Back to Journals » Cancer Management and Research » Volume 14
Bioinformatics Prediction and in vivo Verification Identify SLC7A5 as Immune Infiltration Related Biomarker in Breast Cancer
Authors Zhu Q, Wang J, Shi Y, Zha X, Wang S
Received 4 May 2022
Accepted for publication 20 August 2022
Published 27 August 2022 Volume 2022:14 Pages 2545—2559
DOI https://doi.org/10.2147/CMAR.S370397
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Seema Singh
Qiannan Zhu,1 Jue Wang,1 Yuenian Shi,2 Xiaoming Zha,1 Shui Wang1
1The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210029, People’s Republic of China; 2Nanjing Medical University, Nanjing, Jiangsu, 210029, People’s Republic of China
Correspondence: Shui Wang; Xiaoming Zha, The First Affiliated Hospital of Nanjing Medical University, 300# Guangzhou Road, Nanjing, 210000, People’s Republic of China, Email [email protected]; [email protected]
Purpose: Solute carrier family 7, member 5 (SLC7A5) is reportedly a key gene in various tumors. However, the relationship between SLC7A5 and immune cell infiltration in breast cancer remains elusive. In this study, we investigated the expression levels and clinical value of SLC7A5 using the R package in breast cancer through the TCGA database.
Methods: Public transcript data and clinical information of patients were downloaded from the TCGA database. Prognosis analysis was performed to explore the clinical value of SLC7A5 as well as drug sensitivity prediction and Gene Set Enrichment Analysis (GSEA). We also explored the correlations between SLC7A5 and immune infiltration as well as immune markers.
Results: We observed that high SLC7A5 expression levels were related to poor survival. Our GSEA demonstrated that G protein-coupled receptor-ligand binding, interleukin signaling, neutrophil degranulation, amino acid and derivative metabolism, and Rho GTPase signaling were differentially enriched in the SLC7A5 high-expression phenotype. Patients in the low SLC7A5 expression group showed sensitivity to paclitaxel and doxorubicin.
Conclusion: SLC7A5 could serve as a biomarker for breast cancer prognosis.
Keywords: SLC7A5, breast cancer, immune cell infiltration, chemosensitivity
Introduction
Breast cancer is a significantly common disease worldwide with approximately 1.3 billion new cases and half a million cancer-related deaths per year.1 In general, breast cancer can be divided into three subtypes based on histological differences: endocrine-dependent, HER2-positive, and triple-negative breast cancer (TNBC).2 Among these, TNBC displays the worst prognosis due to its higher heterogeneity. Surgery is still the first-line option for localized breast cancer, and patients with breast cancer receiving surgical excision combined with adjuvant therapy tend to have a satisfactory prognosis.3 Nevertheless, nearly 5% of the patients with breast cancer are metastatic at diagnosis and 20–30% of localized breast cancer would develop secondary metastasis.4 At present, breast cancer diagnosis and treatment are still facing significant challenges due to the high cancer relapses and metastasis rate. Therefore, identifying novel and effective biomarkers associated with the early diagnosis and treatment of patients with breast cancer would be urgent.
Solute carrier family 7, member 5 (SLC7A5), also called LAT1, is largely accepted as an amino acid transporter and reportedly exerts remarkable effects in cancer and developmental processes.5 For instance, Najumudeen et al revealed that SLC7A5 could maintain intracellular amino acid levels through transcriptional and metabolic reprogramming, required for the efficient growth of KRAS-mutant colorectal cancer.6 Ding et al showed that CTLA4 (cytotoxic T-lymphocyte associated protein 4) could positively regulate SLC7A5 expression, responsible for enhanced gastric cancer cell proliferation and invasion ability.7 In lung cancer, Wang et al indicated that lncRNA MINCR facilitated cancer cell growth via the miR-126/SLC7A5 axis, a competitive endogenous RNA mechanism.8 Meanwhile, SLC7A5 was also reported to exert a specific role in immune disease. Loftus et al described that the SLC7A5-mediated amino acid transport was a requirement for c-Myc induction in cytokine-stimulated NK cells.9 Yoon et al indicated that SLC7A5-limited leucine influx could lead to proinflammatory cytokine production in activated human monocytes/macrophages through glycolytic reprogramming.10 Moreover, Nachef et al systematically reviewed the role of SLC1A5 in the tumor immune microenvironment and concluded that SLC1A5 might potentially strengthen antitumor immunity.11 However, the SLC7A5 effect in breast cancer and the tumor microenvironment has not yet been fully investigated.
