Back to Journals » Infection and Drug Resistance » Volume 16
Within-Host Resistance and Virulence Evolution of a Hypervirulent Carbapenem-Resistant Klebsiella pneumoniae ST11 Under Antibiotic Pressure
Authors Zhou C , Zhang H, Xu M, Liu Y, Yuan B, Lin Y, Shen F
Received 19 August 2023
Accepted for publication 7 November 2023
Published 16 November 2023 Volume 2023:16 Pages 7255—7270
DOI https://doi.org/10.2147/IDR.S436128
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Sandip Patil
Cong Zhou,1 Hui Zhang,1 Maosuo Xu,1 Yajuan Liu,1 Baoyu Yuan,2 Yong Lin,1 Fang Shen1
1Department of Clinical Laboratory, Shanghai Fifth People’s Hospital, Fudan University, Shanghai, People’s Republic of China; 2Department of Clinical Laboratory, Shanghai Children’s Hospital, Shanghai Jiao Tong University, Shanghai, People’s Republic of China
Correspondence: Fang Shen; Yong Lin, Department of Clinical Laboratory, Shanghai Fifth People’s Hospital, Fudan University, No. 128, Ruili Road, Minhang District, Shanghai, 200240, People’s Republic of China, Tel +86 18021073261 ; +86 18718168695, Email [email protected]; [email protected]
Background: Hypervirulent carbapenem-resistant Klebsiella pneumoniae (hv-CRKP) has recently aroused an extremely severe health challenge and public concern. However, the underlying mechanisms of fitness costs that accompany antibiotic resistance acquisition remain largely unexplored. Here, we report a hv-CRKP-associated fatal infection and reveal a reduction in virulence due to the acquisition of aminoglycoside resistance.
Methods: The bacterial identification, antimicrobial susceptibility, hypermucoviscosity, virulence factors, MLST and serotypes were profiled.The clonal homology and plasmid acquisition among hv-CRKP strains were detected by XbaI and S1-PFGE. The virulence potential of the strains was evaluated using Galleria mellonella larvae infection model, serum resistance assay, capsular polysaccharide quantification, and biofilm formation assay. Genomic variations were identified using whole-genome sequencing (WGS).
Results: Four K. pneumoniae carbapenemase (KPC)-producing CRKP strains were consecutively isolated from an 86-year-old patient with severe pneumonia. Whole-genome sequencing (WGS) showed that all four hv-CRKP strains belonged to the ST11-KL64 clone. PFGE analysis revealed that the four ST11-KL64 hv-CRKP strains could be grouped into the same PFGE type. Under the pressure of antibiotics, the antimicrobial resistance of the strains increased and the virulence potential decreased. Further sequencing, using the Nanopore platform, was performed on three representative isolates (WYKP586, WYKP589, and WYKP594). Genomic analysis showed that the plasmids of these three strains underwent a large number of breaks and recombination events under antibiotic pressure. We found that as aminoglycoside resistance emerged via acquisition of the rmtB gene, the hypermucoviscosity and virulence of the strains decreased because of internal mutations in the rmpA and rmpA2 genes.
Conclusion: This study shows that ST11-KL64 hv-CRKP can further evolve to acquire aminoglycoside resistance accompanied by decreased virulence to adapt to antibiotic pressure in the host.
