Back to Journals » OncoTargets and Therapy » Volume 14
Diagnosis of NUT Carcinoma Despite False-Negative Next-Generation Sequencing Results: A Case Report and Literature Review
Authors Wang X, Wang J, Luo X, Lu J, Wang L , Li Q, Wang EH
Received 3 July 2021
Accepted for publication 18 August 2021
Published 27 August 2021 Volume 2021:14 Pages 4621—4633
DOI https://doi.org/10.2147/OTT.S327722
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr William C. Cho
Xi Wang, Jinping Wang, Xue Luo, Jinxi Lu, Liang Wang, Qingchang Li, En-Hua Wang
Department of Pathology, The First Affiliated Hospital and College of Basic Medical Sciences, China Medical University, Shenyang, 110001, People’s Republic of China
Correspondence: Liang Wang
Department of Pathology, The First Affiliated Hospital and College of Basic Medical Sciences, China Medical University, Shenyang, 110001, People’s Republic of China
Tel/Fax +86 24 23261638
Email [email protected]
Abstract: Nuclear protein in testis (NUT) carcinoma (NC) is a poorly differentiated malignant tumor with a poor prognosis, which is caused by the NUTM1 gene rearrangement. Positive staining of NUT using immunohistochemistry (IHC) or gene rearrangement of NUTM1 revealed by genetic analysis, such as fluorescence in situ hybridization (FISH) or next-generation sequencing (NGS), are important strategies used for accurate diagnosis. In the current study, we present a case of NC in an 18-year-old man who had a chief complaint of nasal congestion, nasal bleeding, and anosmia. Magnetic resonance imaging revealed a mass in the nasal cavity and nasal septum. The initial pathological diagnosis was basaloid squamous cell carcinoma. Based on the tumor location and abrupt keratinization, further genetic tests were performed, and NC was diagnosed using FISH, which was further verified by IHC. However, neither DNA-based NGS nor RNA-based NGS revealed the NUTM1 gene rearrangement. Using this case as a basis, we have reviewed the related literature, compared the common diagnostic methods of NC, and discussed the advantages and limitations of current tools employed for molecular analysis of the gene fusion.
Keywords: NUTM1, NUT midline carcinoma, FISH, next-generation sequencing, gene rearrangement
Introduction
Nuclear protein in testis (NUT) carcinoma (NC) is defined by the rearrangement of the chromosomal region 15q14 harboring the NUTM1 gene. As a clinically aggressive neoplasm with poor differentiation, NC was previously believed to occur primarily in children and young adolescents. However, with an increasing number of reports, middle-aged or elderly patients have been diagnosed subsequently. NC mainly arises from midline anatomical sites, such as the mediastinum and head. Cases of primary pulmonary NC have recently been recognized. Because of the extremely poor prognosis and short survival time, timely diagnosis is of great significance for clinical treatment and management. NC presented in different regions has different clinical symptoms, and the morphological and immunohistochemical features are not specific. Therefore, the diagnosis of NC is mainly based on squamous differentiation and evidence of gene rearrangement of NUTM1, which leads to the ectopic expression of NUT protein outside the testis. The most common partner gene is BRD4,1 and other related genes, such as BRD3, NSD3, ZNF532, WHSC1L1, and NAP1L4 have also been identified.2–6 The gene rearrangement of NUTM1 can be identified by fluorescence in situ hybridization (FISH), reverse transcription-polymerase chain reaction (RT-PCR), Sanger DNA sequencing, and next-generation sequencing (NGS).7,8
Herein, we present a special case of NC that was diagnosed using FISH and confirmed by immunohistochemistry (IHC). However, the gene rearrangement of NUTM1 was not revealed by either DNA-based NGS or RNA-based NGS. In this case report, we review and discuss the advantages and disadvantages of NGS in the diagnosis of rare entities.
Materials and Methods
Histopathologic and Immunohistochemical Examination
Tissues were fixed in 10% neutral-buffered formalin and embedded in paraffin blocks. Tissue blocks were sectioned into 4-μm sections, affixed onto slides, deparaffinized in xylene, rehydrated with graded alcohols, and immunostained with the following antibodies: cytokeratin (CK), vimentin, p40, p63, S-100, chromogranin A, synaptophysin, and Ki-67 (MaiXin, Fuzhou, China). All samples were counterstained with hematoxylin, dehydrated, and mounted for microscopy observation. After the diagnosis of NC with FISH, it was further confirmed by staining the tissue slide with NUT antibody (clone C52B1, 1:200, Cell Signal Technology, BU, USA).
