Back to Journals » Journal of Inflammation Research » Volume 16
C-Type Lectin Receptors-Triggered Antifungal Immunity May Synergize with and Optimize the Effects of Immunotherapy in Hepatocellular Carcinoma
Authors Xia J, Ding H, Liu S, An R, Shi X, Chen M, Ren H
Received 28 October 2022
Accepted for publication 24 December 2022
Published 5 January 2023 Volume 2023:16 Pages 19—33
DOI https://doi.org/10.2147/JIR.S394503
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Monika Sharma
Jinkun Xia,1,2,* Haoran Ding,1,* Shujun Liu,3 Ran An,1,2 Xiaolei Shi,1– 3 Ming Chen,1 Haozhen Ren1– 3
1Department of Hepatobiliary Surgery, The Affiliated Drum Tower Hospital, Medical School, Nanjing University, Nanjing, People’s Republic of China; 2Institute of Hepatobiliary Surgery, Medical School, Nanjing University, Nanjing, People’s Republic of China; 3Nanjing Drum Tower Hospital Clinical College of Nanjing University of Chinese Medicine, Nanjing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Haozhen Ren; Ming Chen, Email [email protected]; [email protected]
Abstract: Hepatocellular carcinoma (HCC) is one of the most common malignant tumors of the digestive system worldwide, and there is a lack of effective treatment for late-stage HCC. Recent experimental studies have demonstrated that dysfunction of the intestinal flora has a significant impact on hepatocarcinogenesis. The pathophysiological link between the intestine, its microbiota, and the liver has been described as the “gut-liver axis”. Dysbiosis of the intestinal flora and increased permeability of the intestinal wall are closely associated with liver pathology through the immune response. The “gut-liver axis” theory has been applied to the clinical study of the pathogenesis and treatment of HCC. The intestinal fungal community, as part of the gut microbiome, has a significant impact on human health and disease, while relatively little research has been done in HCC. In this study, we performed a comprehensive analysis of the expression and potential biological functions of the fungal recognition receptors C-type lectin receptors (CLRs) (Dectin-1, Dectin-2, Dectin-3, and Mincle) in HCC. We found that CLRs were downregulated in HCC, and their expressions were correlated with the clinical prognosis of HCC patients. Further studies suggested that the expression of CLRs were significantly correlated with immune infiltration and immunotherapy efficacy in HCC. Based on previous studies and our findings, we hypothesize that intestinal fungal communities and CLRs-triggered antifungal immunity have a key role in the pathogenesis of HCC, and these findings may provide new perspectives and targets for HCC immunotherapy.
Keywords: hepatocellular carcinoma, mycobiome, fungus, immunity, C-type lectin receptor, gut-liver axis
Introduction
Hepatocellular carcinoma (HCC) is the most common histologic type of liver cancer, accounting for approximately 80–90% of cases, and is the sixth most common cancer and the third leading cause of cancer-related deaths worldwide.1 HCC often originates in the liver with chronic inflammation induced by diseases such as viral hepatitis and nonalcoholic steatohepatitis (NASH); thus, its microenvironmental composition is very complex and highly heterogeneous among patients.2 Since the 5-year survival rate for liver cancer is only 18%, the search for additional new and effective treatments is ongoing.3 Adverse events with conventional anti-HCC drugs have also raised concerns.4 Recently, the treatment of HCC with immune checkpoint blockade (ICB) has shown promising efficacy and prolonged patient survival.5 However, the high degree of tumor heterogeneity and the frequent emergence of immune resistance limit the benefit of immunotherapy, and as a consequence, most patients do not benefit from ICB therapy.6,7 Therefore, understanding the mechanisms of immune regulation in the HCC microenvironment, identifying predictive biomarkers for checkpoint inhibitor-based immunotherapy, and developing new drug targets may improve the antitumor efficacy of immunotherapy.8
The liver is the largest organ for anabolism and detoxification in the human body. On the one hand, nutrients from the intestine and metabolites of the intestinal microflora enter the liver through the blood into the portal system.9 On the other hand, metabolic products from the liver flow into the intestine through the biliary system, which in turn affects the colonization environment of the intestinal flora.9 This bidirectional enterohepatic exchange system consisting of the intestinal flora with the portal system and bile duct system is usually called the “gut-liver axis” and is the physiological basis of the interaction between the intestinal flora and the liver.10 The intestinal flora is one of the focal points of emerging research in the field of HCC in recent years. Currently, the intestinal flora can be broadly classified into 3 types - beneficial, conditionally pathogenic and pathogenic, and most members of the intestinal flora, such as Bacteroidetes, Eubacterium, Bifidobacterium, Rumex and Clostridium, have a symbiotic relationship with human hosts.11 When the intestinal ecology is disturbed, the intestinal flora is altered qualitatively and/or quantitatively. Such disturbance destroys the intestinal barrier and increases intestinal permeability, changing the body’s metabolic capacity, lowering the organism’s immunity, leading to proinflammatory changes in the liver, and consequently affecting the development of liver fibrosis, cirrhosis, and HCC.12 Many studies have reported that the intestinal flora plays an important role in the progression of HCC. Existing views hold that the immune response to and metabolic disorders caused by the intestinal flora triggered by microbiota-associated molecular patterns (MAMPs), such as leaky gut, intestinal flora translocation, and dysbiosis, are the main mechanisms through which the intestinal flora contribute to the development of HCC via the intestine-hepatic axis.13 Regulation of the intestinal flora with probiotics, rational use of antibiotics, and fecal transplantation can significantly improve the host’s antitumor immune response;14 therefore, reshaping intestinal flora homeostasis has important potential value in delaying the progression of HCC and improving the prognostic effect of HCC treatment.