Keywords: hypervirulent carbapenem-resistant Klebsiella pneumoniae, within-host evolution, plasmids, whole-genome sequencing
Introduction
Klebsiella pneumoniae (KP) is an opportunistic pathogen that causes a wide range of infections, including pneumonia, urinary tract infections, bacteremia and meningitis.1 In recent years, the continuous evolution of hypervirulent or carbapenem-resistant plasmids has led to the emergence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae(hv-CRKP).2,3 These strains are considered real ‘superbugs’ as they are not only hypervirulent but also multidrug resistant, causing severe and often fatal infections.2,4,5 In China, the most prevalent type of hv-CRKP was ST11-KL64.6 The formation mechanism of hv-CRKP can be divided into the following three paths: (i) Carbapenem-resistance plasmids were acquired from K1/K2 hypervirulent Klebsiella pneumoniae(hvKP) and evolved into CR-hvKP. Carbapenem-resistant plasmids are usually conjugative and can be transmitted horizontally among different strains.5,7 (ii) Carbapenem-resistant Klebsiella pneumoniae(CRKP) acquired virulence plasmids and evolved into hv-CRKP. However, virulence plasmids are generally considered non-mobilizable owing to the lack of tra gene locus for plasmid conjugation.8 (iii) KP directly acquired hybrid plasmids encoding carbapenem resistance and hypervirulence, which led to the rapid evolution of KP into hv-CRKP or CR-hvKP.9–13 It seems easier for hvKP strains to obtain carbapenem-resistant plasmids than for CRKP strains to obtain nonconjugative virulence plasmids. But in fact, hv-CRKP strains producing KPC carbapenemase has an overwhelming dominance of CR-hvKP strains.6 Outbreaks caused by hv-CRKP, especially ST11 hv-CRKP, have been reported in hospitals in China.2,14
Aminoglycosides are an important choice for the treatment of life-threatening hv-CRKP infection, and combined with β-lactam drugs can effectively reduce the mortality rate.15,16 Acquired 16S rRNA methylases (16S-RMTases) are the most clinically significant aminoglycoside resistance mechanism.17 Since 16S-RMTase encoding genes were first discovered in Enterobacteriaceae and Pseudomonas aeruginosa in 2003, they have been identified in a variety of gram-negative bacteria worldwide. Until now, we have found eleven 16S RMTase-encoding genes (armA, rmtA to rmtH, npmA, and npmB) that confer high-level resistance to all clinically relevant aminoglycosides.18,19 The 16S-RMTase gene rmtB is primarily found in E. coli and K. pneumoniae worldwide and results in resistance to aminoglycosides with MIC≥512µg/mL.20–22 The 16S-RMTase encoding genes are mostly located within transferable plasmids and/or are associated with mobile genetic elements, such as transposons, integrons, and insertion sequences.23
At present, there are many studies on the formation mechanism of hv-CRKP, but there is still a lack of research on within-host resistance and virulence evolution of hv-CRKP under antibiotic pressure. Here, we report within-host evolution of a series of KPC-2-producing ST11-KL64 hv-CRKP strains collected from an 86-year-old patient with severe pneumonia. In this study, we found for the first time that as aminoglycoside resistance emerged via acquisition of the rmtB gene, the hypermucoviscosity and virulence of the ST11-KL64 hv-CRKP strains decreased due to internal mutations in the rmpA and rmpA2 genes. Tracking the phenotypic and genetic changes of hv-CRKP strains under antibiotic pressure in the host and exploring their evolutionary mechanisms can provide technical support for more effective clinical treatment and nosocomial infection control.
Materials and Methods
Bacterial Strains
A retrospective study was conducted to collect four KPC-producing CRKP strains consecutively isolated from an 86-year-old patient with severe pneumonia. Carbapenem resistance was determined when imipenem or meropenem were resistant by antimicrobial susceptibility test. Aminoglycoside resistance was determined when amikacin or gentamicin were resistant by antimicrobial susceptibility test. In this study, hypervirulent K. pneumoniae was defined as K. pneumoniae carrying virulence-plasmid-associated loci (rmpA, rmpA2, iutA-iucABCD, and iroN).