FISH
Paraffin-embedded tissue blocks were serially sectioned at 4-μm intervals. Hematoxylin eosin-staining of the sections was used to determine the area of the tissue to be targeted for analysis. The slides were deparaffinized in xylene twice for 10 min, dehydrated twice with 100% ethanol, and then pretreated using the ZytoLight FISH-Tissue Implementation Kit (Z-2028-20, ZytoVision, Bremerhaven, Germany). Slides were digested for 36 min in protease solution (0.5 mg/mL) at 37°C and examined with NUT (15q14) Break Apart Rearrangement Probe (LBP Medicine Science & Technology, Guangzhou, China) according to the manufacturer’s instructions. The slides were observed under a ×100 objective magnification using a fluorescence microscope (Leica DM6000, Wetzlar, Germany) equipped with Applied Imaging software (version 4.0; Genetix, England, UK). The samples were considered positive if more than 15% of the 100 scored tumor cells showed split signals. The slides were evaluated independently by two pathologists who were blinded to the case profiles.
NGS
NGS was performed on the Illumina HiSeq/MiSec platform (Illumina, CA, USA). This platform covers 1.28 Mb base sites such as exons, fusion-related introns, variable splicing regions, and microsatellite sites, accounting for 425 genes. The gene panel was shown in Supplementary Table 1. At least 50 ng of DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumor tissue from unstained slides, and peripheral blood was used as a control. The mean exon coverage depth of sequencing was >1000× and >200× for tissue and blood controls, respectively. RNA-NGS was performed on the HiSeq 2000 System (Illumina, CA, USA) using the TruSeq paired-end (PE) 100 bp Kit (Illumina, CA, USA).The detection results included point mutations, insertion/deletion mutations, gene fusion, gene copy number abnormalities, microsatellite status, and tumor mutation burden (TMB).
Clinical Summary
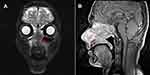
An 18-year-old man presented with nasal bleeding, anosmia, and bilateral nasal congestion for 5 months. Magnetic resonance imaging (MRI) revealed a mass measuring 20 mm in the nasal cavity, affecting the ethmoid sinus, sphenoid sinus, and hard palate bone (Figure 1). A biopsy was performed in the local hospital, and a diagnosis of basaloid squamous cell carcinoma was made. The patient was referred to our hospital for surgical treatment. During the surgery, a small tumor was identified and removed. An intraoperative frozen section was examined and diagnosed as basaloid squamous cell carcinoma. Postoperative pathological assessment of the specimens revealed a malignant epithelial tumor consisting of sheets and nests of atypia basaloid squamous cells. The periphery of the tumor nests was arranged in a palisade pattern. Abundant cytoplasm, frequent mitotic figures, and focal abrupt keratinization with few keratin pearls were evident (Figure 2). The results of IHC staining showed that tumor cells were positive for CK, p40, p63, and negative for vimentin, S-100, chromogranin A, and synaptophysin. The Ki-67 index was >60% (Figure 3).
 |
Figure 3 Immunohistochemistry staining of tumor cells that were positive for CK (A ×200), p40 (B ×200) and NUT (D ×40; inset ×400). Ki-67 index of tumor cells was >60% (C ×200). |
Before the diagnosis of basaloid squamous cell carcinoma was signed out, for such a young patient with a lesion in the midline, NC needed to be ruled out, especially when focal abrupt keratinization was present. Therefore, a FISH test for NUTM1 has been suggested. After providing detailed information, the patient requested genetic analysis by NGS in a third-party inspection institution. Without professional medical consultation, the patient’s guardian ordered the unsuitable DNA-based NGS for this case which might be harboring gene fusions. The NGS result showed a missense mutation of RAD50 (c.2293A > G [p.K765E]) in exon 14 with only 0.7% abundance. The TMB was 1.1 mutations/Mb. No NUTM1 gene translocation or microsatellite instability was identified. To avoid exon-skipping events and better cover gene fusions, we suggested that the patient undergoes a FISH test or an additional RNA-based NGS. The patient requested an RNA-based NGS in the same third-party inspection institution. The RNA-NGS results revealed no other mutations. Finally, the patient requested a FISH test in our hospital, which demonstrated that the proportion of 1 red 1 green 1 fusion signal was 23%, the proportion of 1 red 1 green signal was 10%, and that of 1 red 1 fusion signal was 5% (Figure 4). This indicated that gene rearrangement occurred in NUTM1. Accordingly, the diagnosis of NC was confirmed. Because of the negativity derived from NGS, immunostaining of NUT was performed with the NUT antibody (a generous gift from Dr. Shundong Dai, Department of Pathology, Shanghai Ninth People’s Hospital), and the diffuse strong staining of NUT was consistent with the FISH data (Figure 3D). The patient was treated with conventional radiotherapy and chemotherapy for 4 months after the operation. At present, the patient has no disease progression or recurrence for 12 months.