As part of the gut microbiome, the intestinal fungi have a remarkable impact on human health and disease.15 A human microbiome project study revealed that the most common fungal organisms in the gut of healthy people are Yeast, Malassezia and Candida.16 Compared to the percentage of bacteria, the total percentage of fungi in the intestinal microbial community is only 0.03% to 2%.17 Nevertheless, clinical studies have identified patients with nonalcoholic fatty liver disease and more severe diseases with a specific fecal fungal composition and an elevated systemic immune reaction to Candida albicans.18 Bone marrow immune cells, such as dendritic cells (DCs), macrophages and natural killer T cells, are the first cells to detect alterations in the intestinal flora and to initiate defense against potential invaders through pattern recognition receptors (PRRs).19 Activation of PRRs on cells of the innate immune system induces an inflammatory response to clear pathogens, and C-type lectin receptors (CLRs) are the main PRRs mediating recognition of fungal pathogens.20 The four major family members of CLR are Dectin-1, Dectin-2, Dectin-3 and macrophage-inducible Ca2+-dependent lectin receptor (Mincle). Their recognition of fungi activates the immune response; however, their actions vary by disease, and they can have dual effects.21,22 For example, Dectin-1 signaling in DCs is essential for triggering protective antifungal T helper type 17 (Th17) cells responses and may be clinically important; however, there is evidence that activation of Dectin-1 by galactose lectin-9 on tumor-infiltrating macrophages induces immune escape and accelerates the progression of pancreatic cancer.22,23 These results suggest that the action of CLRs may occur in a tissue-, cell-, or even gene-specific manner.
The fungal community in the liver has been identified to contain a wide distribution of microorganisms with high diversity and heterogeneity,24,25 and it may have key functions in the progression of HCC, potentially providing new targets and strategies for the treatment of HCC. However, relevant studies are lacking. In this article, we report a comprehensive analysis of the correlation between CLRs expression, potential CLRs functions and immune infiltration in HCC to explore possible mechanisms underlying the immunomodulatory functions of fungal community members and CLRs-expressing immune cells. In addition, we discuss the possibility and feasibility of targeting CLRs-mediated antifungal immunity to optimize the efficacy of ICB therapy in HCC.
Materials and Methods
The TNMplot database (https://www.tnmplot.com/) is an open-access online search tool of the Kaplan‒Meier plotter platform that can be used to compare the expression levels of genes in tumor tissues and the respective normal tissues. The data are mainly derived from The Cancer Genome Atlas (TCGA) database. In this article, we used TNMplot to compare the expression levels of Dectin-1 (CLEC7A), Dectin-3 (CLEC4D) and Mincle (CLEC4E) in HCC with those in the corresponding normal tissues (Dectin-2 data were not available). To observe the protein expression levels of CLRs, the Human Protein Atlas (HPA) platform (www.proteinatlas.org) was used to acquire immunohistochemistry (IHC) images of CLRs (Dectin-3 and Mincle) staining in normal liver tissues and liver cancer tissues. The methylation levels of CLRs promoter were obtained from UALCAN (http://ualcan.path.uab.edu/), which is a multi-feature, web-based platform housing interactive resources for profiling cancer omics data. The prognostic value of CLRs gene expression for patients with HCC were analyzed by using Kaplan‒Meier Plotter (www.kmplot.com) on the basis of relevant TCGA cancer data.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) pathway analyses were performed to further explore the functions of CLRs and their coexpressed genes. Differentially expressed genes (DEGs) between groups with high and low CLR expression were displayed in volcano plots generated with the “ggplot” R package. The top 50 most upregulated and the top 50 most downregulated DEGs were selected as coexpressed genes of CLRs. To determine the underlying biological actions of SAA1 in HCC, GO and KEGG analyses were performed with the Assistant for Clinical Bioinformatics (https://www.aclbi.com/static/index.html#/) platform using the ClusterProfiler package in R software based on relevant TCGA cancer data.
The TIMER platform (https://cistrome.shinyapps.io/timer/) can be used to systematically identify the levels of infiltrating immune cells in a wide range of cancer types. In this study, TIMER was applied to determine the correlations between CLR expression and immune cell infiltration in HCC. TISIDB (http://cis.hku.hk/TISIDB/index.php) was used to analyze the correlation of CLR expression with the levels of infiltrating Th1/Th17 cells. To assess the relationship between CLRs expression and immunotherapy efficacy in HCC patients, we extracted data from the TCGA database and divided the registered patients into low and high CLR expression groups, and we predicted the response to immune checkpoint blockade therapy in these groups using the TIDE algorithm (http://tide.dfci.harvard.edu/). All statistical results were calculated by the listed online tools.
