Back to Journals » Neuropsychiatric Disease and Treatment » Volume 15
Association analysis of polymorphisms in STARD6 and near ECHDC3 in Alzheimer’s disease patients carrying the APOE ε4 Allele
Authors Yin J , Feng W, Yuan H, Yuan J , Wu Y , Liu X, Jin C, Cheng Z
Received 6 September 2018
Accepted for publication 14 December 2018
Published 11 January 2019 Volume 2019:15 Pages 213—218
DOI https://doi.org/10.2147/NDT.S186705
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yuping Ning
Jiajun Yin,1 Wei Feng,2 Hongwei Yuan,3 Jianmin Yuan,1 Yue Wu,3 Xiaowei Liu,3 Chunhui Jin,1 Zaohuo Cheng3
1Brain Science Basic Laboratory, The Affiliated Wuxi Mental Health Center with Nanjing Medical University, Wuxi, Jiangsu Province, China; 2Department of Social Prevention and Control, The Affiliated Wuxi Mental Health Center with Nanjing Medical University, Wuxi, Jiangsu Province, China; 3Department of Geriatric Psychiatry, The Affiliated Wuxi Mental Health Center with Nanjing Medical University, Wuxi, Jiangsu Province, China
Background and purpose: Lipid metabolism plays an important role in Alzheimer’s disease (AD), and recent evidence suggests that single nucleotide polymorphisms (SNPs) in the StAR-related lipid transfer domain 6 (STARD6) and near the enzyme enoyl CoA hydratase domain containing 3 (ECHDC3) gene are related to plasma lipid levels or lipid traits in AD.
Materials and methods: To identify whether the variants in or near the STARD6 and ECHDC3 genes contribute to AD susceptibility, we carried out an association analysis of STARD6 rs10164112 and ECHDC3 rs7920721 in combination with the apolipoprotein E (APOE) ε4 allele in a case–control study (278 cases, 509 controls) in China.
Results: We identified that SNP rs10164112 in the STARD6 gene was a risk factor associated with AD and the APOE ε4 carriers (all P<0.05) after Bonferroni correction. However, multivariate logistic regression analysis indicated that only the minor T allele of STARD6 rs10164112 combined with the APOE ε4 allele increased the risk of AD under the additive and dominant models (additive model: P=0.0078, OR=1.988, 95 % CI: 1.198–3.298; dominant model: P=0.0172, OR=2.169, 95% CI: 1.147–4.102).
Conclusion: These results suggest that the rs10164112-T allele is not an independent risk factor for AD patients. However, in combination with the APOE ε4 allele, the rs10164112-T allele has been found to be a risk factor for AD in the Han Chinese population reported in this study.
Keywords: Alzheimer’s disease, STARD6, ECHDC3, APOE, polymorphism, association study
Introduction
Alzheimer’s disease (AD) is the most common type of dementia and is characterized by the interaction of both genetic and environmental factors.1 Amyloid-β (Aβ) peptide and phosphorylated Tau (p-Tau) reflect cerebral amyloid deposition and tau-related neurodegeneration, respectively, and both are potential diagnostic biomarkers for AD.
The ε4 allele of the apolipoprotein E gene (APOE ε4) is the strongest known genetic risk factor for AD, indicating that lipid metabolism may significantly contribute to AD risk and progression.2–6 However, the molecular mechanisms behind APOE ε4 are not fully understood. One potential mechanism connecting AD to lipid metabolism is the fact that APOE also involves the aggregation and clearance of brain Aβ peptide, which is the main component of senile plaques.7 The amyloid-β protein precursors (AβPP), presenilin 1 (PSEN1) and presenilin 2 (PSEN2), have been shown to lead to familial forms of AD and have also been reported to influence Aβ metabolism.8 Thus, it is important that additional AD risk genes are uncovered.
