Back to Journals » International Journal of General Medicine » Volume 15
Comprehensive Analysis of the E2F Transcription Factor Family in Human Lung Adenocarcinoma
Authors Wang Q, Liu J, Cheang I , Li J , Chen T, Li Y, Yu B
Received 7 May 2022
Accepted for publication 28 June 2022
Published 2 July 2022 Volume 2022:15 Pages 5973—5984
DOI https://doi.org/10.2147/IJGM.S369582
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Scott Fraser
Qixun Wang,1,* Jinping Liu,1,* Iokfai Cheang,2,* Jinghang Li,3 Tingzhen Chen,4 Yanxiu Li,4 Bo Yu1
1Department of Cardiovascular Surgery, The First People’s Hospital of Lianyungang, Lianyungang Clinical College of Nanjing Medical University, Lianyungang, 222000, Jiangsu, People’s Republic of China; 2Department of Cardiology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210009, People’s Republic of China; 3Department of Cardiovascular Surgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210009, People’s Republic of China; 4Department of Critical Care Medicine, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210009, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Bo Yu, Department of Cardiovascular Surgery, The First People’s Hospital of Lianyungang, Lianyungang Clinical College of Nanjing Medical University, Lianyungang, 222000, Jiangsu, People’s Republic of China, Email [email protected] Yanxiu Li, Department of Critical Care Medicine, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210009, People’s Republic of China, Email [email protected]
Background: E2F transcription factors (E2Fs), code a family of pivotal transcription factors, have been identified as key regulators in tumor tumorigenesis. However, the function of E2F family in human lung adenocarcinoma (LUAD) have not been fully elucidated.
Methods: Herein, The Cancer Genome Atlas (TCGA) databases, Kaplan-Meier plotter, cBioPortal and TIMER were used to analyze differential expression, prognostic value, genetic alteration and immune cell infiltration of E2Fs in LUAD patients.
Results: The expression levels of E2Fs (E2F1-8) were all significantly upregulated in LUAD tissues compared with normal lung tissues. All eight E2Fs had low rates of gene mutation in LUAD patients from cBioPortal databases. Survival analysis revealed that E2F2 (P=0.038; HR 1.36; 95% CI 1.02– 1.81), E2F7 (P< 0.001; HR 1.78; 95% CI 1.33– 2.39) and E2F8 (P=0.03; HR 1.37; 95% CI 1.02– 1.82) were significantly associated with poor prognosis. Multivariate cox regression analysis found that only E2F7 (P< 0.001; HR 2.72; 95% CI 1.75– 4.25) was an independent prognostic predictor in LUAD after adjusting common clinical parameters. The receiver operating characteristic (ROC) analysis also found that E2F7 had high diagnostic value for LUAD (AUC=0.901). Further analysis found that E2F7 was significantly associated with LUAD immune cell infiltration of B cell, T cell, neutrophil, and myeloid dendritic cell. E2F7 also have positive correlations with immune checkpoint genes including SIGLEC15, CD274, HAVCR2, PDCD1LG2, CTLA4, TIGIT, LAG3 and PDCD1 in LUAD.
Conclusion: Our findings showed various association of E2F7 in LUAD diagnostic and prognostic aspects, which suggested its potential in becoming a novel biomarker.