Justification of the Hypothesis
Fungal Mycobiome-Induced Immunosuppression Drives Carcinogenesis
To discover whether fungi change in cancer patients, researchers classified and studied fungi in more than 17,000 tissue and blood samples from 35 cancer types.24 As expected, fungi (including several types of yeast) appeared in all cancer types studied, and the specific fungal species were related to the type of cancer. For example, breast cancer patients who had Malassezia sphericalis - a fungus naturally found on the skin - had a much lower survival rate than breast cancer patients without this fungus.24 Characterization of the bacteria in these tumors showed that most types of fungi tended to coexist with specific types of bacteria. Different microorganisms coevolve in the human ecosystem, usually relying on the same resources. This suggests that the tumor environment may favor both fungal and bacterial growth, unlike the normal tissue environment, in which fungi and bacteria are in competition for resources.24 Another study revealed that gastrointestinal tumors and lung and breast tumors tend to have fungi of the genera Candida, Blastomyces and Malassezia.26 In addition, higher levels of Candida in gastrointestinal tumor cells were associated with more inflammation-promoting gene activity, higher rates of cancer metastasis, and lower cancer survival rates.26 These two studies provide the clearest link between fungi and cancer, but more research is needed to further understand whether these fungi promote cancer development by triggering inflammation or whether tumors create an environment conducive to fungal survival. Recently, a study revealed that fungi within pancreatic cancers, which were virtually absent in the healthy pancreas, stimulated the secretion of IL-33 and recruited and activated type 2 innate lymphoid cells, thus promoting tumor progression.27 Genetic deletion of IL-33 or antifungal treatment reduced the infiltration of type 2 immune cells and increased survival.27 In addition, C. albicans-dominant commensal fungi were reported to be sensed by the fungal receptor Dectin-1, which increases the number of tumor-associated macrophages and deregulates antitumor T cells, thereby suppressing the antitumor immune response after radiotherapy.28 Fungal-mediated tumor-promoting effects can be reversed when the antifungal drug fluconazole is administered to inhibit commensal fungi.28 Similarly, Candida tropicalis can potently induce the differentiation and proliferation of myeloid-derived suppressor cells (MDSCs), a key immune cell subpopulation with a strong immunosuppressive capacity in the tumor microenvironment.29 MDSCs contribute to the development and progression of colorectal cancer by suppressing the immune function of T cells.29 These results confirm that fungal populations can modulate the immune response to tumors by inducing the differentiation and increasing the activity of immune cells and that antifungal therapy appears may have antitumor effects.
CLRs are Downregulated and are Associated with Poor Prognosis in HCC Patients
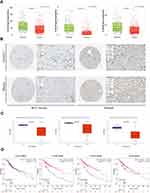
The major CLRs include Dectin-1 (CLEC7A), Dectin-2 (CLEC6A), Dectin-3 (CLEC4D) and Mincle (CLEC4E), which are mainly expressed on myeloid immune cells and activate a series of downstream signaling pathways to initiate innate and adaptive immune responses by recognizing β-glucan and α-mannan, the major components expressed on the fungal cell wall.30 CLRs primarily activate the SYK-CARD9, SYK-PLCG2, or RAF1 pathways, as detailed in.20 With the activation of these signaling pathways, the secretion of interleukin 1β (IL-1β), interleukin 6 (IL-6) and interleukin 23 (IL-23) is induced, and differentiated 4+ (CD4+) T cells are polarized into T helper type 1 (Th1)/T helper type 17 (Th17) cells, resulting in essential antifungal immunity.31 To verify the expression levels of CLRs in HCC, we downloaded the expression data of CLRs in normal and HCC tissues from TNMplot and the Human Protein Atlas (HPA) platform. The results confirmed that Dectin-1, Dectin-3 and Mincle were significantly downregulated in HCC tissues (Dectin-2 data were not available) (Figure 1A and B). Methylation is a widely studied mode of epigenetic regulation that does not alter the gene sequence but can provide heritable changes in gene function and ultimately lead to phenotypic changes.32 Interestingly, the low expression of CLRs seems to be associated with hypomethylation of their promoters in HCC (Figure 1C). In addition, we found that the downregulation of several CLRs was closely associated with poor prognosis of HCC patients, suggesting the involvement of CLRs-mediated immunomodulatory pathways in the progression of HCC (Figure 1D). Reduced or complete absence of CLRs signaling pathway activitys can lead to fungal ecological dysregulation, resulting in increased susceptibility to fungal infections and related diseases.33 This may explain the poor prognosis of HCC patients with low CLR expression.
Enrichment Analysis of CLRs in HCC
Recently, an interesting study discovered a significant increase in Candida albicans in HCC patients by internal transcribed spacer (ITS) sequencing and that tube feeding of Candida albicans in wild-type HCC mice increased the size and weight of tumors.25 However, colonization with Candida albicans did not affect tumor growth in nucleotide oligomerization domain-like receptor family pyrin domain containing 6 (NLRP6) knockout mice.25 NLRP6 acts as a cytoplasmic pattern recognition receptor that identifies molecular patterns associated with microorganisms in the body and protects the host from pathogenic bacteria and viruses,34 and that its tumor-suppressive effects are mediated by fungi suggest an important role for PRRs in HCC progression.
To further explore the potential biological functions and regulatory roles of CLRs in the HCC environment, we performed differential gene analysis based on TCGA data from HCC tissue samples according to methods described previously35 and further performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. The results showed that the most common DEGs included the C-X-C motif chemokine ligand (CXCL) family members CXCL8, CXCL9, CXCL10, CCL5, CCL19, and CCL21 (Figure 2A–C), which largely participate in the induction and chemotaxis of immune cells by serving as immunomodulatory chemokines. Interestingly, the results of the enrichment analysis showed that the four CLRs participate in similar processes, including Th1 and Th2 cell differentiation, Th17 cell differentiation, cytokine‒cytokine receptor interactions, regulation of cell‒cell adhesion, T-cell activation, and regulation of T-cell activation. (Figure 2A–C). These processes promote the activation and migration of innate and adaptive immune cells. Taken together, these results suggest that CLRs may promote immune effects against HCC.