In this study, we explored the association between SCL7A5 expression and immune status in breast cancer based on a large population cohort, including different immune infiltration cells and factors. Moreover, we discovered that SLC7A5 could predict the chemosensitivity and survival status of patients with breast cancer, indicating that SLC7A5 might be a promising clinical biomarker.
Methods
Public Data Acquisition
All the public transcript data and clinical information of patients (including breast cancer and pan-cancer data) were downloaded from the TCGA database (https://portal.gdc.cancer.gov/). Specifically, the expression profile data were an “FPKM” form, collated using the Perl code. The clinical information form was an “xml.” Probe annotation was carried out on the basis of the GENCODE v19 genome assembly. Patients with complete transcript data and clinical information were selected for further analysis. Table 1 shows the clinical characteristics of patients with breast cancer in the TCGA database. Another dataset in external validation dataset METABRIC was used for independent validation. Representative SLC7A5 immunohistochemistry images were obtained from the Human Protein Atlas database (https://www.proteinatlas.org/) to indicate the protein level in breast cancer.
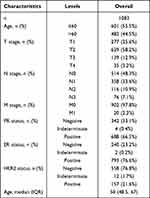 |
Table 1 The Clinical Characteristics of Patients with Breast Cancer in the TCGA Database |
Cell Lines and Culture
All the cell lines used in this study, including the normal human breast epithelial cell line MCF-10A and breast cancer cell lines (MCF-7, SUM1315, and SKBR3), were ordered from the American Type Culture Collection (ATCC, Manassas, VA, USA). Dulbecco’s modified Eagle medium (Gibco, China) supplemented with 10% fetal bovine serum (Gibco, China) and 1% penicillin/streptomycin (Gibco, China) were applied in the cell cultures. MCF-10A cells were cultured following the ATCC recommendations. For cell culture, we used a thermostatic humidified incubator at 37°C containing 5% CO2.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
To extract total RNA from tissues and cells, we used the TRIzol reagent (TaKaRa, Kyoto, Japan), and the RNA concentration was quantified using a NanoDrop 2000 device (Thermo Scientific, USA). A HifairII 1st Strand cDNA Synthesis Kit (Yeasen, China) was used to transcribe total RNA (0.5 µg) to complementary DNA (cDNA). qRT-PCR was conducted in a StepOnePlus Real-time PCR System (Applied Biosystems, USA) using SYBR Green Master Mix (Yeasen, China). Table 2 summarizes the sequences of the primers used in this study. Relative expression values were calculated using the comparative delta CT (2−ΔΔCT) method, and the values were normalized to β-actin.
 |
Table 2 Primers Used for qRT-PCR |
Prognosis Analysis
We performed prognosis analysis using the survival package in the R environment to explore the clinical value of SLC7A5. Patients with breast cancer in TCGA dataset were divided into high and low SLC7A5 groups with the median cutoff. Overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI) were set as the endpoints. The survival records of TCGA breast cancer patients were downloaded from the UCSC Xena data portal (https://xena.ucsc.edu/). We used the Cox regression model in the survival and forest package to calculate and visualize the hazard ratio and Cox p values.
Immune Infiltration and Correlation Analyses
We performed immune infiltration analysis using the single-sample Gene Set Enrichment Analysis (ssGSEA) algorithm to assess the underlying association between SLC7A5 and tumor immune microenvironment. ssGSEA algorithm can quantify 22 reported TIL cells according to the input expression matrix (24138885). We used TISIDB, an integrated repository portal playing a crucial role in seeking the interaction between tumors and the immune system (http://cis.hku.hk/TISIDB), to evaluate the correlation between SLC7A5 and multiple immune-related genes.