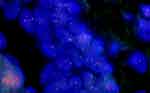 |
Figure 4 Fluorescence in situ hybridization demonstrated the gene rearrangement of NUTM1. Large proportion cells showing 1 red 1 green 1 fusion signal, indicating NUTM1 fused with partner genes. |
Discussion and Literature Review
NC is largely distributed in sheets and nest patterns, which are composed of round cells of the same size. It is similar to small round cell tumors, such as poorly differentiated squamous cell carcinoma, basaloid squamous cell carcinoma, and olfactory neuroblastoma.9 Markers of squamous epithelial differentiation are often expressed. It can be distinguished from other non-epithelial malignant tumors by CK, p63, and p40. Focal abrupt keratinization is an important diagnostic tool. In the current case, the tumor cells were distributed in round nests with an invasive growth pattern, similar to basaloid squamous cell carcinoma. IHC showed positive staining for CK, p40, and p63, making it difficult to differentiate them from basaloid squamous cell carcinoma. In 2020, Kohei et al found that p40, an isoform of p63, could only be focal or patchy positive in NC, and in certain cases completely negative.10 Therefore, even though p40 is recommended for identifying squamous cell carcinoma in clinical practice due to its better specificity, it is not suitable for identifying NC alone. In addition, some NCs showed no squamous differentiation, as reported by Prall et al.11 Immunostaining with the NUT antibody is particularly important. When NUT staining is diffusely positive (≥50), the diagnosis of NC can be rendered even in the absence of FISH results. When NUT staining signals are focally positive (<50%) or negative, and the diagnosis of NC is still highly suspected, FISH or other molecular tests should be performed to further confirm the rearrangement of NUTM1. However, since NUT antibody is far less commonly adopted in clinical detection, this method is not the usual strategy to diagnose NC. IHC targets the protein expression level, which is an indirect result of fusion mutations. In particular, false-positive or non-specific staining in IHC can lead to inaccurate evaluations. Therefore, FISH and NGS serve as better backup methods to reveal the molecular profiles in NC diagnosis. We have summarized different methods of diagnosis of NC in the last 20 years. Among the 68 patients, the presenting locations included the lung and trachea (22 cases), thorax and mediastinum (10 cases), nasal cavity and paranasal sinuses (12 cases), abdominal organs (9 cases), oral cavity and throat (6 cases), salivary glands (4 cases), orbit (2 cases), sellar (1 case), and lymph node (2 cases). Among these 68 cases, twenty-four cases were diagnosed by NUT IHC alone, 9 cases were diagnosed by FISH alone, 1 case was diagnosed by NGS alone, 22 cases were verified by FISH and NUT IHC simultaneously, and 7 cases were diagnosed by FISH and NGS together. Five cases showed positive results for all the three methods. The details of all cases are summarized in Table 1.12–51
 |  |  |  |
Table 1 NUT Carcinoma with Literature Review |
In these 68 cases, the disease occurred in all ages with a median age of 34.5 years (range 2–82 years), and the sex ratio of the incidence was 1:1. The tumor size was 10–100 mm, with a median of 44 mm. Follow-up data were available for 54 patients, of which 14 were alive during the follow-up period. Thirty-eight patients died with a median follow-up duration of 6 months (range, 1–30 months). In the 68 cases, most of the tumor cells were spherical, ovoid, or spindle-shaped with prominent nucleoli and scant cytoplasm. The tumor cells showed proliferation of nest-like, island-like, or cluster distribution. Fifty-eight patients showed abrupt keratinization. These pathological findings were consistent with the morphological features observed in the current case. Gene rearrangement was detected in 29 cases by FISH or NGS. Nineteen cases were BRD4-NUT, four were NSD3-NUT, five were MXD4-NUT, and one was NUT-MAML2. One patient was initially misdiagnosed with squamous cell carcinoma. The patient was sent to chemotherapy and showed no relief.18 Although NC was confirmed by NGS on re-evaluation, the patient quickly died with disease progression. Early accurate diagnosis can help physicians to improve clinical management and avoid misjudgment.
