Back to Journals » Journal of Inflammation Research » Volume 14
Zc3h12d, a Novel of Hypomethylated and Immune-Related for Prognostic Marker of Lung Adenocarcinoma
Authors Yang B, Ji LL , Xu HL, Li XP, Zhou HG, Xiao T, Li XH, Gao ZY, Li JZ, Zhang WD, Wang GS, Li MJ
Received 28 January 2021
Accepted for publication 14 May 2021
Published 15 June 2021 Volume 2021:14 Pages 2389—2401
DOI https://doi.org/10.2147/JIR.S304278
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Ning Quan
Bo Yang,1– 3,* Lin-Lin Ji,1,2,* Hong-Liang Xu,1,* Xiao-Ping Li,3 Hong-Gang Zhou,4 Ting Xiao,4 Xiao-He Li,4 Zhou-Yong Gao,1,2 Jian-Zhong Li,5 Wei-Dong Zhang,3 Guang-Shun Wang,1 Ming-Jiang Li3
1Department of Thoracic Surgery, Tianjin Baodi Hospital, Baodi Clinical College of Tianjin Medical University, Tianjin, 301800, People’s Republic of China; 2State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing, 102206, People’s Republic of China; 3Department of Thoracic Surgery, Tianjin First Central Hospital, School of Medicine, Nankai University, Tianjin, 300192, People’s Republic of China; 4State Key Laboratory of Medicinal Chemical Biology, College of Pharmacy and Tianjin Key, Laboratory of Molecular Drug Research, Nankai University, Tianjin, 300353, People’s Republic of China; 5Department of Thoracic Surgery, The Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, 710004, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Guang-Shun Wang
Department of Thoracic Surgery, Tianjin Baodi Hospital, Baodi Clinical College of Tianjin Medical University, Tianjin, 301800, People’s Republic of China
Email [email protected]
Ming-Jiang Li
Department of Thoracic Surgery, Tianjin First Central Hospital, School of Medicine, Nankai University, Tianjin, 300192, People’s Republic of China
Email [email protected]
Background: Zc3h12d is a negative regulator which plays a crucial role in immune modulation. However, the role of zc3h12d in lung adenocarcinoma (LUAD) remains unclear. We aim to explore the prognostic of zc3h12d and investigate the relationship between zc3h12d expression and immune infiltration in LUAD.
Methods: TIMER site was used to analyze the expression of zc3h12d in LUAD. The zc3h12d protein levels in patient tissue samples were detected by immunohistochemistry staining assays. Meanwhile, based on UALCAN database and samples’ data from our cohort, we explored the relationship of clinicopathological features and zc3h12d expression to determine the clinical effect of zc3h12d in LUAD. Several databases including GEPIA, Kaplan–Meier plotter and our samples’ data were used to explore the prognostic value of zc3h12d in LUAD. Cox regression analysis was established to further evaluate the prognostic value of zc3h12d in LUAD. In addition, zc3h12d promoter methylation was analyzed by UALCAN database. Genetic alteration analysis was observed in the cBioPortal web. GO and KEGG analyses were conducted to elucidate the underlying mechanisms. Finally, the correlation between zc3h12d and tumor-infiltrating immune cells in LUAD was investigated by TIMER database. The B cells level was investigated by flow cytometry analysis of peripheral blood from our LUAD cohort.
Results: Zc3h12d expression was significantly higher in LUAD, compared with adjacent normal tissues. The clinical data from the UALCAN database demonstrated that zc3h12d expression was closely related with cancer stage and nodal metastasis. However, patient sample detection revealed that zc3h12d expression was closely related to pathological N (p = 0.0431) and grade (p = 0.004). Moreover, low zc3h12d expression was associated with poorer overall survival in LUAD. We analyzed the methylation level of zc3h12d in LUAD and found that the methylation levels of zc3h12d promoter in LUAD were significantly reduced. In addition, zc3h12d genetic alterations, including deep deletion, could be found in LUAD. GO and KEGG pathway analysis results indicated that zc3h12d has a certain value in immune infiltration. We investigated the expression of zc3h12d in tumor-immune interactions. It was found that zc3h12d might be associated with the immune infiltration and markers of infiltrating immune cells of LUAD. The results of patient sample detection confirmed that B cells level was significantly lower in the patients with low zc3h12d expression than those in the patients with high zc3h12d expression.