Pathway Enrichment Analysis
We used Gene Set Enrichment Analysis (GSEA) to explore the biological difference between SLC7A5-high and low patients. In detail, the reference pathway set was “REACTOME.” The pathways meeting the criteria of FDR <0.05 were considered statistically significant. The top five enriched pathways were selected for visualization.
Drug Sensitivity Analysis
We implemented the pRRophetic package in the R environment for drug sensitivity prediction, based on the pharmacogenomics database of the Cancer Genome Project cell line data and the Cancer Cell Line Encyclopedia. We used the ridge regression algorithm to estimate the half-maximal inhibitory concentration (IC50) of chemotherapeutic drugs, with 10-fold cross-validated for model stability.
Statistical Analysis
All the statistical analyses were performed using the R software v3.6.2. We used the Wilcoxon rank-sum test to compare the SLC7A5 expression pattern in the different groups. P-values were two sided, and p ˂ 0.05 were considered statistically significant.
Results
SLC7A5 Expression Levels in Different Cancer Types
First, we investigate the SLC7A5 expression levels in various cancer types and corresponding normal controls using the RNA-seq data of multiple malignancies in the TCGA data. We discovered that SLC7A5 expression levels significantly increased in breast, gastric, brain and CNS, colorectal, esophageal, and pancreatic cancer tissues, as well as in leukemia, lymphoma, and melanoma compared with the healthy tissues (Figure 1A). SLC7A5 was prominently overexpressed or underexpressed with pairs of noncancerous adjacent tissues using the Wilcoxon signed-rank test (Figure 1B). SLC7A5 expression in breast cancer cell lines such as SKBR3, SUM1315, and MCF-7 was also higher than that in MCF-10a normal breast cells (Figure 1C). To investigate SLC7A5 expression in breast cancer, we examined both normal and breast tumor tissues using TCGA RNA-sequencing data. The nonpaired analysis included 113 and 1109 normal and breast tumor tissues, respectively. We evaluated 112 paired normal and tumor tissues in the paired analysis. SLC7A5 showed the same expression trend in the breast tissue in the case of both the TCGA and TIMER data. According to both paired and nonpaired analyses, SLC7A5 expression was higher in breast cancer (Figure 1D and E). To confirm the clinical relevance of SLC7A5 expression, we analyzed the expression between SLC7A5 and clinicopathological factors in breast cancer. We calculated the SLC7A5 receiver operating characteristic curve and area under the curve (AUC) values (AUC 0.822, 95% CI [0.792–0.851]) (SFigure 1A). Finally, we also compared the SLC7A5 protein levels using IHC staining based on the THPA database. The SLC7A5 protein expression level in breast cancer tumor tissues was higher than that in normal tissues (SFigure 1B), which has been reported by El-Ansari et al.12 SLC7A5 was also overexpressed in various breast cancer subtypes. The expression of SLC7A5 in each subgroup was significantly different from that in adjacent tissues in TCGA (SFigure 2A). The expression of SLC7A5 in various breast cancer subtypes in METABARIC was consistent with that in TCGA (SFigure 2B). Data analysis showed that somatic mutation TP53 was associated with SLC7A5 expression in TCGA. SlC7A5 is highly expressed in TP53 mutant (SFigure 3). These findings suggested that SLC7A5 upregulation might predict advanced breast cancer malignancy.
Relationship Between SLC7A5 Expression and Survival of Patients with Breast Cancer
Patients with high SLC7A5 expression displayed a poor OS (HR[95% CI] = 1.17[1.23–2.37], p = 0.001), DSS (HR [95% CI] = 2.22[1.41–3.49], p = 0.001) and PFI (HR [95% CI] = 1.76[1.26–2.45], p = 0.001) (Figure 2A). High SLC7A5 expression in N I–III also correlated with bad OS (HR [95% CI] = 2.02[1.34–3.06], p = 0.001), DSS (HR [95% CI] = 2.43[1.44–4.12], p = 0.001) and PFI (HR [95% CI] = 2.08[1.36–3.17], p = 0.001) (Figure 2B). The prognosis of SLC7A5 in different breast cancer subtypes of TCGA was further analyzed. According to the median expression of SLC7A5, it was divided into high expression group and low expression group. Prognosis analysis included 5-year survival. Although there was no statistical significance in all subtypes (p > 0.05), the trend was basically the same: Patients with high SLC7A5 expression had a poor prognosis (SFigure 4). According to the expression of SLC7A5, the cases were divided into high and low expression groups. The impact on the overall prognosis (OS) was evaluated in METABARIC: the prognosis of those with high expression was poor, which was consistent with the results of TCGA (SFigure 5). Univariate and multivariate analyses of prognosis are shown in Supplementary Tables 1–4. These results showed that N stage, and SLC7A5 expression were significantly associated with both OS, DSS and PFI. Our data demonstrate that SLC7A5 expression could predict the prognosis in patients with breast cancer.