Keywords: lung adenocarcinoma, E2F transcription factors, TCGA, prognosis, immune cell infiltration
Introduction
E2Fs, as pivotal transcription factor members, play a critical role in regulating downstream genes transcription, DNA synthesis and cell cycle.1 Mammalian E2F family are generally divided into the following three categories according to its functional and structural characteristics: (1) transcription activators (E2F1, E2F2 and E2F3a), which are transcription activators of E2F target genes. They regulate transcription activity by binding their response elements to the promoters of target genes in the late G1/ S phase of cell cycle.2 (2) The repressors (E2F3b, E2F4, and E2F5) were found to be interacted with E2F inhibitory RB pocket proteins and block E2F target gene transcription.3 This subclass preferentially plays a role of transcriptional inhibition in the quiescent phase and early G1 phase of cell cycle.4 (3) Transcriptional inhibitors (E2F6-E2F8) have similar function with “repressor E2Fs”, but they do not interact with pocket proteins.5 Recent studies have found that the abnormal expression of E2F family members was reported in various tumors and E2Fs may play an important role in promoting or inhibiting cancer by affecting a variety of downstream genes.6–8
Lung cancer is a common malignant tumor with the highest morbidity and mortality rate in the world.9 In 2016, there were about 224,390 newly reported lung cancer cases and 158,080 lung cancer related deaths in the United States. The 5-year survival rate in the United States is less than 17.4%.10 Lung adenocarcinoma (LUAD) is the most common type of lung cancer. The treatment of LUAD mainly includes surgical resection, radiotherapy, chemotherapy, hormone therapy, and molecular targeted therapy. Although the overall outcome of surgical resection is satisfactory in the early stage of LUAD, the diagnosis of LUAD is often in the advance stage due to the early asymptomatic or only with subtle symptoms. In advanced LUAD, despite advances in treatment strategies, curative options are prolonged and limited. Molecular targeted therapy is a promising choice. However, there were lack of effective molecular targets and could not satisfy the therapeutic effect in LUAD. Therefore, therapy targets and potential prognostic biomarkers need to be identified to promote treatments and optimize the prognosis of LUAD.
E2Fs are supposed to have complex and distinct roles in various human tumorigenesis. For example, the increased expression of E2F and EZH2 with the corresponding retinoblastoma (Rb)-E2F-EZH2 axis changes in bladder cancer played a role in regulating tumor development, recurrence and progression.6 Previous research also showed that HBXIP could induce PKM2 expression through transcription factor E2F1 and promote the proliferation of ER+ breast cancer cells.11 E2F2 was overexpressed in glioma and promoted glioma progression via regulating the PI3K/AKT axis.12 E2F3 was overexpressed in gastric cancer by promoting tumor growth and metastasis via H19/miR-194/E2F3 axis.13
Although these studies suggested that the E2F family are involved in various types of cancers. However, a systematical analysis of the prognostic value of the entire E2F family remains poorly investigated in LUAD. Additionally, tumor-infiltrating immune cells are closely associated with clinical outcome of cancers. Herein, we comprehensively assessed the prognostic value of the E2F family and potential relationship with immunotherapy in LUAD, based on updated public resources and integrative bioinformatics analysis.
Methods
The mRNA Expressions of E2Fs in LUAD and Normal Lung Tissues
RNA-seq data and clinical information of 535 LUAD patients were derived from The Cancer Genome Atlas (TCGA) databases. The mRNA expression values of E2Fs in tumor tissue (535 specimens) were compared with adjacent normal tissue (59 specimens) by using nonparametric test of paired samples and unpaired samples. P<0.05 was considered statistical significance (Supplementary Table 1).
cBioPortal Gene Mutation Analysis and Methylation Status of E2Fs in LUAD
The cBioPortal for Cancer Genomics (http://cbioportal.org) provides an open-source platform for exploring, visualizing, and analyzing multidimensional cancer genomics data. Four LUAD datasets from cBioPortal database (Broad, Cell 2012; OncoSG, Nat Genet 2020; MSKCC, Cancer Discov 2017; TCGA, PanCancer Atlas), including 1969 total LUAD samples, were selected for further analyses the mutation of E2Fs in LUAD patients. The mutation types included the missense mutation, splice mutation, truncating mutation, amplification, fusion amplification, and deep deletion. Additionally, the methylation status of E2F family members in LUAD vs normal lung were investigated by Ualcan database (http://ualcan.path.uab.edu/).
Prognostic Value of E2Fs
Patients were divided into high expression group and low expression group according to the median value of E2Fs expression. Kaplan-Meier survival analysis was performed to assess the prognostic value of E2Fs in LUAD. The results were presented as Kaplan-Meier survival plot with the hazard ratio (HR) and log rank. Univariate and multivariate cox regression were further performed to assess the prognostic value of E2Fs and clinical variables. The receiver operating characteristic (ROC) analysis then performed to evaluate the diagnostic value of E2Fs for diagnosing LUAD. We further verified the prognosis value of E2Fs through two other cohorts of GEO databases (GSE13213 and GSE31210).