Conclusion: zc3h12d might be considered as a potential biomarker for determining prognosis and immune-related therapeutic target in LUAD.
Keywords: zc3h12d, lung adenocarcinoma, hypomethylation, tumor microenvironment, prognosis
Introduction
Although surgical resection, chemotherapy, radiotherapy, targeted therapy, and immunotherapy show responses during the treatment of non-small cell lung cancer (NSCLC), the survival rate of the patients remains poor in the 21st century.1 Lung adenocarcinoma (LUAD) represented the most frequent types of NSCLC, which account for up to 40% of all lung cancer cases.2 It is noteworthy that DNA methylation was shown to correlate with the progression of LUAD.3 Recent studies also have shed light on the diverse immune cell infiltration for LUAD.4 Under this background, new prognostic markers and incorporating of methylation and immune information are urgent needs for NSCLC prognostication.
The CCCH-zinc finger protein family consisted of four members: zc3h12a, zc3h12b, zc3h12c and zc3h12d.5 A previous study suggested that CCCH zinc finger motifs bind to DNA, RNA, and proteins.6 Zc3h12d was enriched in spleen, lung, and lymph node.7 Researches showed that zc3h12d exerted important functions in immune modulation and inflammation. Several studies revealed that zc3h12d attenuated the inflammatory factors by T lymphocytes.8 In addition, overexpression of zc3h12d significantly inhibited TLR-induced activation of JNK, ERK and NF-κB in macrophages.9
As a novel tumor suppressor gene p34 in lung cancer, zc3h12d was revealed previously, but the association of patients with LUAD remains unknown. Therefore, the goal of this study was to determine the clinical usefulness of zc3h12d in LUAD patients.
Materials and Methods
Tissue and Immunohistochemistry (IHC)
All tumor specimens with LUAD after surgical resection were collected between January 2018 and December 2019 in this cohort study. None of the patients received chemotherapy, radiotherapy, and/or immunotherapy prior to resection. This study was performed in accordance with the Declaration of Helsinki and was approved by the Committees for Ethical Review of Research involving human subjects at the Tianjin First Central Hospital, School of Medicine, Nankai University (Tianjin, China; approval no. 2018N054KY). Written informed consent was obtained from all participants. The demographic and clinical characteristics of patients are listed in Table 1. All IHC staining was performed as described in the previous study.10 Staining percentage of zc3h12d was categorized as follows: 0 (0%), 1 (0 −10%), 2 (11–50%), 3 (51–70%), and 4 (≥71%) and the staining intensity was stratified as follows: none (-, 0), faint (+, 1, yellow staining), moderate (++, 2, light brown staining), or strong (+++, 3, dark brown staining). Based on the IHC score of 0 to 7 (staining percentage + intensity), zc3h12d in IHC tissue was evaluated and divided into low groups (< 3 points) and high groups (4 −7 points).
 |
Table 1 Relationship Between zc3h12d Expression and Clinicopathology in the LUAD Cohort |
Differential Expression Analysis
The expression difference of zc3h12d in various types of tumors was observed by employing TIMER database (https://cistrome.shinyapps.io/timer/).11 Then, zch3h12d expression in LUAD patients with different clinical features in UALCAN tool was obtained (http://ualcan.path.uab.edu/analysis.html).12
Survival Analysis
The GEPIA (http://gepia.cancer-pku.cn/) and Kaplan–Meier (K-M) Plotter database (http://kmplot.com/analysis/index.php?p=service) platform were used to analyze the prognostic value of zc3h12d.13,14 The cutoff value was used as the expression thresholds to divide the patients into the high- and low-zc3h12d cohorts.