Relationship Between SLC7A5 Expression and Tumor-Infiltrating Immune Cells
Tumor-infiltrating immune cells reportedly influence the prognosis and survival of patients with breast cancer. We thus analyzed the relationship between SLC7A5 expression and infiltrating immune cells in breast cancer. We showed that SLC7A5 expression positively or negatively correlated with almost all immune cells, except for Tcm (Figure 3A and B). Breast cancer with high SLC7A5 expression exhibited more infiltrating immune cells, including Th2 cells, aDC, Th1 cells, Treg, NK CD56dim cells, B cells, macrophages, DC, neutrophils, cytotoxic cells, T cells, Tgd, Tem, and TFH. High SLC7A5 expression negatively correlated with CD8+ T cells, T helper cells, iDC, Th17 cells, pDC, NK CD56bright cells, eosinophils, NK cells, and mast cells. Moreover, high SLC7A5 expression could affect antitumor immune response.
Furthermore, we tested the correlation between SLC7A5 expression and three infiltrating immune cell types: Th1, Th2, and Treg cells. SLC7A5 expression positively correlated with Th1 (r = 0.300, p < 0.001), Th2 (r = 0.400, p < 0.001), and Treg (r = 0.290, p < 0.001) cells (Figure 4A). Considering that Th1, Th2, and Treg cells had a better correlation, we also examined the relationship between their specific cell surface markers and SLC7A5 expression. SLC7A5 expression positively correlated with IFNG (r = 0.213, p < 0.001), TNF (r = 0.187, p < 0.001), STAT1 (r = 0.255, p < 0.001), TBX21 (r = 0.141, p < 0.001), IL13 (r = 0.124, p < 0.001), CCR8 (r = 0.238, p < 0.001), and FOXP3 (r = 0.246, p < 0.001). However, it negatively correlated with GATA3 (r = −0.447, p < 0.001), STAT5A (r = −0.149, p < 0.001), STAT5B (r = −0.218, p < 0.001), STAT6 (r = −0.215, p < 0.001), and TGFB1 (r = −0.141, p < 0.001) (Figure 4B–D).
Correlation Analysis Between SLC7A5 Expression and Immune Control Genes
As a novel therapy, immune checkpoint inhibitors (ICIs) proved promising efficacy in multiple cancers. To investigate the potential of SLC7A5 in immunotherapy, we analyzed the relationship between SLC7A5 expression and immune control genes. We observed that in breast cancer, SLC7A5 expression positively correlated with IDO1, LAG3, IL2RA, PVR, and ULBP1, all of which modulate immune response positively, whereas it negatively correlated with CXCL12. These results indicate that SLC7A5 expression might indeed regulate ICI efficacy in breast cancer (Figure 5A and B).
Correlation Analysis Between SLC7A5 Expression and Chemokines/Chemokine Receptors
To further explore the role of SLC7A5 in immune cell function and migration, we performed an analysis between SLC7A5 expression and chemokines/chemokine receptors. We demonstrated that chemokines, such as CCL7, CCL8, CCL18, CCL20, CXCL5, and CXCL10, were upregulated when SLC7A5 expression increased (Figure 6A). Interestingly, SLC7A5 expression negatively correlated with CCL14, CXCL12, CXCL14, and CX3CR1 (Figure 6A and B). SLC7A5 was related to the expression of chemokines in the validation set METABRIC (STable 4). The correlation between SLC7A5 expression and various chemokines is statistically significant (p < 0.001). These results imply that SLC7A5 expression is involved with chemokine and chemokine receptor expressions, and it could become a potential target to modulate immune cell function.