Correlation of CLRs with Immune Cell Levels, Immune Checkpoint Protein Expression, and Immune Checkpoint Blockade Therapy Efficacy
To analyze the relationship between CLRs expression and immune infiltration in HCC, the TIMER tool was applied. The results confirmed a strong positive correlation between the expression of the four CLRs and the levels of various infiltrating immune cells, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells (Figure 3A). These cells constitute the tumor immune microenvironment and affect ICB efficacy in cancers via crosstalk with tumor cells and stromal cells. As previously described, CLRs-mediated immune signaling exerts bactericidal effects through Th1/Th17 cells. Therefore, we further analyzed the correlation of CLR expression with the levels of Th1/Th17 cells via TISIDB, and the results confirmed their positive correlation (Figure 3B). To further analyze the association of CLRs expression with immune checkpoint protein expression, we selected PDCD1, LAG3, CTLA-4, TIGIT, HAVCR2, CD274 and PDCD1LG2 as key immune checkpoint molecules and analyzed their expression in HCC patients in the CLR-high and CLR-low groups. The results showed that the expression levels of immune checkpoint proteins were significantly higher in the CLR-high group than in the CLR-low group (Figure 4A). Subsequently, we assessed the relationship between CLR expression and the ICB response in HCC using the TIDE platform. A higher TIDE prediction score indicates a higher probability of immune evasion, suggesting that patients are less likely to benefit from ICI therapy. The immune response scores are shown in Figure 4B. All HCC patients with high levels of CLR expression showed higher TIDE scores than HCC patients with low levels of CLRs expression, implying that patients in the high CLR group are more likely to experience immune dysfunction and immune evasion and may not be sensitive to ICB treatment.
Potential Biological Activity and Regulatory Mechanism of CLRs in HCC
The activity status of immune cells in the tumor immune microenvironment determines the ICB efficacy of tumors. Upon stimulation of Dectin-1 with fungal-derived β-glucan ligands, dendritic cells are activated, thereby inducing the expansion of CD8+ T cells and enhancing their differentiation into cytotoxic T cells.36 Dectin-2, Dectin-3 and Mincle have also been shown to induce immune cell differentiation.37–39 Interestingly, Dectin-1 also induces Treg-cell (the main negative regulatory cells of the immune response) differentiation through SYK signaling and subsequent IL-1β secretion.40,41 This suggests that CLRs maintain the immune balance between CD8+ T cells and regulatory T cells. However, how this homeostasis is hijacked by HCC cells and the specific fungal population driving the disruption of immune homeostasis remain unknown.
Under normal conditions, immune checkpoints can maintain immune tolerance by regulating the strength of the autoimmune response.42,43 However, in the case of cancer, tumor cells can use this mechanism to induce T-cell depletion and suppress their function, favoring tumor cell growth and immune escape.44 Multiple factors have been reported to control the expression of immune checkpoint proteins, of which cytokine‒cytokine receptor interactions appear to be essential.45 For example, γ-chain cytokines such as IL-2, IL-7, IL-12, IL-21, and IL-15 can activate downstream signaling upon recognition of IL receptors, thereby increasing the expression of TIM-3 and PD-1 in T cells.46 Immune checkpoint protein expression is also subject to regulation by transforming growth factor beta (TGF-β), interferon alpha (IFN-α) and interferon gamma (IFN-γ).47 Anti-fungal immunity mediated by CLRs involves the production of a series of functional cytokines.48 Dectin-1 signaling has been reported to induce the secretion of IL-12 and tumor necrosis factor alpha (TNF-α) in monocytes, which subsequently induces an inflammatory response.49 In addition, in the context of multiple sclerosis, the combined use of the Dectin-1 agonists yeast polysaccharide and IFN-β enhances the expression of IL-27, which drives the expression of co-suppressed immune checkpoint proteins.50 Similarly, lung expression of Mincle was significantly upregulated following stimulation by the pathogen Orientia tsutsugamushi, thereby promoting sustained IL-27 production and shaping a proinflammatory immune response to infection.51 Although it remains unknown whether CLRs induce the secretion of IL-27 or other cytokines that induce immune checkpoint protein expression in HCC, we can speculate that this potential pathway or a similar pathway may be utilized by HCC cells to promote the expression of multiple immune checkpoint proteins and to facilitate immune tolerance. Subsequently, we analyzed the correlation between the expression of four CLRs and the expression of cytokines regulating immune checkpoint proteins (IL-2, IL-7, IL-12, IL-21, IL-15, IL-27, TGF-β, and IFN-γ) (Table 1). The results showed a significant positive correlation between the expression of the CLRs and the expression of IL-2, IL-7, IL-12, IL-21, IL-15, TGF-β, and IFN-γ but a negative correlation between the CLRs and IL-27.
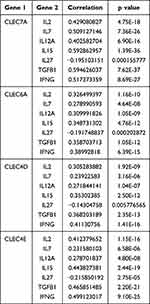 |
Table 1 Correlations of CLR Family Gene Expression and the Expression of Key Cytokines Regulating Immune Checkpoint Proteins in Hepatocellular Carcinoma |
The processes of immune cell transport to tumor sites and mutual contact with tumor cells rely heavily on a subfamily of cytokines called chemokines.52 In addition, chemokine secretion is also associated with the expression of immune checkpoint proteins.53 Guo et al demonstrated that lung-infiltrating dendritic cells and neutrophils can secrete CXCL9 and CXCL10 to recruit plasmacytoid dendritic cells to the lung in response to fungal infection by Aspergillus fumigatus, which is dependent on the activation of Dectin-1.54 This process may also occur in the context of cancer. We found that chemokines accounted for a large proportion of the differentially expressed genes of the CLR-high and CLR-low groups (Figure 2A–C). Therefore, we hypothesize that CLRs regulate the activity and function of immune cells by regulating the secretion and alteration of chemokines, in addition to directly regulating immune cells in the tumor microenvironment. However, more in vivo and in vitro experiments are still needed to validate this hypothesis.