Correlation of E2Fs with Immune Cell Infiltration and Immune Checkpoint Genes in LUAD
The difference of immune cell infiltration between the two expression groups was evaluated with the immunedeconv R package (https://grst.github.io/immunedeconv), which integrates six algorithms (including TIMER, EPIC, MCP-counter, xCell, CIBERSORT, and quanTIseq). The TIMER and CIBERSORT algorithms were used respectively for evaluating immune cell infiltration. The correlation between E2Fs and immune checkpoint genes was analyzed to explore the potential role of E2Fs in immunotherapy.
Immunohistochemistry
In this study, we used paraffin samples from LUAD patients to verified the protein level of E2F7 by Immunohistochemistry (IHC). A total of 40 specimens were obtained from the First People’s Hospital of Lianyungang in China between October 2021 to January 2021, including 30 LUAD tissues and 10 normal lung tissues (14 males and 16 females). The histological evaluation was performed on hematoxylin and eosin (HE) stained sections. The study was approved by the hospital institutional ethic review board (Ethic approval number: LW-20220224001-01). Written informed consent was obtained from all participants or their guardians before the study.
The LUAD tissue sections were immunostained with primary antibody against E2F7 (Proteintech, Cat# 24489-1-AP, China).14 Three-μm sections cut from paraffin-embedded tissue were deparaffinized, rehydrated, and antigen retrieved, then incubated with the primary antibody (1:100) at 37°C. After incubation with primary antibody, the detection of antibodies was accomplished using the streptavidin peroxidase method.
Sections were scored blindly by two independent pathologists. The degree of immunostaining was based on staining intensity and percentage of cells stained. We quantitatively scored tissue sections using the following criteria: (a) percentage of immunoreactive cell (%); and (b) staining intensity: 0 (negative staining), 1 (weak staining), 2 (moderate staining), and 3 (intense staining). Staining results were evaluated using both the percentage of positive staining and intensity of positively stained tumor cells. The Histological-score (H-score) =0×percentage (%) of negative staining+1×percentage (%) of weak staining+2×percentage (%) of moderate staining+3×percentage (%) of intense staining. Statistical data analyses were performed by GraphPad Prism 6.0 (GraphPad Software, USA). The data are presented as the mean ± standard deviation. The protein level of E2F7 in tumor tissue were compared with adjacent normal tissue by using Man Whitney U unpaired nonparametric test, and P<0.05 was considered significant.
The Association Between E2F7 Expression and the Levels of Common Chemokines and Cytokines
We analyzed the association between E2F7 expression and the levels of common chemokines and cytokines (MMPs, TIMPs, IL6, IL10, CXCL17 and CCL24) in LUAD by using starbase database (https://starbase.sysu.edu.cn/index.php).
Results
The mRNA Expression of E2Fs in LUAD and Normal Lung Tissues
The mRNA expression of E2Fs factors between LUAD and normal lung tissues groups showed that the expression levels of E2F1-E2F8 were all significantly higher in LUAD patients with both paired samples and unpaired samples nonparametric tests. (Figure 1).
cBioPortal Gene Mutation Analysis and Methylation Status of E2Fs in LUAD
We analyzed the genetic alterations of E2Fs in LUAD patients by using the cBioPortal online tool. The result showed that E2F1, E2F2, E2F3, E2F4, E2F5, E2F6, E2F7, and E2F8 were altered in 1%, 1.6%, 1%, 1.1%, 4%, 0.9%, 3%, and 1.4% of the queried 1969 LUAD samples (Broad, Cell 2012; OncoSG, Nat Genet 2020; MSKCC, Cancer Discov 2017; TCGA, PanCancer Atlas) respectively. The main mutation type were missense mutation and amplification. All the E2F family demonstrated a low rates of gene mutation in LUAD patients (Figure 2). Also, the methylation status of E2F family members in LUAD vs normal lung showed that the E2F1, E2F3, E2F5 and E2F6 were significantly different in LUAD vs normal lung (Supplementary Figure 1).
Prognostic Value of E2Fs
Kaplan-Meier survival analysis found that the expression of E2F2 (P=0.038; HR 1.36; 95% CI 1.02–1.81), E2F7 (P<0.001; HR 1.78; 95% CI 1.33–2.39), and E2F8 (P=0.03; HR 1.37; 95% CI 1.02–1.82) were significantly associated with worse prognosis of LUAD (Figure 3). And the expression of E2F1, E2F3, E2F4, E2F5, and E2F6 did not show significance (P>0.05).