Methylation Analysis
To evaluate association between zc3h12d expression and methylation levels, the promoter methylation in LUAD with different conditions was analyzed in the UALCAN database.12 And the DNA methylation sites of zc3h12d in TCGA were investigated via the MethSurv platform (https://biit.cs.ut.ee/methsurv/).15
Genetic Alteration Analysis
The results of the mutations and copy number alterations (CNAs) of the zc3h12d from all TCGA tumors were determined in the cBioPortal web (https://www.cbioportal.org/).16 The overall survival (OS), disease-free survival (DFS), progression-free survival (PFS), and disease-specific survival (DSS) differences for the TCGA datasets with or without zc3h12d genetic alteration in LUAD patients were also obtained.
Zc3h12d-Related Gene Set Enrichment Analysis
Furthermore, UALCAN database was used to select the relate gene set of zc3h12d.12 The biological processes (BP), cellular components (CC), molecular function (MF) and KEGG pathway were visualized using the Metascape database (http://metascape.org/gp/index.html#/main/step1).17
Immune Infiltration Analysis
To reveal the immune infiltration of zc3h12d in cancer, the correlation between zc3h12d and the immune infiltrates across all TCGA tumors were estimated by the TIMER database.11 Then, the associations of zc3h12d and mentioned immune cell infiltration gene markers in LUAD were explored.
Measurement of B Cells
Blood samples were collected before treatment initiation. B cells level was examined by flow cytometry analysis of peripheral blood from our LUAD cohort.
Statistical Analysis
Paired t-test and unpaired t-test were used to analyze the expression of zc3h12d in different groups. Distinctions between zc3h12d expression and clinicopathologic characteristics were evaluated by χ2 test or Fisher’s exact test. The survival curves of patient were evaluated by the Log rank test. A Cox regression model was applied for the univariate and multivariate analyses of survival. Correlations of zc3h12d with immune infiltration and type markers of immune cells were analyzed using Spearman correlation. The differences in mean values between the groups were analyzed using the Mann‑Whitney U-test. P-value < 0.05 was considered statistically significant. All analyses and graphics were analyzed using SPSS ver. 26.0 (IBM, USA) and GraphPad Prism 8.0 (GraphPad Software, USA).
Results
Zc3h12d is Upregulated in LUAD and Closely Correlated with Clinical Characteristics
The expression of zc3h12d in different tumor tissues was detected by the TIMER database. As shown in Figure 1A, the zc3h12d expression was significantly higher in breast invasive carcinoma (BRCA), cervical and endocervical cancer (CESC), esophageal carcinoma (ESCA), head and neck cancer (HNSC), kidney renal clear cell carcinoma (KIRC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), and stomach adenocarcinoma (STAD) compared with adjacent normal tissues. However, lower expression was observed in colon adenocarcinoma (COAD), kidney chromophobe (KICH), kidney renal papillary cell carcinoma (KIRP), and pancreatic adenocarcinoma (PAAD) compared with the corresponding control tissues.
Next, the correlations between zc3h12d and clinical features of LUAD patients were examined. As shown in Figure 1B, the expression of zc3h12d was much higher in LUAD tissues than in normal tissues. Moreover, zc3h12d expression was closely related with cancer stage and nodal metastasis. Compared to the normal tissue, zc3h12d expression was augmented regardless of race, gender, aged (41–60, 61–80, and 81–100), and TP53 mutation status.
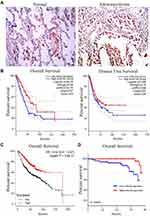
To further examine zc3h12d protein expression in LUAD tissues, the IHC was used to assess the zc3h12d protein expression. Results showed that zc3h12d was predominantly localized in cytoplasm. The expression of zc3h12d was significantly higher in LUAD compared to the uninvolved tissues (Figure 2A).
In addition, to validate the association between zc3h12d protein expression and clinical variables, tumor specimens were analyzed. It was found that zc3h12d expression was closely related to pathological N (p = 0.0431) and grade (p = 0.004). In comparison, other clinical items, such as age (p = 0.288), gender (p = 0.350), smoking status (p = 0.272), T (p = 0.799) and TNM stage (p = 0.090) had no association with zc3h12d expression.