In the last 2 years, studies have identified two single nucleotide polymorphisms (SNPs) that are associated with AD susceptibility, including rs7920721 (closest gene, ECHDC3; enzyme enoyl CoA hydratase domain containing 3) and rs10164112 (STARD6; steroidogenic acute regulatory-related lipid transfer domain 6); the genes have also been shown to be involved in plasma lipid levels or other lipid traits.9–11 Desikan et al9 demonstrated the pleiotropic effects of rs7920721 near the ECHDC3 gene for AD, plasma C-reactive protein and lipid levels. The rs7920721 SNP is adjacent to ECHDC3, a gene that encodes the enzyme enoyl CoA hydratase domain containing 3; furthermore, the expression of ECHDC3 was shown to be altered in the brains of AD patients when compared with those of the brains of controls.9 Subsequently, a recent, large, transethnic genome-wide association study (GWAS) identified rs7920721 to be specifically associated with persons lacking the APOE ε4 allele.10 The risk locus rs7920721 was also identified through proxy case–control analysis and meta-analysis within a published GWAS of AD.12,13 In addition, increased ECHDC3 expression was reported in patients with acute myocardial infarction, and ECHDC3 is known to be involved in fatty acid biosynthesis in mitochondria.14 Furthermore, Kim et al11 reported that STARD6 rs10164112 was significantly correlated with AD. The steroidogenic acute regulatory (StAR)-related lipid transfer (START) domain is a conserved sequence of approximately 210 amino acids that bind lipids, including sterols.15,16 In humans, there are 15 START domain proteins, STARD1–STARD15, which can be classified into six subfamilies.17,18 START proteins are also implicated in the nonvesicular transport of lipids and sterols, lipid metabolism, and cell signaling events.17,18 STARD6 behaves similar to StAR proteins, which control the rate-limiting step of neurosteroid synthesis in which cholesterol is transferred from the outer mitochondrial membrane to the inner membrane.19,20 Accordingly, Létourneau et al21 published on the detailed structural and dynamical data obtained by STARD6 in steroid transport. Nevertheless, to date, no study has evaluated both SNPs rs7920721 and rs10164112 among AD patients in a Han Chinese population.
The SNPs rs7920721 and rs10164112 and the APOE genotypes are known to be related to lipid metabolism, which plays a central role in AD. Therefore, this study was designed to explore these two recently reported SNPs in combination with the APOE ε4 allele in terms of the risk of AD in the Han Chinese population.
Materials and methods
Subjects
Patients with AD were recruited from the Wuxi Mental Health Center and diagnosed according to the criteria of the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA). Participants with a family history of AD, neurological or psychiatric illnesses or dementia were excluded. The healthy controls without memory complaints or cognition dysfunctions were confirmed to be neurologically normal according to their medical history, general examinations and Mini-Mental State Examination (MMSE). All participants were of the Han Chinese ethnic population and were geographically from Wuxi city in Jiangsu province. According to self-reported data, individuals not born in Jiangsu or whose family members were not born in Jiangsu were excluded from the study. This study was approved by the ethics committee of the Wuxi Mental Health Center, and, prior to enrollment, signed informed consent was obtained from each participant or guardian. The study protocols were performed according to the principles of the Declaration of Helsinki.
The study sample included 278 AD patients (118 males and 160 females; mean age =72.77 years at recruitment, SD=9.09) and 509 unrelated healthy controls (249 males and 260 females; mean age =71.78 years at recruitment, SD=6.27), which were drawn from a population of people of Han descent (Table 1).
  | Table 1 Demographic characteristics of the study participants |
DNA extraction and SNP genotyping
Blood samples were collected from all participants using K2EDTA tubes, and DNA was extracted from whole blood using a blood genotyping DNA extraction kit (Tiangen Biotech, Beijing, China). DNA samples were stored at −80°C until genotype analysis, which used the ligase detection reaction–PCR (LDR-PCR) method from Shanghai Biowing Applied Biotechnology Co., Ltd. (Shanghai, China) (www.biowing.com.cn).22,23 Genomic DNA extracted from the clinical samples was first subjected to multiplexed PCR to obtain a PCR product including rs7920721 and rs10164112. The DNA sequencer was then used to detect the LDR reaction products.
Statistical and bioinformatic analyses
PLINK v1.9 (http://pngu.mgh.harvard.edu/purcell/plink/) and SPSS 24.0 software (IBM Corporation, Armonk, NY, USA) were used to perform all statistical analyses, including association studies, Hardy–Weinberg equilibrium (HWE) tests and the calculation of genotype and allele frequencies between patients with AD and healthy controls.24 P-values of allelic and genotypic tests were calibrated using Bonferroni correction and were considered statistically significant if they were less than 0.05. A power calculation of our sample size was carried out using PS software (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize).25,26 ORs and 95% CI were calculated to assess the influence of any difference in heterogeneity.
