski J, Chyczewska E, Chyczewski L. Prognostic molecular markers in non-small cell lung cancer. Lung Cancer. 2001;34(Suppl 2):S53–S58. doi:10.1016/S0169-5002(01)00345-2
5. Sy SM, Wong N, Lee TW, et al. Distinct patterns of genetic alterations in adenocarcinoma and squamous cell carcinoma of the lung. Eur J Cancer. 2004;40(7):1082–1094. doi:10.1016/j.ejca.2004.01.012
6. McDoniels-Silvers AL, Nimri CF, Stoner GD, Lubet RA, You M. Differential gene expression in human lung adenocarcinomas and squamous cell carcinomas. Clin Cancer Res. 2002;8(4):1127–1138.
7. Avila-Moreno F, Lopez-Gonzalez JS, Galindo-Rodriguez G, Prado-Garcia H, Bajana S, Sanchez-Torres C. Lung squamous cell carcinoma and adenocarcinoma cell lines use different mediators to induce comparable phenotypic and functional changes in human monocyte-derived dendritic cells. Cancer Immunol Immunother. 2006;55(5):598–611. doi:10.1007/s00262-005-0060-3
8. Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21(3):283–296. doi:10.1016/j.ccr.2012.03.003
9. Zwaka TP, Thomson JA. Differentiation of human embryonic stem cells occurs through symmetric cell division. Stem Cells. 2005;23(2):146–149. doi:10.1634/stemcells.2004-0248
10. Kim RJ, Nam JS. OCT4 expression enhances features of cancer stem cells in a mouse model of breast cancer. Lab Anim Res. 2011;27(2):147–152. doi:10.5625/lar.2011.27.2.147
11. Chang CJ, Chien Y, Lu KH, et al. Oct4-related cytokine effects regulate tumorigenic properties of colorectal cancer cells. Biochem Biophys Res Commun. 2011;415(2):245–251. doi:10.1016/j.bbrc.2011.10.024
12. Hu T, Liu S, Breiter DR, Wang F, Tang Y, Sun S. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res. 2008;68(16):6533–6540. doi:10.1158/0008-5472.CAN-07-6642
13. Chiou SH, Wang ML, Chou YT, et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010;70(24):10433–10444. doi:10.1158/0008-5472.CAN-10-2638
14. Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3(5):330–338. doi:10.1038/nrc1074
15. Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer. 2011;71(1):3–10. doi:10.1016/j.lungcan.2010.08.022
16. Li X, Jia L, Jia X, et al. Enrichment of Oct3/4-positive cells from a human bronchial epithelial cell line. Apmis. 2013;121(7):612–625. doi:10.1111/apm.12028
17. Xu G, Yang L, Zhang W, Wei X. All the tested human somatic cells express both Oct4A and Its Pseudogenes but Express Oct4A at much lower levels compared with its pseudogenes and human embryonic stem cells. Stem Cells Dev. 2015;24(13):1546–1557. doi:10.1089/scd.2014.0552
18. Babaie Y, Herwig R, Greber B, et al. Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2007;25(2):500–510. doi:10.1634/stemcells.2006-0426
19. Yamagata K, Ueda J, Mizutani E, Saitou M, Wakayama T. Survival and death of epiblast cells during embryonic stem cell derivation revealed by long-term live-cell imaging with an Oct4 reporter system. Dev Biol. 2010;346(1):90–101. doi:10.1016/j.ydbio.2010.07.021
20. Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20(24):3139–3155. doi:10.1038/sj.onc.1204341
21. Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11(7):2611–2617. doi:10.1002/j.1460-2075.1992.tb05326.x
22. Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi:10.1016/0092-8674(92)90611-F
23. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi:10.1016/S0092-8674(00)81656-6
24. Gerhauser C, Heilmann K, Pudenz M. Genome-Wide DNA Methylation Profiling in Dietary Intervention Studies: a User’s Perspective. Current Pharmacol Rep. 2015;1(1):31–45. doi:10.1007/s40495-014-0001-y
25. Smith ZD, Chan MM, Mikkelsen TS, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484(7394):339–344. doi:10.1038/nature10960
26. Scholer HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344(6265):435–439. doi:10.1038/344435a0
27. Rosner MH, Vigano MA, Ozato K, et al. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345(6277):686–692. doi:10.1038/345686a0
28. Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7(1):36–41. doi:10.1016/j.stem.2010.06.012
29. Mei Q, Xue G, Li X, et al. Methylation-induced loss of miR-484 in microsatellite-unstable colorectal cancer promotes both viability and IL-8 production via CD137L. J Pathol. 2015;236(2):165–174. doi:10.1002/path.4525
30. Sun C, Li S, Yang C, et al. MicroRNA-187-3p mitigates non-small cell lung cancer (NSCLC) development through down-regulation of BCL6. Biochem Biophys Res Commun. 2016;471(1):82–88. doi:10.1016/j.bbrc.2016.01.175
31. Zhang T, Cai X, Li Q, et al. Hsa-miR-875-5p exerts tumor suppressor function through down-regulation of EGFR in colorectal carcinoma (CRC). Oncotarget. 2016;7(27):42225–42240. doi:10.18632/oncotarget.9944
32. Sun CC, Li SJ, Li DJ. Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. Oncotarget. 2016;7(24):35960–35978. doi:10.18632/oncotarget.8482
33. Li J, Wang Y, Luo J, et al. miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells. FEBS Lett. 2012;586(20):3761–3765. doi:10.1016/j.febslet.2012.09.016
34. Xu C, Li S, Chen T, et al. miR-296-5p suppresses cell viability by directly targeting PLK1 in non-small cell lung cancer. Oncol Rep. 2016;35(1):497–503. doi:10.3892/or.2015.4392
35. Chen X, Xu B, Han X, et al. Effect of bisphenol A on pluripotency of mouse embryonic stem cells and differentiation capacity in mouse embryoid bodies. Toxicol in Vitro. 2013;27(8):2249–2255. doi:10.1016/j.tiv.2013.09.018
36. Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi:10.1038/nature07299
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.