Univariate cox regression analysis revealed that T stage, N stage, M stage, E2F2, E2F7, E2F8 were significantly associated with prognosis of LUAD (P<0.05). In further multivariate cox regression analysis adjusting with common clinical parameters (including gender, age, smoking, T stage, N stage and M stage), only T stage (P=0.015; HR 1.73; 95% CI 1.11–2.69), N stage (P<0.001; HR 2.34; 95% CI 1.67–3.28) and E2F7 (P<0.001; HR 2.72; 95% CI 1.75–4.25) remained independent association with the prognosis of LUAD (Table 1).
 |
Table 1 The Prognostic Value of E2Fs and Clinical Variables in LUAD in Univariate and Multivariate Cox Regression Model |
With the significant founding on E2F7, further ROC analysis showed that E2F7 had a high diagnostic value for lung adenocarcinoma (AUC=0.901) (Figure 4). To further verify the prognosis value of E2F7, analysis through two other different cohorts of GEO databases (GSE13213 and GSE31210) were conducted. Results found that in both datasets, E2F7 were both significantly associated with prognosis of LAUD. The Cox regression results were shown as follow in GSE13213 (P<0.001; HR 1.39; 95% CI 1.15–1.69) and GSE31210 (P<0.00001; HR 2.04; 95% CI 1.49–2.78).
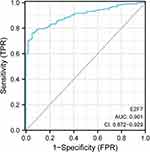 |
Figure 4 The ROC curve of E2F7 for diagnosis lung adenocarcinoma. The results were presented as AUC and 95% CI. |
Correlation of E2Fs with Immune Cell Infiltration and Immune Checkpoint Genes in LUAD
Immune cell level is associated with proliferation and progression of cancer cell. Current study used the TIMER and CIBERSORT database to explore the correlation between E2Fs and immune cell infiltration. The TIMER immune cell infiltration analysis found that expression level of E2F7 was significantly associated with B cell, CD8+ T cells, neutrophil, and myeloid dendritic cell infiltration in LUAD (Figure 5). The CIBERSORT immune cell infiltration analysis found that the expression level of E2F7 was correlated with B cells, CD8+ T cells, Cytotoxic cells, dendritic cell, Neutrophils, T cells infiltration in LUAD.
Results also showed that E2F7 have positive expression relations with common immune checkpoint genes including SIGLEC15 (r=0.25), CD274 (r=0.32), HAVCR2 (r=0.13), PDCD1LG2 (r=0.28), CTLA4 (r=0.15), TIGIT (r=0.18), LAG3 (r=0.24) and PDCD1 (r=0.16) in LUAD (Figure 6).
Protein Expression Level of E2F7 in LUAD Tissues by Immunohistochemistry
The protein expression level of E2F7 by immunohistochemistry showed that the expression of E2F7 was significantly increased in LUAD tissues compared to that in normal lung tissues (Figure 7), which was consistent with the mRNA level of E2F7.
The Association Between E2F7 Expression and the Levels of Common Chemokines and Cytokines
The association between E2F7 expression and the levels of common metastasis related factors (MMPs and TIMPs) in LUAD were further explored. The result showed that the expression of E2F7 had significant co-expression relationship with the level of MMP1, MMP10, MMP11, MMP12, TIMP2, and TIMP4 (Supplementary Figure 2).
Between the E2F7 expression and the levels of common chemokines and cytokines (IL6, IL10, CXCL17 and CCL24) in LUAD, the result showed that the level of E2F7 also had significant co-expression relationship with the expression of IL6, IL10, CXCL17 and CCL24 (Supplementary Figure 3).
Discussion
In this study, we comprehensively analyzed the expression and prognosis value, mutation, and immune cell infiltration of E2Fs in LUAD patients based on integrative bioinformatics analysis. While the expression levels of E2Fs (E2F1-8) were all significantly upregulated in LUAD tissues, E2F7 (P<0.001; HR 2.72; 95% CI 1.75–4.25) was an independent prognostic predictor in LUAD after adjustment, and also had a high diagnostic value for LUAD (AUC=0.901). Immune cell infiltration might be the participating pathway of E2F7 in LUAD.