Decreased Zc3h12d Expression is a Predictor of Poor Prognosis in LUAD
The prognostic value of zc3h12d expression in LUAD was investigated by GEPIA analysis. Results showed that OS was significantly positively correlated with zc3h12d expression in LUAD patients (hazard ratio [HR] = 0.52, log-rank p = 2e-05). However, the DFS in relation to zc3h12d expression was not significant (HR = 0.75, log-rank p = 0.066) (Figure 2B). Kaplan–Meier plotter database showed that high zc3h12d expression was also associated with better prognosis of OS in LUAD (HR = 0.52, log-rank p = 3.6e-07) (Figure 2C). Moreover, analysis of prognostic significance of tumor specimens (n = 87) revealed that patients with low zc3h12d expression had shortened OS in LUAD (p = 0.0039) (Figure 2D).
Cox regression model was performed to evaluate the prognostic potential of zc3h12d expression. Zc3h12d expression was a significant predictor of OS in univariate analysis (HR = 7.140, 95% CI 1.542–33.064, p = 0.012) and multivariate analysis (HR = 12.845, 95% CI 2.093–78.828, p = 0.006) (Table 2).
 |
Table 2 Univariate and Multivariate Analyses of Factors Associated with OS in LUADs Using Cox Regression |
DNA Methylation Analysis Data
The heat map of DNA methylation levels of zc3h12d in LUAD were analyzed and displayed in Figure 3A. Furthermore, compared with normal tissues, the methylation levels of zc3h12d promoter in LUAD was significantly reduced. We observed that zc3h12d promoter methylation levels of the cancer stage, race, gender, age, smoking status, nodal metastasis, and TP53 mutation status were lower than normal in LUAD (Figure 3B).
Genetic Alteration Analysis Data
As shown in Figure 4A, there was the highest genetic alteration of zc3h12d (8.33%) in diffuse large B-cell lymphoma (DLBC). It was noteworthy that all LUAD cases with genetic alteration (0.53%) had copy number deletion of zc3h12d. In addition, the analysis of the prognostic value of LUAD patients between the zc3h12d altered group and the unaltered group showed better prognosis in disease-free (p = 4.506e-06) and progression-free (p = 1.950e-05) survival, but not overall (p = 0.272) and disease-specific (p = 0.396) survival (Figure 4B).
Zc3h12d is Involved in Immune Activation and Proliferation Inhibition in LUAD
Obtain the potential zc3h12d-related gene set by UALCAN database. A total of 931 genes was finally obtained (Supplementary Table 1). GO analysis results indicated that BP category of target genes were significantly enriched in “lymphocyte activation”, “adaptive immune response”, and “immune response-regulating signaling pathway”. CC category were mainly enriched in “side of membrane”, “immunological synapse”, and “plasma membrane protein complex”. MF category of target genes were mainly related to “cytokine receptor activity”, “GTPase binding”, and ‘non-membrane spanning protein tyrosine kinase activity”. KEGG pathway analysis revealed that the target genes were mainly associated with “Th1 and Th2 cell differentiation”, “chemokine signaling pathway”, “primary immunodeficiency”, “T cell receptor signaling pathway”, “B cell receptor signaling pathway”, and “NF-kappa B signaling pathway” (Figure 5). Based on this perspective, the zc3h12d in LUAD patients is deemed important.
 |
Figure 5 Functional enrichment of zc3h12d. The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enriched terms colored according to P-values. |
Relationship Between Zc3h12d and Tumor-Infiltrating Immune Cells in LUAD
Tumor-infiltrating lymphocytes were an independent predictor of immune surveillance in determining the survival in various cancers. Therefore, we investigated whether zc3h12d expression was correlated with immune infiltration levels in LUAD. The results showed that zc3h12d was positive correlation with B cells (r = 0.325, p = 1.30e-13), CD8 + T cells (r = 0.173, p = 1.18e-04), CD4 + T cells (r = 0.399, p = 2.59e-20), neutrophils (r = 0.343, p = 4.52e-15), dendritic cells (r = 0.27, p = 1.13e-09), Tregs (r = 0.117, p = 9.14e-03), and monocyte (r = 0.204, p = 5.01e-06) immune infiltration levels of LUAD (Figure 6A).