 |
Figure 6 Correlation analysis of SLC7A5 expression with classic marker genes from (A) chemokine receptors and (B) chemokines in breast cancer samples. |
Gene Sets Enriched in SLC7A5 Expression Phenotype
By analyzing the GO and KEGG enrichment collection, we performed a GSEA to identify SLC7A5-related signaling pathways in these two groups. We found that FIVE signaling pathways, including chromatin-modifying enzymes, cellular senescence, epigenetic regulation of gene expression, estrogen-dependent gene expression, and chromosome maintenance, were significantly enriched in the high-SLC7A5-expression group (Figure 7A). Figure 7B shows the different gene expression levels between the low- and high-SLC7A5-expression groups. These results shed light on the potential SLC7A5 effector mechanism in modulating breast cancer development and host immune response.
Drug Sensitivity Prediction
Although chemotherapy and immunotherapy sensitivity or resistance is associated with clinical outcomes in breast cancer, we subsequently explored the clinical significance of different SLC7A5 expression groups. We used the ridge regression model to predict individual drug sensitivity. Three immunosuppressants commonly used in breast cancer, CTLA4, PD-1, and PD-L1, showed higher sensitivity in the low-SLC7A5-expression group than in the high-SLC7A5-expression group (p < 0.01) (Figure 8A). Two commonly used chemotherapeutic drugs in breast cancer therapy, paclitaxel and doxorubicin, showed higher sensitivities in the low-SLC7A5-expression group than in the high-SLC7A5-expression group (p = 4.60×10−3 and <0.01 for paclitaxel and doxorubicin, respectively) (Figure 8B).
Discussion
Breast cancer is a major public health concern worldwide.13 Along with the advent of the genomic era, the exploration of effective breast cancer biomarkers offers novel and potential therapeutic targets.
Our study explored the SLC7A5 expression pattern in breast cancer and its association with the immune system and drug sensitivity. SLC7A5 is a key amino acid transport-related molecule, indispensable in various physiological and pathophysiological processes.14 Baronas et al showed that SLC7A5 could regulate the Kv1.2 voltage-gated potassium channels and participate in action potential generation and propagation in the central nervous system, thereby affecting epilepsy progression.15 Moreover, the protein transport process regulated by SLC7A5 could significantly influence the nutrient and xenobiotics absorption and distribution, including those of drugs.16 Meanwhile, SLC7A5 can recognize multiple molecules with pharmacological implications and have anomalous expression patterns in most cancers, making it a promising drug target.17 For instance, Okunushi et al developed a novel anticancer drug targeting SLC7A5 based on HT-29 human colon cancer cells, showing satisfactory preincubation inhibitory effects.18 Multiple researchers reported the role of SLC7A5 in malignancies, as well as in classical premalignant pathways.5 Quan et al indicated that SLC7A5 was aberrantly expressed in vascular endothelial cells and was a pivotal player of tumor angiogenesis, which could regulate VEGF-A signal transduction.19 Li et al revealed that SLC7A5 promoted breast cancer proliferation through the Akt/mTORC1 signaling pathway.20 A growing body of evidence shows the important role of SLC7A5 in human diseases, especially in cancers. Our study indicated that SLC7A5 expression was remarkably higher in breast cancer tissues than in normal tissues, correlating with immune cell infiltration and worse prognosis in patients with breast cancer. Meanwhile, we found that SLC7A5 present a diverse expression pattern in different breast cancer subtype. Also, in all the subtypes of breast cancer, SLC7A5 showed a tight correlation with patients’ prognosis, indicating that SLC7A5 could guide the identification and therapy option of breast cancer, which might be an underlying therapeutic target.