It should be noted that in one case, the result of NUT IHC was positive, while the result of FISH was negative.23 This may be related to the chromoplexy (complex chromosomal rearrangement in DNA). During cell proliferation, normal cells can be transformed into NUT cancer cells through NUTM1 rearrangement, which arises from multiple rearrangements in a single catastrophic event. Due to this complexity, FISH may generate a false-negative result. After transcription, this rearrangement can become a simple fusion transcript, which can be easily detected by IHC at the protein level.52 In addition to genetic chromoplexy, FISH results can also be affected by other factors, such as lack of adequate tumor cells on the slide, poor fluorescence signal caused by improper sample pre-treatment, signal loss, and the situation in which the labeled probe either does not hybridize with intended target sequences or hybridizes with nonspecific sequences that cause FISH failure.53,54 Meanwhile, when complex rearrangement patterns are detected, even if the nucleus shows atypical fusion signals, it may not meet the evaluation criteria in FISH and may be considered as negative.55,56 For instance, subtle and narrow split failing to meet the established criteria for a positive FISH score will be interpreted as negative.57 This type of subtle split often occurs in the event of chromosome deletion and inversion.58 When the probe does not cover the abnormal signal area, false-negative results are also generated.59 Although chromosome breakage occurred, they did not produce functional fusion products or lost expression in the process of disease progression, producing false-positive results.60 The atypical rearrangement of the 5ʹ probe asymmetric division or the deletion of the 5ʹ (telomeric) part may also lead to false positives in some tumor tissues.61,62 Moreover, partial FISH probes are DNA fragments that easily aggregate to form polymers, producing false-positive results. Another limitation of FISH is that it can only show whether there is gene fusion, but it cannot identify the fusion partner and location of the fracture point.60 In some cases, tumor cells were presented as small isolated islands in a background of reactive cells, leading to nuclear overlapping and crush artifacts.62 Thus, it is difficult to obtain accurate FISH results using fluorescence microscopy. In such situations, NGS is a better tool for genetic analysis.
NGS, also known as high-throughput sequencing, has evolved from Sanger sequencing. It can read a large number of DNA or RNA sequences at a single time at a lower cost. Currently, NGS can be divided into two types: DNA-based and RNA-based methods. DNA-NGS detects the cDNA obtained directly from DNA, while RNA-NGS detects the cDNA that was obtained using RNA reverse transcription. DNA-based NGS detects both exons and introns, and RNA-based NGS detects exons only. Although this difference is very small, it can result in a significant difference in the diagnosis results. With the widespread application of these two methods in clinical diagnosis, their limitations have been gradually discovered. Rearrangement and fusion of genes always occur in very long and complex intron regions. It is difficult to design probes to cover them comprehensively. Some splice site mutations can also cause abnormal transcription, which can easily lead to inconsistency between the fusion information detected and the real fusion information in the transcriptome. Song et al found that when the content of guanine and cytosine in the capture region of the probe was lower and the content of thymine and adenine repeat region was higher, the possibility of false negatives in DNA-NGS was higher.63 In addition, fusion detected at the DNA level may not be successfully transcripted. Some of the rare fusions detected by DNA-NGS fail to produce abnormal transcripts and proteins. Most of the gene analysis based on the DNA level focuses on the genes at the break point within the gene; however, for gene fusion, the more common are “genic-intergenic” and “intergenic- intergenic” fusions.64 Direct detection of RNA can focus on coding sequences rather than on introns. Besides, McEvoy et al have found that some cases harboring MXI1-NUT fusion detected by RNA-NGS were showed negative results for DNA NGS. This may be due to the low complexity of DNA sequence in the gene breakpoint.65 Li et al proposed that before using targeted treatments for patients, in addition to the detection results of DNA-NGS, orthogonal analysis for validation at the RNA or protein level is essential to provide more accurate treatment.66,67 However, the results of RNA-NGS are highly dependent on RNA quality. In practice, RNA can be degraded easier than DNA. The disadvantage of RNA-NGS is that, in certain cases, it is difficult to obtain enough RNA of the sufficient quality. These might be potential reasons why, for the current case, NUTM1 rearrangement was not identified by RNA-NGS in the third-party inspection institution but confirmed by FISH and IHC in our hospital. Unfortunately, after several rounds of repeat sectioning, the tissues left in the paraffin block was not enough for further verification by another round of RNA-NGS. Davies et al found that both RNA-NGS and DNA-NGS have the tendency of producing false-negative results in detecting gene fusion when validated with other methods.55 Therefore, to obtain an accurate diagnosis in rare cases, NGS results need to be verified by other molecular analyses tools.