Potential Antitumor Strategies Targeting CLRs or Fungal Communities
Differences in the composition of the microbiota are strongly associated with survival and sensitivity to immunotherapy in patients with malignancies.55,56 Considering that targeting a single immune checkpoint protein, such as PD-L1 or PD-1, has limited clinical efficacy against tumors, co-targeting multiple immune checkpoint proteins may be a better option. Furthermore, given that microbiota characteristics are highly heterogeneous among individuals, it is possible that interfering with common fungal-mediated signaling pathways could be an effective antitumor strategy. Interestingly, CLR expression is strongly correlated with the expression of almost all immune checkpoint proteins and may regulate the expression of immune checkpoint proteins by regulating multiple cytokines. Therefore, in HCC patients with high expression of CLRs, combined inhibition of cytokines and other immune checkpoint molecules may be a more comprehensive antitumor strategy.
Based on the critical immunomodulatory function of the gut microbiota, regulation and remodeling of the commensal microbiota through probiotic administration and microbial transplantation have emerged as potential strategies that can synergize with ICB therapy. Intriguingly, Kapitan et al found that fungal and bacterial abundances in the gut were negatively correlated and that disruption of the bacterial microbiota was a prerequisite for fungal overgrowth.57 In addition, commensal bacteria and fungi have distinct regulatory roles in the antitumor immune response to radiotherapy. Excessive administration of antibiotics suppresses the abundance of commensal bacteria and leads to an overgrowth of fungi, mainly C. albicans, which suppresses the antitumor immune response after radiotherapy.28 Therefore, how to coordinate antifungal treatment and antibiotic treatment has become a common research topic. Notably, the long-term use of antibacterial drugs can lead to the development of fungal resistance. The speed of antimicrobial drug development is insufficient to overcome the emergence of fungal drug resistance. In-depth study of the molecular mechanisms of CLRs can provide new ideas and strategies for the development of broad-spectrum antibacterial drugs, antitumor drugs and immunosuppressive drugs.
Recently, the immunomodulatory mechanisms of in vivo transplantation of specific fungal communities has been explored.29 In future studies, there is a need to identify pathogenic or beneficial fungal species in HCC and their immune functions and to use in vivo transplantation to characterize the biological functions and effects of specific fungal community members on the therapeutic efficacy of ICB. Once we have a clearer understanding of the role of fungal species in cancer, therapies or probiotics can be developed to target and control fungal populations, thus helping to inhibit cancer progression. In addition, it is important to note that the roles of CLRs expressed on different immune cells may not be the same and may even be contradictory.58 Therefore, if the tumor antigen-presenting function of CLRs is to be exploited to enhance tumor immunity in humans, it is not enough to stimulate the activity of all CLRs in the host by simply injecting adjuvants; instead, the treatment needs to be specific to different cell types. For example, DC vaccines can be administered with agonists in vitro to enhance the ability of the DCs to present tumor antigens. Additionally, given the broad role of CLRs in the defense against various infectious diseases caused by viral pathogens,59 combining oncolytic viruses with CLR-mediated antifungal therapies is a potential strategy for HCC treatment.
A Summary of Our Hypothesis
In intestine-associated lymphoid tissues, immune cells defend against or tolerate intestinal fungi, metabolites, and toxins. Upon disruption of homeostasis, potential antigens from commensal fungi, ingested fungi or fungal-derived metabolites in the gut penetrate the intestinal mucosal barrier and are transferred through the portal vein to the liver, where they affect its function. Immune cells in the liver initiate an immune response against enteric fungi to maintain homeostasis; however, this balance is destabilized in the liver cancer environment. Based on the findings of previous literature and our bioinformatics results, we propose that disturbed fungal community homeostasis and a disrupted gut-liver axis contribute to HCC progression and the poor prognosis of HCC patients, mainly mediated by low expression of CLRs. CLRs can modulate the expression of immune checkpoint proteins by inducing the secretion of cytokines such as IL-27 and/or chemokine expression, thus influencing the efficacy of ICB therapy. Based on this, we hypothesized that the efficacy of immunotherapy in HCC could be enhanced by the CLR-mediated antifungal immune response induced by antifungals as well as the transplantation of fungal colonies (Figure 5).
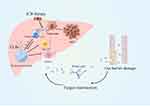 |
Figure 5 Graphical hypothesis of our study generated by FigDraw. |
Discussion
In recent years, research on cancer and microorganisms has mainly focused on the intestinal tract, which contains more bacteria, viruses and fungi than any other part of the human body. However, the roles of fungi, as a minority group of the intestinal flora, in cancer progression have long been overlooked or underestimated, and the effects of cancer-related fungi have not been thoroughly studied. Fungi are eukaryotes and are thus more complex microorganisms than viruses and bacteria. Their cells are more similar to animal cells. The existence and specific roles of fungi in most human cancers are promising research topics. Several recent studies have revealed fungal enrichment in cancer, but the underlying mechanisms remain poorly understood. CLRs, pattern recognition receptors for fungal microbiota elements, are highly expressed in myeloid immune cells and are involved in both nonspecific and specific immunity against tumors. To date, many CLRs have been identified in humans, and different CLRs perform a variety of different functions.60 The majority of CLRs can positively regulate immune cell function after recognizing bound antigens and play important roles in anti-infection, anti-allergic and antitumor responses. However, some pathogens can also escape from the immune response by targeting CLRs on the surface of immune cells.61 The ligands associated with CLR recognition and binding still need to be further explored and refined. Whether different CLRs expressed on the surface of myeloid immune cells trigger synergistic or antagonistic intracellular signaling pathways after recognizing the binding of the same antigen or recognizing different antigens of the same pathogen, thus playing positive or negative roles in the antitumor response, remains to be determined. Understanding these issues is essential for further understanding the integrated regulation of immune cell function by different types of CLRs in specific diseases and for improving the understanding of the network of interactions between different CLRs.