Bacterial Identification and Antimicrobial Susceptibility Testing
Bacterial identification was performed using VITEK-2 compact automated microbiology analyzer (BioMérieux, France). MALDI-TOF mass spectrometry (Bruker Daltonics, Germany) was used to recheck the identification of the bacterial strains. Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used as controls for bacterial identification. Antimicrobial susceptibility testing was performed using the broth microdilution method of the Clinical and Laboratory Standards Institute(CLSI). The results were interpreted according to 2021 CLSI breakpoints24 for all antimicrobial agents except tigecycline and polymyxin B, which were interpreted using the interpretative criteria of the Food and Drug Administration (FDA) and the European Committee on Antimicrobial Susceptibility Testing(EUCAST), respectively. Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used as controls for antimicrobial susceptibility testing.
Definition of Hypermucoviscous Phenotype
The string test was used to detect the hypermucoviscous phenotype. All strains were inoculated on 5% sheep blood agar plates and cultured overnight at 37°C. The bacterial colony was gently pulled upward with an inoculation loop and repeated for 3 times.2 If viscous strings were formed three times and their lengths were longer than 5 mm, the string test was positive.
The mucoviscosity of the test strains was evaluated using a sedimentation assay.25 Briefly, cultures were maintained in LB broth overnight and then subcultured to an OD600 of 0.2 in fresh medium, followed by incubation at 37°C. The cultures were normalized to OD of 1.0 mL−1 after 6 hours and centrifuged at 1000g for 5 min. The supernatant was gently removed without disturbing the pellet to measure the absorbance (OD600). Each assay was repeated three times.
Whole Genome Sequencing and Bioinformatics Analysis
The genomic DNA of four strains was extracted using a QIAamp DNA mini kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. Genomic DNA was subjected to library preparation using a NEXTflex Rapid DNA Sequencing Kit. Fragmented genomic DNA was sequenced using the 150-bp paired-end Illumina NovaSeq platform (Illumina, San Diego, CA, United States). Furthermore, the genomic DNA of three representative isolates (WYKP586, WYKP589, and WYKP594) out of the four hv-CRKP isolates was sequenced on the long-read Oxford Nanopore MinION platform (Nanopore Technologies, Oxford, United Kingdom). The experimental procedures were performed according to the standard protocol provided by Oxford Nanopore Technologies, and included sample quality testing, library construction, library quality testing, and library sequencing. Libraries were constructed using the SQK-LSK109 ligation kit (Nanopore Technologies, Oxford, United Kingdom). EXP-FLP002 kit was used as the Flow Cell Priming (Nanopore Technologies, Oxford, United Kingdom). De novo assembly was conducted using Unicycler version 0.4.8.26 Finally, error correction of the assembled genome sequences was performed using the Pilon version 1.22. Gene annotation was performed using the Prokka version 1.12.27
Mobile genetic elements(MGEs), antibiotic resistance, and virulence genes were predicted using VRProfile2.28 Plasmid incompatible types and conjugative transfer-related modules of plasmids were also predicted using VRProfile2.28 Multilocus sequence typing (MLST) and capsule serotypes were identified according to the BIGSdb-Pasteur (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef&page=sequenceQuery). Core genome MLST(cgMLST) analysis was performed using SeqSphere+ version 9.0.8 (Ridom GmbH, Muenster, Germany).29 The alignment of plasmids at different evolutionary stages was visualized using the BLAST Ring Image Generator (BRIG) version 0.95 and Easyfig version 2.2.5.30,31
Pulsed-Field Gel Electrophoresis(PFGE)
Clonal homology and plasmid acquisition among hv-CRKP strains were detected using XbaI and S1-PFGE. XbaI and S1-PFGE were performed as previously described with some modifications.32 Bacterial DNA was digested with XbaI and S1-nuclease(Sangon, Shanghai, China), respectively. PFGE was performed using the CHEF Mapper system (Bio-Rad Laboratories, Hercules, CA, USA) with a running time of 19 h and pulse time of 6–36 s. Bands were stained with 4SGelred (Sangon Biotech, Shanghai, China) prior to visualization under UV light. PFGE results were interpreted according to the criteria described by Tenover et al.33 Salmonella enterica serotype Braenderup H9812 was used as the molecular size marker. Strains with fewer than three different fragments were considered the same clone.33
Serum Resistance Assay
We performed a serum resistance assay to determine in vitro virulence. The serum resistance assay was performed as described previously with some modifications.34 Bacterial suspensions containing 1×106 CFU/mL were collected from the mid-log phase cultures. Briefly, 10μL of bacterial suspension was added to 90μL of healthy human serum to obtain a bacterial suspension concentration of approximately 1×105 CFU/mL. The mixture was then incubated at 37°C and 300 rpm for 3 h, and colony counts were performed at 0, 1, 2, and 3 h using the serial dilution method. Each experiment was performed in triplicates.