Cohen et al introduced two diagnostic models combining DNA-NGS and RNA-NGS: parallel and sequential approaches.68 The parallel approach enables the simultaneous detection of DNA-NGS and RNA-NGS. In the sequential approach, DNA-NGS was first detected if the result was negative or atypical fusion, then RNA-NGS was essential. Through these two modes, the defects associated with the two detection methods can be avoided to a certain extent to improve the accuracy of diagnosis.
Conclusion
According to the current case, we found that even when combining DNA-NGS and RNA-NGS, it can still lead to false-negative results. This indicates that we cannot fully rely on one or two diagnostic methods, especially when facing rare cases. It is of great significance to recognize the advantages and disadvantages of each detection method and to avoid them consciously so that the accuracy of diagnosis can be assured.
Data Sharing Statement
The datasets supporting the conclusions of this article are included within the article.
Ethical Approval and Consent to Participate
Ethical approval for this study was obtained from the institutional ethic review boards of the First Affiliated Hospital of China Medical University. Writing consent to participate was provided by all patients for the present research.
Patient Consent for Publication
Informed consents were obtained from the patients for the publication of their cases and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by grants from Liaoning Technology Research Fund for Social Development and Industrialization to Liang Wang (2017225010).
Disclosure
The authors declare that they have no competing interests.
References
1. French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63(2):304–7.
2. Alekseyenko AA, Walsh EM, Zee BM, et al. Ectopic protein interactions within BRD4-chromatin complexes drive oncogenic megadomain formation in NUT midline carcinoma. Proc Natl Acad Sci U S A. 2017;114(21):E4184–E4192. doi:10.1073/pnas.1702086114
3. Suzuki S, Kurabe N, Ohnishi I, et al. NSD3-NUT-expressing midline carcinoma of the lung: first characterization of primary cancer tissue. Pathol Res Pract. 2015;211(5):404–408. doi:10.1016/j.prp.2014.10.013
4. Cheng Z, Luo Y, Zhang Y, et al. A novel NAP1L4/NUTM1 fusion arising from translocation t(11;15)(p15;q12) in a myeloid neoplasm with eosinophilia and rearrangement of PDGFRA highlights an unusual clinical feature and therapeutic reaction. Ann Hematol. 2020;99(7):1561–1564. doi:10.1007/s00277-020-04000-x
5. Bishop JA, Westra WH. NUT midline carcinomas of the sinonasal tract. Am J Surg Pathol. 2012;36(8):1216–1221. doi:10.1097/PAS.0b013e318254ce54
6. French CA, Rahman S, Walsh EM, et al. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov. 2014;4(8):928–941. doi:10.1158/2159-8290.CD-14-0014
7. Thompson LDR, Franchi A. New tumor entities in the 4th edition of the World Health Organization classification of head and neck tumors: nasal cavity, paranasal sinuses and skull base. Virchows Arch. 2018;472(3):315–330. doi:10.1007/s00428-017-2116-0
8. Haack H, Johnson LA, Fry CJ, et al. Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am J Surg Pathol. 2009;33(7):984–991. doi:10.1097/PAS.0b013e318198d666
9. Bishop JA, French CA, Ali SZ. Cytopathologic features of NUT midline carcinoma: a series of 26 specimens from 13 patients. Cancer Cytopathol. 2016;124(12):901–908. doi:10.1002/cncy.21761
10. Kohei M, Kashima J, Yatabe Y. The isoform matters in NUT carcinoma: a diagnostic pitfall of p40 immunohistochemistry. J Thorac Oncol. 2020;15(10):e176–e178. doi:10.1016/j.jtho.2020.07.017
11. Prall OWJ, Thio N, Yerneni S, Kumar B, McEvoy CR. A NUT carcinoma lacking squamous differentiation and expressing TTF1. Pathology. 2021;53(5):663–666. doi:10.1016/j.pathol.2020.09.027
12. Satoe N, Saito K, Motoi N, et al. P63-negative pulmonary NUT carcinoma arising in the elderly: a case report. Diagn Pathol. 2020;15(1):134. doi:10.1186/s13000-020-01053-4
13. Ciftci E, Demirsoy U, Anik Y, Gorur G, Corapcioglu F, Demir H. Staging and evaluation of neoadjuvant chemotherapy response with (1)(8) F-FDGPET/CT in NUT-midline carcinoma in a child: a case report and review of the literature. Rev Esp Med Nucl Imagen Mol. 2015;34(1):53–55.