Although studies have demonstrated that the E2F family of transcription factors correlates with the progression and metastases of gastric, colorectal, and lung cancers, the function of E2Fs in LUAD patients is still largely unknown. The E2F transcription factors are mainly responsible for the control of cell mitosis and regulation of cell cycle. The abnormal expression of E2Fs were observed in several kinds of human cancers, including gastrointestinal tumor, ovarian cancer, breast cancer and hepatocellular carcinoma.7,15–17 Recently, more studies have reported the important role of E2Fs in lung cancer.
The E2F1 and E2F2 were considered as transcription activators. Many studies have found that E2F1 and E2F2 expressions are significantly increased in lung cancer and play a variety of roles in the occurrence and development of lung cancer.18 Wang et al recently reported that E2F1, by activating the transcription of NELL2 gene, could enhance the migration, viability, and invasion of non-small cell lung cancer (NSCLC) cells as well as influence tumor progression and metastasis of NSCLC.19 E2F2 could promote the lung cancer cell proliferation and invasion through Lnc-NNT-AS1/miR-3666/E2F2 axis.20 Miao et al21 reported that the expression of E2F2 could be regulated by transcription factor FLI1, the decrease of E2F2 resulted in arrest of lung cancer cell cycle and inhibition of lung cancer proliferation. In our study, both the expression of E2F1 and E2F2 were significantly increased in LUAD. However, only the expression of E2F2 had a significant association with worse prognosis of LUAD (P=0.038; HR 1.36; 95% CI 1.02–1.81), which did not exist after adjusting for gender, age, tumor stage and other clinical parameters.
The E2F3, E2F4 and E2F5 were acted as repressors and found to interact with E2F inhibitory RB pocket proteins and restrict E2F target gene transcription. The mRNA and protein expression levels of E2F3 were both upregulated in NSCLC tumor tissues according to a recent study, and the inhibited expression of E2F3 gene could lead to the decrease of tumor cell metastasis and viability.22 It has been reported that the expression of E2F4 and E2F5 were significantly increased in lung cancer, but there are few studies on their mechanism in lung cancer progress. In our study, the expression of E2F3, E2F4 and E2F5 were also significantly increased in LUAD, but there was no significant correlation between their expression and the prognosis of LUAD.
E2F6, E2F7 and E2F8 were classified as transcriptional inhibitors. Previous study has demonstrated that increase in the expression of E2F6 which conferred with tumor aggressiveness in diffuse subtype of gastric adenocarcinoma [10.1007/s10238-016-0443-0]. E2F6 was also upregulated in lung cancer blood samples.23 Yuan et al found E2F6 could repress the transcription of LINC01436 under normal oxygen condition, and this repression effect was reversed under the condition of microenvironment hypoxia and then lead to growth, invasion and migration of lung cancer cell.24 E2F7 was highly expressed in LUAD tissues and involved in the progression of NSCLC through regulating LncRNA-SNHG19.25 E2F7 also played an important role in the Circ-AASDH/miR-140-3p/E2F7 axis and participated in regulating LUAD tumor growth.26 A recent study reported that E2F8 expression was found to be significantly upregulated in lung cancer tissues and cells by immunofluorescence staining, and knockdown E2F8 could inhibit lung cancer cell proliferation and tumor growth.8 In our study, the expression of E2F6, E2F7 and E2F8 were also significantly increased in LUAD. But there was no significant correlation between the expression of E2F6 and the prognosis of LUAD. E2F8 had a significant association with worse prognosis of LUAD (P=0.03; HR 1.37; 95% CI 1.02–1.82) in Kaplan-Meier analysis, which disappeared after adjusting for clinical parameters. Multivariate cox regression analysis revealed that only E2F7 (P<0.001; HR 2.72; 95% CI 1.75–4.25) remained an independent prognostic predictor of LUAD after adjusting common clinical parameters and with a powerful diagnostic value to predicter LUAD (AUC=0.901) in ROC analysis. This result implied that E2F7 had the potential to become a novel biomarker of LUAD.