To further validate these results, the relationships between zc3h12d expression and immune marker genes of different immune cells were explored by TIMER database, including B cells, CD8+ T cells, neutrophils, macrophages, dendritic cells, tregs, monocytes, NK cells, Th1 cells, Th2 cells, and Th17 cells in LUAD. Interestingly, we found that the levels of most immune markers were significantly correlations with zc3h12d expression in LUAD (Table 3).
 |
Table 3 Correlation Analysis Between zc3h12d and Immune Cell Type Markers in TIMER Database |
As the tumor immune infiltration and immune marker of B cells exhibited significant correlations with zc3h12d. Then, B cells were selected for further study. The result confirmed that B cells level were significantly lower in patients with low zc3h12d expression than patients with high zc3h12d expression (p = 0.0009) (Figure 6B). These findings might provide an explanation for the differences in patient survival.
Discussion
To enhance the prediction of LUAD patients’ prognosis, DNA methylation was demonstrated with vital prognostic value.18 DNA methylation on specific sites could regulate corresponding gene expression.19 In addition to the DNA methylation, the host immune response had emerged as a promising therapeutic option with the potential to improve OS in LUAD patients.20
Zc3h12a plays a critical role in T cell mediated immunosuppression. However, transformed follicular lymphoma (TFL, zc3h12d), an RNase belonging to the same family of Regnase-1 (zc3h12a), was originally reported as a putative tumor suppressor gene.21 Although zc3h12d had not been extensively studied, the anti-tumor properties were confirmed in cell experiments. Subsequently, the same role of zc3h12d in the 3ʹUTRs was revealed, as zc3h12a.6 Simultaneously, the relationship between the primary human memory T lymphocytes and zc3h12d expression was reported as well.22
Even though the zc3h12d correlated with survival in endometrial cancer patients.21 Here, we report that zc3h12d may be considered as a potential biomarker for determining prognosis and immune-related therapeutic target in LUAD.
High zc3h12d expression in LUAD is observed compared with adjacent normal tissues and immunohistochemistry shows that zc3h12d is mainly localized in cytoplasmic granules. Then, various clinic factors of zc3h12d expression are integrated in LUAD patients. Database analysis results show that zc3h12d expression is widely related with disease stage, ethnicity, gender, age, lymph node status, TP53 mutation status and smoking status in LUAD patients. However, sample detection of patients reveals that zc3h12d expression is closely related to pathological N (p = 0.0431) and grade (p = 0.004). In comparison, other clinicopathological items, including age, gender, smoking status, T and TNM stage show that it has no statistical association with zc3h12d protein expression. We find low zc3h12d expression is closely related with poorer OS in LUAD. In addition, univariate and multivariate analyses indicates that lower expression of zc3h12d may be defined as a risk factor which affects the OS of LUAD patients.
Hypomethylation is one of the important components of epigenetic alterations of tumorigenesis and progression.23 Recent study has demonstrated that the promoter hypomethylation might be served as a prognostic indicators and drug target in lung cancer.24 In the course of our investigations, the promoter methylation levels of zc3h12d are significantly decreased. And the hypomethylation of zc3h12d may cause a significant increasing in zc3h12d expression.
It has been proved that genomic mutation is closely associated with tumorigenesis.25 Therefore, a genetic alteration analysis is conducted. Result indicates that genetic alteration of zc3h12d in NSCLC (1.15% frequency) such as deep deletion, and mutation could be found. Intriguingly, all LUAD cases with genetic alteration (0.53% frequency) have copy number deletion of zc3h12d. Considering all the above results, zc3h12d might play an important role in the disease progression and survival prognosis of patients with LUAD.
Similar with zc3h12a, zc3h12d is also a negative regulator of cytokine expression. In cell experiments, zc3h12d is proved as a novel negative feedback regulator of TLR signaling and macrophage activation.26 And zc3h12d recognizes the same 3ʹUTR-dependent regulation of the turnover of mRNAs encoding interleukin-6 (IL-6), tumor necrosis factor (TNF), and immediate early response 3 gene (IER3), as zc3h12a.6 The role of zc3h12d in pathogenic immune responses is also described. Recently, a strong correlation between zc3h12d expression and human T lymphocytes is reported.8 To further identify the mechanism of zc3h12d in LUAD, the zc3h12d-related gene set is obtained by employing UALCAN database. GO and KEGG pathway analysis results indicate that zc3h12d has a certain value in immune infiltration, such as the inhibition of cell proliferation in LUAD.