Galleria Mellonella Infection Model
The virulence potential of the strains was evaluated using a Galleria mellonella larval infection model. The Galleria mellonella injection protocol was performed as previously described, with some modifications.35 Healthy Galleria mellonella larvae were purchased from Tianjin Huiyude Biotechnology Co., Ltd. (Tianjin, China) weighing about 300 mg. Ten Galleria mellonella larvae were in each treatment group. The mid-log phase culture was washed and diluted in phosphate-buffered saline (PBS) to obtain a solution of 106 CFU/mL. For each group, the Galleria mellonella larvae were inoculated with 10 µL of at a concentration of 106 CFU/mL WYKP586, WYKP587, WYKP589 or WYKP594. Galleria mellonella larvae were injected with 10 µL of bacterial suspension using a microsample syringe. The inoculated Galleria mellonella larvae were placed in an incubator at 37°C for 72 h, and the survival of the larvae in each group was recorded every 4 h. The positive control was the ST23 K1 hvKP strain NTUH-K2044, and the negative control was Klebsiella pneumoniae ATCC700603 and phosphate-buffered saline (PBS).Each assay was repeated three times. Survival curves were drawn according to the survival conditions of the larvae using the GraphPad Prism 7 software.
Biofilm Formation Assay
Biofilm formation was performed as described previously in 96-well microtiter plates.36 Briefly, 200µL of a mid-log phase bacterial suspension (2x107 CFU/mL) was added to 96-well microtiter plates and incubated overnight at 37°C. Next, all the cultures were removed and washed twice with phosphate-buffered saline. A crystal violet solution (0.2%) was used for staining for 30 min and the cells were washed three times. The crystal violet bound to the biofilm was completely dissolved by adding 200µL of 95% ethanol, and its absorbance (OD595) was measured. Each assay was repeated three times.
Capsular Polysaccharide Quantification
Capsular polysaccharides were quantified as previously described with slight modifications.37 Briefly, 1.2 mL of sodium tetraborate/sulfuric acid was added to 500µL of the mid-log phase bacterial suspension (1x106 CFU/mL), placed in an ice bath, incubated for 5 min at 100°C, and then left on ice for 10 min. After cooling, 20µL of 1.5 mg/mL m-hydroxydiphenyl was added to the mixture, stirred well, and bubbles were removed. After a 5-min incubation at 25°C, absorbance (OD540) was measured. The uronic acid content was determined with reference to a standard curve of uronic acid and was expressed in mg/L. The standard curve was drawn by the following steps: 500ul ddH2O was put into the EP tube, and 0 μL, 2.5μL, 5μL, 7.5 μL, 10 μL, 12.5 μL, and 15 μL uronic acid standard solution were added to it, respectively. The experimental procedures were performed as above, and finally absorbance (OD540) was measured. Each assay was repeated three times.
Statistical Analysis
Data are presented as the mean ± standard deviation. GraphPad Prism 7 (GraphPad Software, San Diego, CA)) was used to calculate significance using Student’s t-test or analysis of variance (ANOVA). Statistical significance was set at P <0.05.