Tumor microenvironment (TME) could affect tumor progression and recurrence. Immune cells in TME have been shown to harbor either tumor-promoting or tumor-suppressing activities, which are considered as a significant determining factor of the response to immunotherapy. However, there are few reports about the relationship between E2Fs and immune cell infiltrations. Shom et al reported that E2Fs could mediate the anti-tumor immunity effects of CDK4/6 restrainers through regulating the function of T cells.27 Oshi et al reported recently that ER-positive/HER2-negative breast cancer patients with higher E2Fs score were shown higher breast cancer invasiveness and lower reactivity to neoadjuvant chemotherapy. They also found E2Fs score was highly negative correlated with the therapeutic responsiveness of immune checkpoint inhibitors.28 In our study, the immune cell infiltration analysis showed that E2F7 was significantly associated with B cell, T cell, neutrophil and myeloid dendritic cell infiltration in LUAD. We also found E2F7 had positive correlations with the expression of immune checkpoint genes in LUAD, including SIGLEC15, CD274, HAVCR2, PDCD1LG2, CTLA4, TIGIT, LAG3 and PDCD1. These results suggested that E2F7 may participate in mediating the immune cell infiltration, and has the potential to further explore the value as an immunotherapy target of LUAD.
There were some limitations to our study. All the data analyzed of E2Fs in LUAD were retrieved from online databases. Further cell experiments and clinical cohort study on gene expression level, protein activity, and genetic alteration aspects are required to validate our findings.
Conclusion
Our study comprehensively investigated the expression, prognosis value, mutation, and potential relationship with immunological aspects of E2Fs in LUAD by bioinformatics analysis. All E2Fs were highly expressed in LUAD tumor tissues, but only E2F7 was an independent prognostic predictor of LUAD after adjusting with common clinical parameters. Further analysis found that E2F7 was significantly associated with immune cell infiltration and immune checkpoint genes in LUAD. Results showed that E2F7 may become a novel biomarker of prognosis and might provide additional evident for further research in improving treatment and for LUAD patients.
Abbreviations
LUAD, Lung adenocarcinoma; TF, transcription factor; ROC, receiver operating characteristic; NSCLC, non-small cell lung cancer.
Notes
The authors have completed the STROBE reporting checklist. All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethical Statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the First People’s Hospital of Lianyungang and informed consent was taken from all individual participants.
Author Contributions
(I) Conception and design: Jinping Liu, Yanxiu Li, Bo Yu.
(II) Execution and Administrative support: Tingzhen Chen, Yanxiu Li, Bo Yu.
(III) Provision of study materials or patients: Iokfai Cheang, Yanxiu Li.
(IV) Acquisition of data: Qixun Wang, Jinghang Li,
(V) Data analysis and interpretation: Qixun Wang, Jinping Liu, Iokfai Cheang
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
All authors have completed the ICMJE uniform disclosure form. The authors have no conflicts of interest to declare.
References
1. Gaubatz S, Lindeman GJ, Ishida S, Jakoi L, Nevins JR. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell. 2000;6(3):729–735. doi:10.1016/S1097-2765(00)00071-X
2. Cam H, Dynlacht BD. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell. 2003;3(4):311–316. doi:10.1016/S1535-6108(03)00080-1
3. Ren B, Cam H, Takahashi Y. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Develop. 2002;16(2):245–256. doi:10.1101/gad.949802
4. Nevins JR. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258(5081):424–429. doi:10.1126/science.1411535
5. Carvajal LA, Hamard PJ, Tonnessen C, Manfredi JJ. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 2012;26(14):1533–1545. doi:10.1101/gad.184911.111
6. Santos M, Martínez-Fernández M, Dueñas M. In vivo disruption of an Rb–E2F–Ezh2 signaling loop causes bladder cancer. Cancer Res. 2014;74(22):6565–6577. doi:10.1158/0008-5472.CAN-14-1218
7. Tang H, Liu P, Yang L, et al. miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol Cancer Ther. 2014;13(12):3185–3197. doi:10.1158/1535-7163.MCT-14-0243
8. Park SA, Platt J, Lee JW. E2F8 as a novel therapeutic target for lung cancer. J Natl Cancer Inst. 2015;107:djv151.
9. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
10. Mao Y, Yang D, He J, Krasna MJ. Epidemiology of lung cancer - sciencedirect. Surg Oncol Clin N Am. 2016;25(3):439–445. doi:10.1016/j.soc.2016.02.001
11. Liu BW, Wang TJ, Li LL, et al. Oncoprotein HBXIP induces PKM2 via transcription factor E2F1 to promote cell proliferation in ER-positive breast cancer. Acta Pharmacol Sin. 2018;40:530–538.