14. Mao N, Liao Z, Wu J, et al. Diagnosis of NUT carcinoma of lung origin by next-generation sequencing: case report and review of the literature. Cancer Biol Ther. 2019;20(2):150–156. doi:10.1080/15384047.2018.1523852
15. Hsieh MS, French CA, Liang CW, Hsiao CH. NUT midline carcinoma: case report and review of the literature. Int J Surg Pathol. 2011;19(6):808–812. doi:10.1177/1066896909353600
16. Harms A, Herpel E, Pfarr N, et al. NUT carcinoma of the thorax: case report and review of the literature. Lung Cancer. 2015;90(3):484–491. doi:10.1016/j.lungcan.2015.10.001
17. Nakamura H, Tsuta K, Tsuda H, et al. NUT midline carcinoma of the mediastinum showing two types of poorly differentiated tumor cells: a case report and a literature review. Pathol Res Pract. 2015;211(1):92–98. doi:10.1016/j.prp.2014.07.006
18. Oliveira LJC, Gongora ABL, Latancia MT, et al. The first report of molecular characterized BRD4-NUT carcinoma in Brazil: a case report. J Med Case Rep. 2019;13(1):279. doi:10.1186/s13256-019-2213-6
19. Harada Y, Koyama T, Takeuchi K, Shoji K, Hoshi K, Oyama Y. NUT midline carcinoma mimicking a germ cell tumor: a case report. BMC Cancer. 2016;16(1):895. doi:10.1186/s12885-016-2944-3
20. D’Ambrosio L, Palesandro E, Moretti M, et al. Alpha-fetoprotein elevation in NUT midline carcinoma: a case report. BMC Cancer. 2017;17(1):266. doi:10.1186/s12885-017-3262-0
21. Suzuki S, Kurabe N, Minato H, et al. A rare Japanese case with a NUT midline carcinoma in the nasal cavity: a case report with immunohistochemical and genetic analyses. Pathol Res Pract. 2014;210(6):383–388. doi:10.1016/j.prp.2014.01.013
22. Li YY, Liu Y, Ke XX, Lu Y. The primary pulmonary NUT carcinomas and some uncommon somatic mutations identified by next-generation sequencing: a case report. AME Case Rep. 2020;4:24. doi:10.21037/acr-19-168
23. McLean-Holden AC, Moore SA, Gagan J, et al. NUT carcinoma in a patient with unusually long survival and false negative FISH results. Head Neck Pathol. 2020;15(2):698–703.
24. Kakkar A, Antony VM, Irugu DVK, Adhikari N, Jain D. NUT midline carcinoma: a series of five cases, including one with unusual clinical course. Head Neck Pathol. 2018;12(2):230–236. doi:10.1007/s12105-017-0858-2
25. Van Treeck BJ, Thangaiah JJ, Torres-Mora J, et al. NUTM1-rearranged colorectal sarcoma: a clinicopathologically and genetically distinctive malignant neoplasm with a poor prognosis. Mod Pathol. 2021;34(8):1547–1557. doi:10.1038/s41379-021-00792-z
26. Chen M, Yang J, Lv L, et al. Comprehensive genetic profiling of six pulmonary NUT carcinomas with a novel micropapillary histological subtype in two cases. Hum Pathol. 2021;115:56–66. doi:10.1016/j.humpath.2021.02.004
27. Saik WN, Da Forno P, Thway K, Khurram SA. NUT carcinoma arising from the parotid gland: a case report and review of the literature. Head Neck Pathol. 2020. doi:10.1007/s12105-020-01254-91
28. D’Souza JN, Notz G, Bogdasarian RN, et al. Orbital involvement by NUT midline carcinoma. Ophthalmic Plast Reconstr Surg. 2015;31(6):e147–150. doi:10.1097/IOP.0000000000000179
29. Cho HJ, Lee HK. Lung nuclear protein in testis carcinoma in an elderly Korean woman: a case report with cytohistological analysis. Thorac Cancer. 2020;11(6):1724–1727. doi:10.1111/1759-7714.13438
30. Arimizu K, Hirano G, Makiyama C, Matsuo M, Sasaguri T, Makiyama A. NUT carcinoma of the nasal cavity that responded to a chemotherapy regimen for Ewing’s sarcoma family of tumors: a case report. BMC Cancer. 2018;18(1):1134. doi:10.1186/s12885-018-5087-x
31. Andreasen S, French CA, Josiassen M, Hahn CH, Kiss K. NUT carcinoma of the sublingual gland. Head Neck Pathol. 2016;10(3):362–366. doi:10.1007/s12105-015-0672-7