Results
Medical History of Infection
An 86-year-old male patient was admitted to our hospital on June 9, 2021. The patient had a history of cerebral infarction, hypertension, and diabetes for several years. Three days prior, the patient accidentally fell and landed on the waist and buttocks. He was unconscious of fever for one day. He was later admitted to the hospital and was diagnosed with severe pneumonia, pulmonary embolism, and cerebral infarction. After admission, the patient was treated using endotracheal intubation and ventilator-assisted ventilation. Symptomatic treatments such as anti-infection, atomization, and expectoration were actively performed. The first strain of hv-CRKP (named WYKP586) was isolated from a sputum specimen and the second strain of hv-CRKP (named WYKP587) was isolated from a tracheal tube specimen on June 17. The third strain of hv-CRKP (named WYKP589) was isolated from the sputum specimens on June 21. Amikacin was administered intravenously at a dose of 400mg every 12 hours. After treatment, the patient’s vital signs stabilized and his cough and expectoration improved. The patient was discharged on June 23.
The patient was readmitted to the hospital on June 25, 2021, and was diagnosed with septic shock, severe pneumonia, and cerebral infarction. He was admitted to the hospital because of fever with chest tightness, shortness of breath, and confusion for one day. The patient’s condition did not improve after emergency treatments such as physical cooling, endotracheal intubation, ventilator-assisted ventilation, active anti-infection, and atomization. The fourth strain of hv-CRKP (named WYKP594) was isolated from a sputum specimen on June 25. On June 26, the patient’s condition deteriorated, and he suffered cardiorespiratory arrest in the afternoon and was declared clinically dead.
Clinical and strain details, timelines, body temperature, indicators of bacterial infection, and antibiotic use are summarized in Figure 1.
 |
Figure 1 Medical history of the patient with hv-CRKP infection. Abbreviations: WBC, white blood cell count; CRP, C-reactive protein; PCT, procalcitonin |
Antimicrobial Resistance and Virulence Phenotypes of Hv-CRKP Strains
Four hv-CRKP strains from the same patient at different evolutionary stages showed similar drug resistance phenotypes, which were resistant to most antibiotics, but sensitive to trimethoprim/sulfamethoxazole, tigecycline, and polymyxin B. The first isolated hv-CRKP (WYKP586) was sensitive to aminoglycosides such as amikacin and gentamicin. The remaining three hv-CRKP strains (WYKP587, WYKP589, and WYKP594) were resistant to amikacin and gentamicin (Table 1). The resistance of the strains increased under antibiotic pressure. The first three strains of hv-CRKP (WYKP586, WYKP587, and WYKP589) were positive in the string test, and the length of the viscous string was >100 mm, 10 mm, and 5 mm respectively and the last isolated strain of hv-CRKP (WYKP594) was negative in the string test, as shown in Figure S1. Under antibiotic pressure, the length of the viscous string decreases. The sedimentation assay showed the mucoviscosity of the strains decreased with antibiotic treatment (Figure 2E).
 |
Table 1 Antimicrobial Resistance Phenotypes of Hv-CRKP Strains |
We estimated the virulence potential of these four hv-CRKP strains by infecting Galleria mellonella larvae with an inoculum of 1×106 CFU. The 48h survival rate of Galleria mellonella larvae infected with the classic Klebsiella pneumoniae ATCC700603 and PBS was 80% and 100%, respectively. The 36h survival rate of Galleria mellonella larvae infected with the hvKP reference strain NTUH-K2044 was 0. The 48h survival rate of Galleria mellonella larvae infected with WYKP586 was 0, while the 48h survival rate of Galleria mellonella larvae infected with WYKP587, WYKP589, and WYKP594 were 10%, 20%, and 40%, respectively, as shown in Figure 2A. The virulence potential of these four hv-CRKP strains was higher than that of the classical strain ATCC700603 but lower than that of the hvKP reference strain NTUH-K2044. Following antibiotic treatment, the virulence of these strains decreased.