Our results suggest that immune modulation mediated by low CLR expression in HCC may be involved in intestinal fungus-mediated immune homeostasis and may improve ICB therapy outcome. Our hypothesis highlights the long-underestimated or neglected role of antifungal immunity in the clinical treatment of HCC and may provide new drug targets for HCC patients. In addition, we revealed potential CLR-mediated immunomodulatory processes and antitumor strategies in HCC, ie, CLRs may regulate the transcription and expression of immune checkpoint proteins by modulating the expression of chemokines and their receptors as well as cytokines. These finding may offer directions for future research. However, our hypothesis needs to be further confirmed by animal and clinical experiments.
Future studies should address several questions. The first concern is to explore the specific fungal community patterns and changes in these patterns in the different stages of HCC to identify the most critical functional fungi in the HCC immunomodulatory network. Such information can be gained by using advanced sequencing technologies and conducting a series of clinical trials. Second, the current understanding of the exact mechanisms underlying the roles of CLRs on the immune cell surface and antifungal immunity in cancer is still limited. Therefore, exploring the combined effects of different CLRs on the regulation of immune cell functions, the relevant specific mechanisms, and the interactions of different CLRs with antitumor pathways could support their use as new therapeutic drug targets and thus advance antitumor therapy research. In addition, the human body is home to a large number of bacteria, fungi, and viruses. Previous studies have often considered only one of these types of microbe in relation to human health, while in reality, they interact with each other throughout the body. It is also significant to understand how these microbes mutually interact with each other and influence the development of cancer. Moreover, existing fungal databases have fewer total sequences than bacterial databases. In addition, the accuracy of the classification information is insufficient, the data management is lacking, and fungus polymorphisms have caused different studies to identify the same ITS sequence as two different fungi, all of which increase research error. Therefore, improving the fungal databases by improving the information and using more accurate fungal identification methods is key for future microbiological research.
Data Sharing Statement
The data used to support the results of this study are available from TCGA (https://portal.gdc.cancer.gov), the Assistant for Clinical Bioinformatics database (https://www.aclbi.com/static/index.html#/), and the TIMER platform (https://cistrome.shinyapps.io/timer/). Further inquiries can be directed to the corresponding authors.
Ethical Approval
All data for our study were obtained from public databases, as shown in the Material and Methods. Nevertheless, we need to state that all of our studies involving human data were performed in accordance with the Declaration of Helsinki and with the approval of the Committee on Ethics of Affiliated Drum Tower Hospital of Nanjing University Medical School. All patients who donated tissues have provided informed consent.
Acknowledgments
The authors would like to acknowledge the technical assistance provided by the staff of the Department of Hepatobiliary Surgery, The Affiliated Drum Tower Hospital of Nanjing University Medical School, Nanjing, China. We would like to thank SpringNature for English language editing. The graphical figure was generated by Figdraw (www.figdraw.com).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was funded by the National Natural Science Foundation of China (82270646, 81872359), Jiangsu Provincial Key Research and Development (BE2020752), Key Scientific Research Project of Jiangsu Provincial Health Commission (ZDA2020002), the Nanjing health science and technology development project for Distinguished Young Scholars (JQX19002), the Fundamental Research Funds for the Central Universities (0214-14380510), Project of Modern Hospital Management and Development Institute, Nanjing University and Aid project of Nanjing Drum Tower Hospital Health, Education & Research Foundation (NDYG2022057), funding for Clinical Trials from the Affiliated Drum Tower Hospital, Medical School of Nanjing University (2022-LCYJ-PY-35), the Chen Xiao-ping Foundation for the Development of Science and Technology of Hubei Province, China (CXPJJH121001-2021073).
Disclosure
The authors have declared that no competing interests exist.