32. Bjornstrup LR, Reibel J, Kiss K, Schioedt M. NUT carcinoma presenting in the palate - a case report. APMIS. 2017;125(10):933–936. doi:10.1111/apm.12710
33. Boleto G, Perotin JM, Launois C, et al. Nuclear protein in testis carcinoma of the mediastinum: a case report. J Med Case Rep. 2017;11(1):152. doi:10.1186/s13256-017-1328-x
34. Cao J, Chen D, Yang F, Yao J, Zhu W, Zhao C. NUT midline carcinoma as a primary lung tumor: a case report. J Thorac Dis. 2017;9(12):E1045–E1049. doi:10.21037/jtd.2017.11.50
35. Chai P, Zhou C, Jia R, Wang Y. Orbital involvement by NUT midline carcinoma: new presentation and encouraging outcome managed by radiotherapy combined with tyrosine kinase inhibitor: a case report. Diagn Pathol. 2020;15(1):2. doi:10.1186/s13000-019-0922-1
36. Cho YA, Choi YL, Hwang I, Lee K, Cho JH, Han J. Clinicopathological characteristics of primary lung nuclear protein in testis carcinoma: a single-institute experience of 10 cases. Thorac Cancer. 2020;11(11):3205–3212. doi:10.1111/1759-7714.13648
37. den Bakker MA, Beverloo BH, van den Heuvel-eibrink MM, et al. NUT midline carcinoma of the parotid gland with mesenchymal differentiation. Am J Surg Pathol. 2009;33(8):1253–1258. doi:10.1097/PAS.0b013e3181abe120
38. Dragoescu E, French C, Cassano A, Baker S
39. Baine MJ, Elkhatib SK, Neilsen BK, Sleightholm RL, Zhen W. A 47-year-old woman with nuclear protein in testis midline carcinoma masquerading as a sinus infection: a case report and review of the literature. J Med Case Rep. 2019;13(1):57. doi:10.1186/s13256-019-2015-x
40. Gokmen-Polar Y, Kesler K, Loehrer PJ
41. Klijanienko J, Le Tourneau C, Rodriguez J, Caly M, Theocharis S. Cytological features of NUT midline carcinoma arising in sino-nasal tract and parotid gland: report of two new cases and review of the literature. Diagn Cytopathol. 2016;44(9):753–756. doi:10.1002/dc.23506
42. Kuroda S, Suzuki S, Kurita A, et al. Cytological features of a variant NUT midline carcinoma of the lung harboring the NSD3-NUT fusion gene: a case report and literature review. Case Rep Pathol. 2015;2015:572951.
43. Chen L, Ma Y, Feng J, Xiao X. NUT midline carcinoma in the mediastinum in a ten-year-old boy. Arch Bronconeumol. 2018;54(10):539–541. doi:10.1016/j.arbres.2017.09.010
44. Mills AF, Lanfranchi M, Wein RO, et al. NUT midline carcinoma: a case report with a novel translocation and review of the literature. Head Neck Pathol. 2014;8(2):182–186. doi:10.1007/s12105-013-0479-3
45. Policarpio-Nicolas ML, de Leon EM, Jagirdar J. Cytologic findings of NUT midline carcinoma in the hilum of the lung. Diagn Cytopathol. 2015;43(9):739–742. doi:10.1002/dc.23291
46. Rutt AL, Poulik J, Siddiqui AH, et al. NUT midline carcinoma mimicking tonsillitis in an eight-year-old girl. Ann Otol Rhinol Laryngol. 2011;120(8):546–549. doi:10.1177/000348941112000810
47. Shehata BM, Steelman CK, Abramowsky CR, et al. NUT midline carcinoma in a newborn with multiorgan disseminated tumor and a 2-year-old with a pancreatic/hepatic primary. Pediatr Dev Pathol. 2010;13(6):481–485. doi:10.2350/09-10-0727-CR.1
48. Ball A, Bromley A, Glaze S, French CA, Ghatage P, Kobel M. A rare case of NUT midline carcinoma. Gynecol Oncol Case Rep. 2012;3:1–3.
49. Ueki H, Maeda N, Sekimizu M, Yamashita Y, Moritani S, Horibe K. A case of NUT midline carcinoma with complete response to gemcitabine following cisplatin and docetaxel. J Pediatr Hematol Oncol. 2014;36(8):e476–e480. doi:10.1097/MPH.0000000000000082
50. Zhang H, Liu MH, Zhang J, et al. Successful treatment of a case with NUT midline carcinoma in the larynx and review of the literature. Clin Case Rep. 2020;8(1):176–181. doi:10.1002/ccr3.2568
51. Zhou L, Yong X, Zhou J, Xu J, Wang C. Clinicopathological analysis of five cases of NUT midline carcinoma, including one with the gingiva. Biomed Res Int. 2020;2020:9791208.