12. Zhang L, Liu Z, Dong Y, Kong L. E2F2 drives glioma progression via PI3K/AKT in a PFKFB4-dependent manner. Life Sci. 2021;276:119412. doi:10.1016/j.lfs.2021.119412
13. Qin W, Shi Y, Zhu D, Chen Y, Cui J. H19/miR-194/E2F3 regulating loop promotes gastric cancer growth and metastasis; 2020.
14. Yang C, Zhang ZC, Liu TB, Xu Y, Xia BR, Lou G. E2F1/2/7/8 as independent indicators of survival in patients with cervical squamous cell carcinoma. Cancer Cell Int. 2020;20:500. doi:10.1186/s12935-020-01594-0
15. Zhan L, Zhang Y, Wang W, Song E, Fan Y, Wei B. E2F1: a promising regulator in ovarian carcinoma. Tumor Biol. 2016;37(3):2823–2831. doi:10.1007/s13277-015-4770-7
16. Evangelou K, Havaki S, Kotsinas A. E2F transcription factors and digestive system malignancies: how much do we know? World J Gastroenterol. 2014;20(29):10212–10216. doi:10.3748/wjg.v20.i29.10212
17. Lü Y, Zhang J, Li L, Li S, Yang Z. Carcinogenesis effects of E2F transcription factor 8 (E2F8) in hepatocellular carcinoma outcomes: an integrated bioinformatic report. Biosci Rep. 2020;40(2). doi:10.1042/BSR20193212
18. Sun C, Zhou Q, Hu W, Li S-J, Zhang F. Transcriptional E2F1/2/5/8 as potential targets and transcriptional E2F3/6/7 as new biomarkers for the prognosis of human lung carcinoma. Aging. 2018;10(5):973–987. doi:10.18632/aging.101441
19. Wang Y, Li M, Zhang L, Chen Y, Zhang S. m6A demethylase FTO induces NELL2 expression by inhibiting E2F1 m6A modification leading to metastasis of non-small cell lung cancer. Mol Ther Oncolytics. 2021;21:367–376. doi:10.1016/j.omto.2021.04.011
20. Huang J, Luo X, Li Z, Lang B. LncRNA NNT-AS1 regulates the progression of lung cancer through the NNT-AS1/miR-3666/E2F2 axis. Eur Rev Med Pharmacol Sci. 2020;24(1):238–248. doi:10.26355/eurrev_202001_19916
21. Miao B, Bauer A, Hufnagel K, et al. The transcription factor FLI1 promotes cancer progression by affecting cell cycle regulation. Int J Cancer. 2020;147(1):189–201. doi:10.1002/ijc.32831
22. Wu L, Wan S, Li J, et al. Expression and prognostic value of E2F3 transcription factor in non-small cell lung cancer. Oncol Lett. 2021;21(5):411. doi:10.3892/ol.2021.12672
23. Barh D, Jain N, Tiwari S, et al. A novel in silico reverse-transcriptomics-based identification and blood-based validation of a panel of sub-type specific biomarkers in lung cancer. BMC genom. 2013;14(Suppl 6):S5. doi:10.1186/1471-2164-14-S6-S5
24. Yuan S, Xiang Y, Wang G, et al. Hypoxia-sensitive LINC01436 is regulated by E2F6 and acts as an oncogene by targeting miR-30a-3p in non-small cell lung cancer. Mol Oncol. 2019;13(4):840–856. doi:10.1002/1878-0261.12437
25. Zhao G, Ning Z, Wang R. viaLncRNA SNHG19 promotes the development of non-small cell lung cancer mediating miR-137/E2F7 axis. Front Oncol. 2021;11:630241. doi:10.3389/fonc.2021.630241
26. Wang Y, Wo Y, Lu T, et al. Circ-AASDH functions as the progression of early stage lung adenocarcinoma by targeting miR-140-3p to activate E2F7 expression. Transl Lung Cancer Res. 2021;10(1):57–70. doi:10.21037/tlcr-20-1062
27. Goel S, DeCristo MJ, Watt AC, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548(7668):471–475. doi:10.1038/nature23465
28. Oshi M, Takahashi H, Tokumaru Y, et al. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9(7):1643. doi:10.3390/cells9071643
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