As important components of the tumor microenvironment, immune cells would have a significant influence on the malignant behavior of cancer cells through cell interaction.21 Our results showed that SLC7A5 expression positively correlated with Th2, Treg, and CD56− NK cells, as well as macrophages, yet negatively correlated with mast, NK, CD56+ NK, fv and CD8+ T cells. A study conducted by Zhao et al showed that the balance of Th1/Th2 cells could affect breast cancer growth (Th2 cells led to promoting and Th1 cells led to inhibition).22 Ni et al indicated that CD73+γδ1 cells were the major Treg population in breast cancer, resulting in immunosuppressive functions to facilitate disease progression.23 Moreover, Bai et al revealed that Treg cells exert a negative role in antitumor immunity against breast cancer, which could reduce patient survival rates.24 M2 macrophages are reportedly involved in the formation of an immunosuppressive microenvironment.25 In breast cancer, Chen et al revealed that tumor recruited M2 macrophages could secrete CHI3L1 protein, further activating the MAPK signaling pathway and promoting breast cancer metastasis.26 Moreover, the positive correlation with multiple immune-killing cells could also partly affect the cancer-promoting effect of SLC7A5. NK cell-induced cytotoxicity can kill target tumor cells in a cytokine-dependent manner, making it NK cell a promising therapeutic direction in malignancies.27 Meanwhile, Zhang et al showed that breast cancer neoantigen could be identified by CD8+ T cells and activate antitumor immunity.27 The results indicated that SLC7A5 might affect tumor microenvironment remodeling to exert its cancer-promoting effect.
Our GSEA showed that GPCR ligand binding, interleukin signaling, neutrophil degranulation, the metabolism of amino acids and their derivatives, and RHO GTPase signaling were significantly enriched in the high-SLC7A5-expression group. Majumder et al indicated that EP4 could be a therapeutic target for aggressive human breast cancer, exerting its role through GPCR ligand binding.28 Furthermore, Ashok et al indicated that GPCR-associated PAR1 and PI3K kinase might be a promising direction for breast cancer drug development.29 Interleukin signaling also plays an important role in breast cancer. Siersbæk et al found that IL6/STAT3 signaling could hijack estrogen receptor α enhancers to drive breast cancer metastasis.30 Rho GTPases, a family of the Ras GTPase superfamily, are also key regulators of the actin cytoskeleton.31 Studies have shown that Rho GTPase participates in the development and progression of a series of cancers, including breast cancer.31,32
Drug analysis indicated that patients with high SLC7A5 expression were insensitive to paclitaxel and doxorubicin. Immune response and cells in the tumor environment reportedly affect the efficacy of cancer therapy.33 Kubo et al found that paclitaxel might enhance NK cell cytotoxicity against breast cancer cells through increasing perforin levels.34 Vicari et al indicated that paclitaxel could hamper the activity of Treg cells to enhance the anticancer effect of a TLR9 agonist.35 Moreover, based on both in vitro and in vivo models, Hwang et al revealed that NK cells could have treatment effects on doxorubicin-sensitive and doxorubicin-resistant breast cancer cells.36 Considering the correlation between SLC7A5 expression and multiple immune cells, this might explain the difference between paclitaxel and docetaxel sensitivity in patients with different SLC7A5 levels. These results indicated that SLC7A5 might represent a breakthrough for patients with breast cancer with paclitaxel and doxorubicin resistance.
Even though our present study was exclusively based on effective data analysis, it also has certain limitations. First, the included population mainly comprised patients from Western countries. Therefore, the conclusions are inevitably affected by race bias. Second, further experimental exploration should be performed to validate our results through cell- and tissue-based results.
Conclusion
In summary, our study suggests that breast cancer with high SLC7A5 expression levels is correlated with poor survival and enhanced infiltration of many immune cells. In addition, SLC7A5 might indeed regulate ICI efficacy in breast cancer and become a potential target to modulate immune cell function. SLC7A5 may play a prominent role in immune cell infiltration and serve as a prognosis biomarker for breast cancer.