References
1. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. doi:10.1038/s41572-020-00240-3
2. Nault JC, Villanueva A. Biomarkers for hepatobiliary cancers. Hepatology. 2021;73(Suppl 1):115–127. doi:10.1002/hep.31175
3. Sonbol MB, Riaz IB, Naqvi SAA, et al. Systemic therapy and sequencing options in advanced hepatocellular carcinoma: a systematic review and network meta-analysis. JAMA Oncol. 2020;6(12):e204930. doi:10.1001/jamaoncol.2020.4930
4. Rizzo A, Nannini M, Novelli M, et al. Dose reduction and discontinuation of standard-dose regorafenib associated with adverse drug events in cancer patients: a systematic review and meta-analysis. Ther Adv Med Oncol. 2020;12:1758835920936932. doi:10.1177/1758835920936932
5. Donisi C, Puzzoni M, Ziranu P, et al. Immune checkpoint inhibitors in the treatment of HCC. Front Oncol. 2021;10:601240. doi:10.3389/fonc.2020.601240
6. Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19(3):151–172. doi:10.1038/s41571-021-00573-2
7. Rizzo A, Ricci AD, Gadaleta-Caldarola G, et al. First-line immune checkpoint inhibitor-based combinations in unresectable hepatocellular carcinoma: current management and future challenges. Expert Rev Gastroenterol Hepatol. 2021;15(11):1245–1251. doi:10.1080/17474124.2021.1973431
8. Rizzo A, Ricci AD, Di Federico A, et al. Predictive biomarkers for checkpoint inhibitor-based immunotherapy in hepatocellular carcinoma: where do we stand? Front Oncol. 2021;11:803133. doi:10.3389/fonc.2021.803133
9. Visschers RG, Luyer MD, Schaap FG, et al. The gut-liver axis. Curr Opin Clin Nutr Metab Care. 2013;16(5):576–581. doi:10.1097/MCO.0b013e32836410a4
10. Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 2020;72(3):558–577. doi:10.1016/j.jhep.2019.10.003
11. Benson AK. The gut microbiome-an emerging complex trait. Nat Genet. 2016;48(11):1301–1302. doi:10.1038/ng.3707
12. Li J, Sung CYJ, Lee N, et al. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc Natl Acad Sci U S A. 2016;113(9):E1306–E1315. doi:10.1073/pnas.1518189113
13. Schwabe RF, Greten TF. Gut microbiome in HCC - mechanisms, diagnosis and therapy. J Hepatol. 2020;72(2):230–238. doi:10.1016/j.jhep.2019.08.016
14. Milosevic I, Vujovic A, Barac A, et al. Gut-liver axis, gut microbiota, and its modulation in the management of liver diseases: a review of the literature. Int J Mol Sci. 2019;20(2):395. doi:10.3390/ijms20020395
15. Hallen-Adams HE, Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8(3):352–358. doi:10.1080/21505594.2016.1247140
16. Nash AK, Auchtung TA, Wong MC, et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome. 2017;5(1):153. doi:10.1186/s40168-017-0373-4
17. Huffnagle GB, Noverr MC. The emerging world of the fungal microbiome. Trends Microbiol. 2013;21(7):334–341. doi:10.1016/j.tim.2013.04.002
18. Demir M, Lang S, Hartmann P, et al. The fecal mycobiome in non-alcoholic fatty liver disease. J Hepatol. 2022;76(4):788–799. doi:10.1016/j.jhep.2021.11.029
19. Brubaker SW, Bonham KS, Zanoni I, et al. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. 2015;33(1):257–290. doi:10.1146/annurev-immunol-032414-112240
20. Casadevall A. Immunity to invasive fungal diseases. Annu Rev Immunol. 2022;40(1):121–141. doi:10.1146/annurev-immunol-101220-034306
21. Wang T, Pan D, Zhou Z, et al. Dectin-3 deficiency promotes colitis development due to impaired antifungal innate immune responses in the gut. PLoS Pathog. 2016;12(6):e1005662. doi:10.1371/journal.ppat.1005662
22. Daley D, Mani VR, Mohan N, et al. Dectin 1 activation on macrophages by galectin 9 promotes pancreatic carcinoma and peritumoral immune tolerance. Nat Med. 2017;23(5):556–567. doi:10.1038/nm.4314
23. Zhao Y, Chu X, Chen J, et al. Dectin-1-activated dendritic cells trigger potent antitumour immunity through the induction of Th9 cells. Nat Commun. 2016;7(1):12368. doi:10.1038/ncomms12368
24. Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. 2022;185(20):3789–3806 e17. doi:10.1016/j.cell.2022.09.005
25. Liu Z, Li Y, Li C, et al. Intestinal candida albicans promotes hepatocarcinogenesis by up-regulating NLRP6. Front Microbiol. 2022;13:812771. doi:10.3389/fmicb.2022.812771
26. Dohlman AB, Klug J, Mesko M, et al. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell. 2022;185(20):3807–3822 e12. doi:10.1016/j.cell.2022.09.015
27. Alam A, Levanduski E, Denz P, et al. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. 2022;40(2):153–167 e11. doi:10.1016/j.ccell.2022.01.003
28. Shiao SL, Kershaw KM, Limon JJ, et al. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell. 2021;39(9):1202–1213 e6. doi:10.1016/j.ccell.2021.07.002
29. Wang T, Fan C, Yao A, et al. The adaptor protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity. 2018;49(3):504–514 e4. doi:10.1016/j.immuni.2018.08.018
30. Tang J, Lin G, Langdon WY, et al. Regulation of C-type lectin receptor-mediated antifungal immunity. Front Immunol. 2018;9:123. doi:10.3389/fimmu.2018.00123
31. Zielinski CE, Mele F, Aschenbrenner D, et al. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484(7395):514–518. doi:10.1038/nature10957
32. Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2(5):657–669. doi:10.2217/epi.10.44
33. Netea MG, Joosten LAB, van der Meer JWM, et al. Immune defence against Candida fungal infections. Nat Rev Immunol. 2015;15(10):630–642. doi:10.1038/nri3897
34. Li R, Zhu S. NLRP6 inflammasome. Mol Aspects Med. 2020;76:100859. doi:10.1016/j.mam.2020.100859
35. Gu C, Chen J, Dang X, et al. Hippo pathway core genes based prognostic signature and immune infiltration patterns in lung squamous cell carcinoma. Front Oncol. 2021;11:680918. doi:10.3389/fonc.2021.680918
36. Leibundgut-Landmann S, Osorio F, Brown GD, et al. Stimulation of dendritic cells via the dectin-1/Syk pathway allows priming of cytotoxic T-cell responses. Blood. 2008;112(13):4971–4980. doi:10.1182/blood-2008-05-158469
37. Carter RW, Thompson C, Reid DM, et al. Induction of CD8+ T cell responses through targeting of antigen to Dectin-2. Cell Immunol. 2006;239(2):87–91. doi:10.1016/j.cellimm.2006.05.001
38. Preite NW, Feriotti C, Souza de lima D, et al. The syk-coupled C-type lectin receptors Dectin-2 and Dectin-3 are involved in paracoccidioides brasiliensis recognition by human plasmacytoid dendritic cells. Front Immunol. 2018;9:464. doi:10.3389/fimmu.2018.00464
39. Shiga M, Miyazaki J, Tanuma K, et al. The liposome of trehalose dimycolate extracted from M. bovis BCG induces antitumor immunity via the activation of dendritic cells and CD8+ T cells. Cancer Immunol Immunother. 2021;70(9):2529–2543. doi:10.1007/s00262-021-02870-2
40. Karnam A, Bonam SR, Rambabu N, et al. Wnt-beta-catenin signaling in human dendritic cells mediates regulatory T-cell responses to fungi via the PD-L1 pathway. mBio. 2021;12(6):e0282421. doi:10.1128/mBio.02824-21
41. Patel D, Gaikwad S, Challagundla N, et al. Spleen tyrosine kinase inhibition ameliorates airway inflammation through modulation of NLRP3 inflammosome and Th17/Treg axis. Int Immunopharmacol. 2018;54:375–384. doi:10.1016/j.intimp.2017.11.026
42. Chihara N, Madi A, Kondo T, et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature. 2018;558(7710):454–459. doi:10.1038/s41586-018-0206-z
43. Zhang Y, Zheng J. Functions of immune checkpoint molecules beyond immune evasion. Adv Exp Med Biol. 2020;1248:201–226.