In the NSCLC tumor microenvironment (TME), the infiltrating immune cells account for a large proportion, including T cells, B cells, and macrophages, etc.27,28 In vivo-isolated human CD4+ T cells experiments, it is found that deficiency of zc3h12d could increase pro-inflammatory phenotype of T lymphocytes.8 In the present study, our data shows that there is a positive relationship between zc3h12d and infiltration level of B cells, CD8 + T cells, CD4 + T cells, neutrophils, dendritic cells, Tregs, and monocyte in LUAD. In addition, tumor-associated-B cells in NSCLC is related to a favorable outcome.29 Our results of patient sample detection confirm that B cells level is significantly lower in the patients with low zc3h12d expression than patients with high zc3h12d expression. As such, zc3h12d has significantly positively correlation with the immune cell-type markers in LUAD. Notably, markers of T helper cells, such as STAT1, TBX21, STAT5A, and IL17A also show positive association with zc3h12d. Some studies have revealed that tumor infiltration by Th1 CD4+ T cells, Th2 CD4+ T cells, and Th17 CD4+ T cells can mediate regression of advanced solid tumors.30 Thus, these evidences provide a potential mechanism of zc3h12d in tumor immune microenvironment for researchers.
Conclusions
In summary, there is a close correlation of zc3h12d with LUAD. Low zc3h12d expression is closely related with poorer prognosis in LUAD. Besides, zc3h12d may promote tumor-induced immune response activation and immune infiltration in LUAD and inhibit the proliferation of lung cancer to play an anticancer role. Although our results offer compelling evidence of zc3h12d in LUAD, further studies should be verified in multicenter, large-sample in future.
Declarations
The authors declare no support from any organizations for the submitted work. The design of the study, the analyses and the writing of the manuscript were solely the responsibility of the authors.
Data Sharing Statement
The data of the current research are available from the corresponding author on a reasonable request.
Consent for Publication
Not applicable
Acknowledgments
We thank Prof. Chang-Qing Sun, and Hui Wang from the department of Science and Education of Tianjin Baodi Hospital of Tianjin Medical University for their kind technical assistance. Bo Yang, Lin-Lin Ji and Hong-Liang Xu should be regarded as co-first authors.
Disclosure
The authors declare that there are no conflicts of interest.
References
1. Schneider BJ, Ismaila N, Aerts J, et al. Lung cancer surveillance after definitive curative-intent therapy: ASCO Guideline. J Clin Oncol. 2020;38(7):753–766. doi:10.1200/JCO.19.02748
2. Aizemaiti R, Wu Z, Tang J, et al. Heat shock factor 5 correlated with immune infiltration serves as a prognostic biomarker in lung adenocarcinoma. Int J Med Sci. 2021;18(2):448–458. doi:10.7150/ijms.51297
3. Zhao W, Rong Z, Wang W, et al. Methylation biomarkers with discriminating ability are potential therapeutic targets in lung adenocarcinoma. Epigenomics. 2020. doi:10.2217/epi-2019-0142
4. Li Z, Ding B, Xu J, et al. Relevance of STK11 mutations regarding immune cell infiltration, drug sensitivity, and cellular processes in lung adenocarcinoma. Front Oncol. 2020;10:580027. doi:10.3389/fonc.2020.580027
5. Huang S, Qi D, Liang J, et al. The putative tumor suppressor Zc3h12d modulates toll-like receptor signaling in macrophages. Cell Signal. 2012;24(2):569–576. doi:10.1016/j.cellsig.2011.10.011
6. Wawro M, Kochan J, Krzanik S, et al. Intact NYN/PIN-Like domain is crucial for the degradation of inflammation-related transcripts by ZC3H12D. J Cell Biochem. 2017;118(3):487–498. doi:10.1002/jcb.25665
7. Zhang H, Wang WC, Chen JK, et al. ZC3H12D attenuated inflammation responses by reducing mRNA stability of proinflammatory genes. Mol Immunol. 2015;67(2Pt B):206–212. doi:10.1016/j.molimm.2015.05.018
8. Emming S, Bianchi N, Polletti S, et al. A molecular network regulating the proinflammatory phenotype of human memory T lymphocytes. Nat Immunol. 2020;21(4):388–399. doi:10.1038/s41590-020-0622-8
9. Liang J, Wang J, Azfer A, et al. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem. 2008;283(10):6337–6346. doi:10.1074/jbc.M707861200
10. Wang W, Zhang Z, Yang B, et al. [Down-regulation of Zc3h12d expression in lung tissues of rats with acute lung injury induced by intestinal ischemia-reperfusion]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2014;30(4):355–359.
11. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.CAN-17-0307
12. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
13. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
14. Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi:10.1371/journal.pone.0082241
15. Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277–288. doi:10.2217/epi-2017-0118
16. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi:10.1126/scisignal.2004088
17. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
18. Li R, Yang YE, Yin YH, et al. Methylation and transcriptome analysis reveal lung adenocarcinoma-specific diagnostic biomarkers. J Transl Med. 2019;17(1):324. doi:10.1186/s12967-019-2068-z
19. Koch A, Joosten SC, Feng Z, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15(7):459–466. doi:10.1038/s41571-018-0004-4
20. García-González J, Ruiz-Bañobre J, Afonso-Afonso FJ, et al. PD-(L)1 Inhibitors in combination with chemotherapy as first-line treatment for non-small-cell lung cancer: a pairwise meta-analysis. J Clin Med. 2020;9(7):2093. doi:10.3390/jcm9072093
21. Wakahashi S, Kawakami F, Wakahashi K, et al. Transformed Follicular Lymphoma (TFL) predicts outcome in advanced endometrial cancer. Cancer Epidemiol Biomarkers Prev. 2018;27(8):963–969. doi:10.1158/1055-9965.EPI-17-0762
22. Minagawa K, Wakahashi K, Kawano H, et al. Posttranscriptional modulation of cytokine production in T cells for the regulation of excessive inflammation by TFL. J Immunol. 2014;192(4):1512–1524. doi:10.4049/jimmunol.1301619
23. Imperatori A, Sahnane N, Rotolo N, et al. LINE-1 hypomethylation is associated to specific clinico-pathological features in Stage I non-small cell lung cancer. Lung Cancer. 2017;108:83–89. doi:10.1016/j.lungcan.2017.03.003
24. Noguera-Uclés JF, Boyero L, Salinas A, et al. The Roles of Imprinted SLC22A18 and SLC22A18AS gene overexpression caused by promoter cpg island hypomethylation as diagnostic and prognostic biomarkers for non-small cell lung cancer patients. Cancers (Basel). 2020;12(8):2075. doi:10.3390/cancers12082075
25. Li Y, Yuan J. Role of deubiquitinating enzymes in DNA double-strand break repair. J Zhejiang Univ Sci B. 2021;22(1):63–72. doi:10.1631/jzus.B2000309
26. Huang WQ, Yi KH, Li Z, et al. DNA methylation profiling reveals the change of inflammation-associated ZC3H12D in Leukoaraiosis. Front Aging Neurosci. 2018;10:143. doi:10.3389/fnagi.2018.00143
27. Sun J, Xie T, Jamal M, et al. CLEC3B as a potential diagnostic and prognostic biomarker in lung cancer and association with the immune microenvironment. Cancer Cell Int. 2020;20:106. doi:10.1186/s12935-020-01183-1
28. Liu F, Wu H. CC chemokine receptors in lung adenocarcinoma: the inflammation-related prognostic biomarkers and immunotherapeutic targets. J Inflamm Res. 2021;14:267–285. doi:10.2147/JIR.S278395
29. Yuan Q, Sun N, Zheng J, et al. Prognostic and immunological role of FUN14 domain containing 1 in pan-cancer: friend or foe? Front Oncol. 2020;10(9):1502. doi:10.3389/fonc.2019.01502
30. Davies SI, Barrett J, Wong S, et al. Robust production of merkel cell polyomavirus oncogene specific T cells from healthy donors for adoptive transfer. Front Immunol. 2020;11:592721. doi:10.3389/fimmu.2020.592721
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.