Multivariate logistic regression was carried out to analyze the association between the SNP and susceptibility to AD after adjusting for age, gender and the APOE ε4 allele in the additive, dominant or recessive models. We defined various genetic models such as A2A2 vs (A1A1+ A1A2) for dominant, (A1A2+ A2A2) vs A1A1 for recessive and A1A1 vs A1A2 vs A2A2 for additive (A1: minor allele; A2: major allele).
Results
In this study, we analyzed the data from 278 patients with AD and 509 unrelated healthy controls. The demographic and clinical characteristics of the participants are summarized in Table 1. There were no significant differences in age (P=0.106) or gender (P=0.106) between the two groups. However, there were significant differences in MMSE scores, marital status, education level and APOE genotype between the AD and control groups (P<0.001). One copy of the APOE ε4 allele was a significant risk factor for AD (P<0.001, OR =2.422, 95% CI=2.718–3.414).
Genotype distributions were in HWE for rs7920721 and rs10164112 in both the AD and control groups (P>0.05). The total genotyping rate of all individuals was 100%. In addition, the power of this study for both SNPs were estimated to be ~70% using PS software (assumption condition: α=0.05, P0=0.25, n=556, m=1.83, ?=1.339).
The results of the association study suggested that SNP rs10164112 within STARD6 was significantly associated with the risk of AD after Bonferroni correction (Pallele =0.0254, OR =1.339, 95% CI: 1.064–1.685; Pgenotype =0.0460) (Table 2). Subsequently, according to APOE ε4 status, we stratified our data to investigate whether APOE ε4 influenced the relationship between SNPs and AD susceptibility risk. The allele and genotype frequencies of STARD6 rs10164112 were significantly different between AD patients and healthy controls in the APOE ε4 carriers (ε4+: Pallele =0.0016, Pgenotype =0.0072) (Table 2). However, there were no significant differences in the genotype and allele frequencies of the rs7920721 SNP closest to ECHDC3 between the AD and control groups.
Multivariate logistic regression analyses were applied to re-evaluate the relation between STARD6 rs10164112 and AD susceptibility risk in subjects with APOE ε4 status while adjusting for age and gender under different genetic models (Table 3). In APOE ε4 carriers, the minor T allele of STARD6 rs10164112 strongly increased the risk of AD under the additive and dominant models (the additive model: P=0.0078, OR =1.988, 95% CI: 1.198–3.298; the dominant model: P=0.0172, OR =2.169, 95% CI: 1.147–4.102).
Discussion
To our knowledge, this is the first study to associate the T allele of rs10164112 within STARD6 with an increased risk for AD in the Han Chinese population. There were significant differences in the allele and genotype distributions of the STARD6 rs10164112 polymorphism between the AD and control groups in subjects with the APOE ε4 allele. Thus, it is possible to suggest that the risk of AD increases with the combination of the T allele of rs10164112 and APOE ε4 alleles (ORADD =1.988, ORDOM =2.169) in the Han Chinese population.
It was initially reported by Kim et al11 that STARD6 rs10164112 was significantly correlated with AD using multivariate logistic regression models (P≤0.0014, OR ≤0.037). Although the genotype and allele frequencies had a statistically significant correlation between STARD6 rs10164112 and AD within the total sample of this study, the logistic regression models indicated no significant correlation in the total samples. In addition, inconsistent from the results reported by Kim et al,11 the frequency of the rs10164112 “T” allele in the AD patients was significantly higher than that in the controls in this study. Moreover, when we compared the allele frequency of STARD6 rs10164112 from different ethnic groups using dbSNP data from the 1,000 Genomes Project, the C allele frequency was 0.39, as shown in Figure 1, as it consists of different ethnic groups; however, it ranged from 0.06 to 0.70. The Han Chinese population (1,000 genomes) frequency is similar to what was described in the manuscript. Thus, our allele frequency analysis of these data for STARD6 rs10164112 revealed substantial differences when compared to other ethnic groups (Figure 1). The differences in the rs10164112 polymorphism profiles suggested that populations from different geographies might exhibit genetic heterogeneity for AD. Therefore, in studies involving the association between the STARD6 gene polymorphism and AD, racial and geographic aspects should be taken into account. Additional limitations of this study that should be considered are that our results may be influenced by the lack of statistical power (~70%) due to a limited sample size.