Ethics Approval
Ethics approval was received from the Institutional Review Board (IRB) of The First Affiliated Hospital of Nanjing Medical University (2022-QT-08).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi:10.3322/caac.21708
2. Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 2010;4(3):192–208. doi:10.1016/j.molonc.2010.04.004
3. Wöckel A, Albert US, Janni W, et al. The screening, diagnosis, treatment, and follow-up of breast cancer. Dtsch Arztebl Int. 2018;115(18):316–323. doi:10.3238/arztebl.2018.0316
4. Tosello G, Torloni MR, Mota BS, et al. Breast surgery for metastatic breast cancer. Cochrane Database Syst Rev. 2018;3(3):Cd011276. doi:10.1002/14651858.CD011276.pub2
5. Kandasamy P, Gyimesi G, Kanai Y, et al. Amino acid transporters revisited: new views in health and disease. Trends Biochem Sci. 2018;43(10):752–789. doi:10.1016/j.tibs.2018.05.003
6. Najumudeen AK, Ceteci F, Fey SK, et al. The amino acid transporter SLC7A5 is required for efficient growth of KRAS-mutant colorectal cancer. Nat Genet. 2021;53(1):16–26. doi:10.1038/s41588-020-00753-3
7. Ding K, Tan S, Huang X, et al. GSE1 predicts poor survival outcome in gastric cancer patients by SLC7A5 enhancement of tumor growth and metastasis. J Biol Chem. 2018;293(11):3949–3964. doi:10.1074/jbc.RA117.001103
8. Wang J, Ding M, Zhu H, et al. Up-regulation of long noncoding RNA MINCR promotes non-small cell of lung cancer growth by negatively regulating miR-126/SLC7A5 axis. Biochem Biophys Res Commun. 2019;508(3):780–784. doi:10.1016/j.bbrc.2018.11.162
9. Loftus RM, Assmann N, Kedia-Mehta N, et al. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat Commun. 2018;9(1):2341. doi:10.1038/s41467-018-04719-2
10. Yoon BR, Oh YJ, Kang SW, et al. Role of SLC7A5 in metabolic reprogramming of human monocyte/macrophage immune responses. Front Immunol. 2018;9:53. doi:10.3389/fimmu.2018.00053
11. Nachef M, Ali AK, Almutairi SM, et al. Targeting SLC1A5 and SLC3A2/SLC7A5 as a potential strategy to strengthen anti-tumor immunity in the tumor microenvironment. Front Immunol. 2021;12:624324. doi:10.3389/fimmu.2021.624324
12. El-Ansari R, Craze ML, Alfarsi L, et al. The combined expression of solute carriers is associated with a poor prognosis in highly proliferative ER+ breast cancer. Breast Cancer Res Treat. 2019;175(1):27–38. doi:10.1007/s10549-018-05111-w
13. Tray N, Taff J, Adams S. Therapeutic landscape of metaplastic breast cancer. Cancer Treat Rev. 2019;79:101888. doi:10.1016/j.ctrv.2019.08.004
14. Lamothe SM, Kurata HT. Slc7a5 alters Kvβ-mediated regulation of Kv1.2. J Gen Physiol. 2020;152(7). doi:10.1085/jgp.201912524
15. Baronas VA, Yang RY, Morales LC, et al. Slc7a5 regulates Kv1.2 channels and modifies functional outcomes of epilepsy-linked channel mutations. Nat Commun. 2018;9(1):4417. doi:10.1038/s41467-018-06859-x
16. César-Razquin A, Snijder B, Frappier-Brinton T, et al. A call for systematic research on solute carriers. Cell. 2015;162(3):478–487. doi:10.1016/j.cell.2015.07.022
17. Scalise M, Scanga R, Console L, et al. Chemical approaches for studying the biology and pharmacology of membrane transporters: the histidine/large amino acid transporter SLC7A5 as a benchmark. Molecules. 2021;26(21):6562. doi:10.3390/molecules26216562
18. Okunushi K, Furihata T, Morio H, et al. JPH203, a newly developed anti-cancer drug, shows a preincubation inhibitory effect on L-type amino acid transporter 1 function. J Pharmacol Sci. 2020;144(1):16–22. doi:10.1016/j.jphs.2020.06.006