44. Dyck L, Mills KHG. Immune checkpoints and their inhibition in cancer and infectious diseases. Eur J Immunol. 2017;47(5):765–779. doi:10.1002/eji.201646875
45. Schreiber G, Walter MR. Cytokine-receptor interactions as drug targets. Curr Opin Chem Biol. 2010;14(4):511–519. doi:10.1016/j.cbpa.2010.06.165
46. Curdy N, Lanvin O, Laurent C, et al. Regulatory mechanisms of inhibitory immune checkpoint receptors expression. Trends Cell Biol. 2019;29(10):777–790. doi:10.1016/j.tcb.2019.07.002
47. Terawaki S, Chikuma S, Shibayama S, et al. IFN-alpha directly promotes programmed cell death-1 transcription and limits the duration of T cell-mediated immunity. J Immunol. 2011;186(5):2772–2779. doi:10.4049/jimmunol.1003208
48. Pedro ARV, Lima T, Fróis-Martins R, et al. Dectin-1-mediated production of pro-inflammatory cytokines induced by yeast beta-glucans in bovine monocytes. Front Immunol. 2021;12:689879. doi:10.3389/fimmu.2021.689879
49. Gringhuis SI, den Dunnen J, Litjens M, et al. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-kappaB activation through Raf-1 and Syk. Nat Immunol. 2009;10(2):203–213. doi:10.1038/ni.1692
50. Sweeney CM, Lonergan R, Basdeo SA, et al. IL-27 mediates the response to IFN-beta therapy in multiple sclerosis patients by inhibiting Th17 cells. Brain Behav Immun. 2011;25(6):1170–1181. doi:10.1016/j.bbi.2011.03.007
51. Fisher J, Card G, Liang Y, et al. Orientia tsutsugamushi selectively stimulates the C-type lectin receptor mincle and type 1-skewed proinflammatory immune responses. PLoS Pathog. 2021;17(7):e1009782. doi:10.1371/journal.ppat.1009782
52. Palomino DC, Marti LC. Chemokines and immunity. Einstein. 2015;13(3):469–473. doi:10.1590/S1679-45082015RB3438
53. Xiu W, Luo J. CXCL9 secreted by tumor-associated dendritic cells up-regulates PD-L1 expression in bladder cancer cells by activating the CXCR3 signaling. BMC Immunol. 2021;22(1):3. doi:10.1186/s12865-020-00396-3
54. Guo Y, Kasahara S, Jhingran A, et al. During aspergillus infection, monocyte-derived DCs, neutrophils, and plasmacytoid DCs enhance innate immune defense through CXCR3-dependent crosstalk. Cell Host Microbe. 2020;28(1):104–116 e4. doi:10.1016/j.chom.2020.05.002
55. Nejman D, Livyatan I, Fuks G, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973–980. doi:10.1126/science.aay9189
56. Riquelme E, Zhang Y, Zhang L, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795–806 e12. doi:10.1016/j.cell.2019.07.008
57. Kapitan M, Niemiec MJ, Steimle A, et al. Fungi as part of the microbiota and interactions with intestinal bacteria. Curr Top Microbiol Immunol. 2019;422:265–301. doi:10.1007/82_2018_117
58. Chiffoleau E. C-type lectin-like receptors as emerging orchestrators of sterile inflammation represent potential therapeutic targets. Front Immunol. 2018;9:227. doi:10.3389/fimmu.2018.00227
59. Kalia N, Singh J, Kaur M. The role of dectin-1 in health and disease. Immunobiology. 2021;226(2):152071. doi:10.1016/j.imbio.2021.152071
60. Nikolakopoulou C, Willment JA, Brown GD. C-type lectin receptors in antifungal immunity. Adv Exp Med Biol. 2020;1204:1–30.
61. Yan H, Kamiya T, Suabjakyong P, et al. Targeting C-type lectin receptors for cancer immunity. Front Immunol. 2015;6:408. doi:10.3389/fimmu.2015.00408
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.