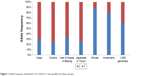  | Figure 1 Allele frequency distribution of rs10164112 among different ethnic groups. |
Conclusion
The rs10164112-T allele of the STARD6 gene was not an independent risk factor for AD. However, this allele, combined with the APOE ε4 allele, leads to an increased risk for AD in the studied Han Chinese population. When compared with the results of previous research, inconsistent results were obtained. Thus, in the future, the data need to be replicated and tested using large-scale genetic studies involving different ethnic groups.
Acknowledgments
We thank the AD patients and healthy subjects for their cooperation in the study. This work was supported by the key R&D Program of Jiangsu Province from Jiangsu Province Science and Technology Department (BE2015615).
Author contributions
JJY and ZC conceived and designed the experiments. JJY, WF and JMY performed the experiments. JJY and WF analyzed the data. WF, HY, YW and XL contributed reagents/materials/analysis tools. JJY and CJ wrote the paper. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Tanzi RE. The genetics of Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2(10):a006296. | ||
Bertram L, Tanzi RE. Thirty years of Alzheimer’s disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9(10):768–778. | ||
Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(5):1977–1981. | ||
Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. | ||
Poirier J, Davignon J, Bouthillier D, Kogan S, Bertrand P, Gauthier S. Apolipoprotein E polymorphism and Alzheimer’s disease. Lancet. 1993;342(8873):697–699. | ||
Strittmatter WJ, Saunders AM, Goedert M, et al. Isoform-specific interactions of apolipoprotein E with microtubule-associated protein tau: implications for Alzheimer disease. Proc Natl Acad Sci U S A. 1994;91(23):11183–11186. | ||
Beffert U, Aumont N, Dea D, Lussier-Cacan S, Davignon J, Poirier J. Apolipoprotein E isoform-specific reduction of extracellular amyloid in neuronal cultures. Brain Res Mol Brain Res. 1999;68(1–2):181–185. | ||
Scheuner D, Eckman C, Jensen M, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2(8):864–870. | ||
Desikan RS, Schork AJ, Wang Y, et al. Polygenic overlap between C-reactive protein, plasma lipids, and Alzheimer disease. Circulation. 2015;131(23):2061–2069. | ||
Jun GR, Chung J, Mez J, et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement. 2017;13(7):727–738. | ||
Kim YJ, Paik JW, Kang WS, et al. Genetic association of STARD6 polymorphisms with Alzheimer’s disease in a Korean population. J Neurol Sci. 2016;366:100–101. | ||
Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–1458. | ||
Liu JZ, Erlich Y, Pickrell JK. Case-control association mapping by proxy using family history of disease. Nat Genet. 2017;49(3):325–331. | ||
Eicher JD, Wakabayashi Y, Vitseva O, et al. Characterization of the platelet transcriptome by RNA sequencing in patients with acute myocardial infarction. Platelets. 2016;27(3):230–239. | ||
Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci. 1999;24(4):130–132. | ||
Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 2001;43(2):134–144. | ||
Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005;118(Pt 13):2791–2801. | ||
Clark BJ. The mammalian START domain protein family in lipid transport in health and disease. J Endocrinol. 2012;212(3):257–275. | ||
Lavaque E, Sierra A, Azcoitia I, Garcia-Segura LM. Steroidogenic acute regulatory protein in the brain. Neuroscience. 2006;138(3):741–747. | ||
Bose HS, Whittal RM, Ran Y, Bose M, Baker BY, Miller WL. StAR-like activity and molten globule behavior of StARD6, a male germ-line protein. Biochemistry. 2008;47(8):2277–2288. | ||
Létourneau D, Bédard M, Cabana J, Lefebvre A, Lehoux JG, Lavigne P. STARD6 on steroids: solution structure, multiple timescale backbone dynamics and ligand binding mechanism. Sci Rep. 2016;6:28486. | ||
Yin J, Jia N, Liu Y, et al. No association between FOXP2 rs10447760 and schizophrenia in a replication study of the Chinese Han population. Psychiatr Genet. 2018;28(2):1–23. | ||
Yi P, Chen Z, Zhao Y, et al. PCR/LDR/capillary electrophoresis for detection of single-nucleotide differences between fetal and maternal DNA in maternal plasma. Prenat Diagn. 2009;29(3):217–222. | ||
Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. | ||
Dupont WD, Plummer WD. Power and sample size calculations for studies involving linear regression. Control Clin Trials. 1998;19(6):589–601. | ||
Dupont WD, Plummer WD. Power and sample size calculations. A review and computer program. Control Clin Trials. 1990;11(2):116–128. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