19. Quan L, Ohgaki R, Hara S, et al. Amino acid transporter LAT1 in tumor-associated vascular endothelium promotes angiogenesis by regulating cell proliferation and VEGF-A-dependent mTORC1 activation. J Exp Clin Cancer Res. 2020;39(1):266. doi:10.1186/s13046-020-01762-0
20. Li Y, Wang W, Wu X, et al. SLC7A5 serves as a prognostic factor of breast cancer and promotes cell proliferation through activating AKT/mTORC1 signaling pathway. Ann Transl Med. 2021;9(10):892. doi:10.21037/atm-21-2247
21. Fujii SI, Shimizu K. Immune networks and therapeutic targeting of iNKT cells in cancer. Trends Immunol. 2019;40(11):984–997. doi:10.1016/j.it.2019.09.008
22. Zhao X, Liu J, Ge S, et al. Saikosaponin A inhibits breast cancer by regulating Th1/Th2 balance. Front Pharmacol. 2019;10:624. doi:10.3389/fphar.2019.00624
23. Ni C, Fang QQ, Chen WZ, et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal Transduct Target Ther. 2020;5(1):41. doi:10.1038/s41392-020-0129-7
24. Bai F, Zhang P, Fu Y, et al. Targeting ANXA1 abrogates Treg-mediated immune suppression in triple-negative breast cancer. J Immunother Cancer. 2020;8(1):e000169. doi:10.1136/jitc-2019-000169
25. Locati M, Curtale G, Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev Pathol. 2020;15:123–147. doi:10.1146/annurev-pathmechdis-012418-012718
26. Chen Y, Zhang S, Wang Q, et al. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J Hematol Oncol. 2017;10(1):36. doi:10.1186/s13045-017-0408-0
27. Wu SY, Fu T, Jiang YZ, et al. Natural killer cells in cancer biology and therapy. Mol Cancer. 2020;19(1):120. doi:10.1186/s12943-020-01238-x
28. Majumder M, Nandi P, Omar A, et al. EP4 as a therapeutic target for aggressive human breast cancer. Int J Mol Sci. 2018;19(4):1019. doi:10.3390/ijms19041019
29. Ashok SR, Shivananda MK, Manikandan A, et al. Discovery and synthesis of 2-amino-1-methyl-1H-imidazol-4(5H)-ones as GPCR ligands; an approach to develop breast cancer drugs via GPCR associated PAR1 and PI3Kinase inhibition mechanism. Bioorg Chem. 2019;86:641–651. doi:10.1016/j.bioorg.2019.02.048
30. Siersbæk R, Scabia V, Nagarajan S, et al. IL6/STAT3 signaling hijacks estrogen receptor α enhancers to drive breast cancer metastasis. Cancer Cell. 2020;38(3):412–23.e9. doi:10.1016/j.ccell.2020.06.007
31. Humphries B, Wang Z, Yang C. Rho GTPases: big players in breast cancer initiation, metastasis and therapeutic responses. Cells. 2020;9(10):2167. doi:10.3390/cells9102167
32. Burbelo P, Wellstein A, Pestell RG. Altered Rho GTPase signaling pathways in breast cancer cells. Breast Cancer Res Treat. 2004;84(1):43–48. doi:10.1023/B:BREA.0000018422.02237.f9
33. Galluzzi L, Zitvogel L, Kroemer G. Immunological mechanisms underneath the efficacy of cancer therapy. Cancer Immunol Res. 2016;4(11):895–902. doi:10.1158/2326-6066.CIR-16-0197
34. Kubo M, Morisaki T, Matsumoto K, et al. Paclitaxel probably enhances cytotoxicity of natural killer cells against breast carcinoma cells by increasing perforin production. Cancer Immunol Immunother. 2005;54(5):468–476. doi:10.1007/s00262-004-0617-6
35. Vicari AP, Luu R, Zhang N, et al. Paclitaxel reduces regulatory T cell numbers and inhibitory function and enhances the anti-tumor effects of the TLR9 agonist PF-3512676 in the mouse. Cancer Immunol Immunother. 2009;58(4):615–628. doi:10.1007/s00262-008-0586-2
36. Hwang MH, Li XJ, Kim JE, et al. Potential therapeutic effect of natural killer cells on doxorubicin-resistant breast cancer cells in vitro. PLoS One. 2015;10(8):e0136209. doi:10.1371/journal.pone.0136209
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