52. Lee JK, Louzada S, An Y, et al. Complex chromosomal rearrangements by single catastrophic pathogenesis in NUT midline carcinoma. Ann Oncol. 2017;28(4):890–897. doi:10.1093/annonc/mdw686
53. McLeer-Florin A, Moro-Sibilot D, Melis A, et al. Dual IHC and FISH testing for ALK gene rearrangement in lung adenocarcinomas in a routine practice. J Thorac Oncol. 2012;7(2):348–354. doi:10.1097/JTO.0b013e3182381535
54. Camidge DR, Kono SA, Flacco A, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin Cancer Res. 2010;16(22):5581–5590. doi:10.1158/1078-0432.CCR-10-0851
55. Davies KD, Le AT, Sheren J, et al. Comparison of molecular testing modalities for detection of ROS1 rearrangements in a cohort of positive patient samples. J Thorac Oncol. 2018;13(10):1474–1482. doi:10.1016/j.jtho.2018.05.041
56. Camidge DR, Skokan M, Kiatsimkul P, et al. Native and rearranged ALK copy number and rearranged cell count in non-small cell lung cancer: implications for ALK inhibitor therapy. Cancer. 2013;119(22):3968–3975. doi:10.1002/cncr.28311
57. Rogers TM, Russell PA, Wright G, et al. Comparison of methods in the detection of ALK and ROS1 rearrangements in lung cancer. J Thorac Oncol. 2015;10(4):611–618. doi:10.1097/JTO.0000000000000465
58. Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16(5):1561–1571. doi:10.1158/1078-0432.CCR-09-2845
59. Ren S, Hirsch FR, Varella-Garcia M, et al. Atypical negative ALK break-apart FISH harboring a crizotinib-responsive ALK rearrangement in non-small-cell lung cancer. J Thorac Oncol. 2014;9(3):e21–23. doi:10.1097/JTO.0000000000000013
60. Heydt C, Ruesseler V, Pappesch R, et al. Comparison of in situ and extraction-based methods for the detection of ROS1 rearrangements in solid tumors. J Mol Diagn. 2019;21(6):971–984. doi:10.1016/j.jmoldx.2019.06.006
61. Dacic S, Villaruz LC, Abberbock S, Mahaffey A, Incharoen P, Nikiforova MN. ALK FISH patterns and the detection of ALK fusions by next generation sequencing in lung adenocarcinoma. Oncotarget. 2016;7(50):82943–82952. doi:10.18632/oncotarget.12705
62. Sholl LM, Weremowicz S, Gray SW, et al. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol. 2013;8(3):322–328. doi:10.1097/JTO.0b013e31827db604
63. Song Z, Lu C, Xu CW, Zheng Z. Noncanonical gene fusions detected at the DNA level necessitate orthogonal diagnosis methods before targeted therapy. J Thorac Oncol. 2021;16(3):344–348. doi:10.1016/j.jtho.2020.12.006
64. Yun JW, Yang L, Park HY, et al. Dysregulation of cancer genes by recurrent intergenic fusions. Genome Biol. 2020;21(1):166. doi:10.1186/s13059-020-02076-2
65. McEvoy CR, Holliday H, Thio N, et al. A MXI1-NUTM1 fusion protein with MYC-like activity suggests a novel oncogenic mechanism in a subset of NUTM1-rearranged tumors. Lab Invest. 2021;101(1):26–37. doi:10.1038/s41374-020-00484-3
66. Li W, Liu Y, Li W, Chen L. Intergenic breakpoints identified by DNA sequencing confound targetable kinase fusion detection in NSCLC. J Thorac Oncol. 2020;15(7):1223–31.
67. Li W, Guo L, Liu Y. Potential unreliability of uncommon ALK, ROS1, and RET genomic breakpoints in predicting the efficacy of targeted therapy in NSCLC. J Thorac Oncol. 2021;16(3):404–18.
68. Cohen D, Hondelink LM, Solleveld-Westerink N. Optimizing mutation and fusion detection in NSCLC by sequential DNA and RNA sequencing. J Thorac Oncol. 2020;15(6):1000–1014. doi:10.1016/j.jtho.2020.01.019
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.