We used a serum resistance assay to estimate the virulence potential of the four hv-CRKP strains. The hvKP reference strain NTUH-K2044 showed a slight increase in colony counts in the first 2 h, which decreased by the third hour. These four hv-CRKP strains showed a decrease in colony counts within 3 h of the serum resistance assay, but the difference was that the later the isolates were isolated, the faster the colony counts decreased, as shown in Figure 2B. Under antibiotic pressure, the ability of these strains to resist immune killing by serum decreased with antibiotic treatment.
We further evaluated the virulence potential of these strains using capsular polysaccharide quantification and biofilm formation. With antibiotic treatment, capsular polysaccharide quantification and biofilm formation decreased, as shown in Figure 2C and D. Compared with biofilm formation, the use of antibiotics had a more serious effect on the decrease in capsular polysaccharide quantification.
Genotypic Analysis of Hv-CRKP Strains
According to the Whole-genome sequencing(WGS) analysis, these four hv-CRKP strains belonged to ST11 and had the same K type of KL64. According to the WGS data, antimicrobial resistance and virulence genes of the four ST11-KL64 hv-CRKP strains were determined (Table S1 and Figure S2). Seven antimicrobial resistance genes, blaKPC-2, blaSHV-12, CatA2, sul2, tet(A), blaLAP-2 and qnrS1, were identified across the four strains, whereas two genes were variable (Table S1 and Figure S2). Two antimicrobial resistance genes, rmtB and blaTEM-1B, were present in WYKP587, WYKP589, and WYKP594 but were absent in WYKP586. Virulence factors related to iron acquisition, regulator of mucoid phenotype, serum resistance adherence, anti-phagocytosis, secretion systems, and efflux pumps were found, which revealed that all these strains belonged to the hypervirulent CRKP ST11-KL64 type (Table S1 and Figure S2).
PFGE and Phylogenetic Analysis of Hv-CRKP
The four ST11-KL64 hv-CRKP strains had almost identical antibacterial susceptibility profiles and belonged to the ST11-KL64 type. PFGE analysis revealed that the four ST11-KL64 hv-CRKP strains could be grouped into the same PFGE type (Figure 3A). Four ST11-KL64 hv-CRKP strains were clustered into the same cgMLST group, with fewer than 12 allelic differences, and were named Cluster 1(Figure 3C). This suggested that the four ST11-KL64 hv-CRKP strains were the same clonal strains at different evolutionary times in the same patient.
Comparative Analysis of Plasmids in ST11-KL64 Hv-CRKP
S1-PFGE analysis revealed that the four hv-CRKP strains, despite being members of the same clone, harbored different numbers of plasmids (Figure 3B). S1-PFGE results showed that strain WYKP586 harbored two plasmids, strain WYKP587 and strain WYKP589 harbored three plasmids, and strain WYKP594 harbored four plasmids (Figure 3B). Because strains WYKP587 and WYKP589 harbored the same number of plasmids, only one of the two strains (WYKP589), as well as WYKP586 and WYKP594, were subjected to long-read sequencing. However, the number of plasmids detected by WGS in these three strains differed from that detected by S1-PFGE. The complete genome of WYKP586 was found to contain a chromosome of 5.5 Mbp in size, and 4 plasmids ranging from 5.6 to 302.2 Kbp. The complete genome of WYKP589 contained a chromosome 5.5 Mbp in size and five plasmids ranging from 5.6 to 217.9 Kbp. The complete genome of WYKP594 was found to contain a chromosome of 5.5 Mbp in size, and five plasmids ranging from 5.6 to 217.7 Kbp (Table S1). This phenomenon may be attributed to the detection limit of S1-PFGE. It is well known that owing to technical limitations, plasmids with sizes less than 30kb may be missed by S1-PFGE. The three hv-CRKP strains used in this study contained plasmids less than 30kb (Table S1). Using the available clinical data from the patient, we reconstructed a timeline of the evolution of these three hv-CRKP strains, showing their plasmid content (Figure 4). Under antibiotic pressure, the plasmids of these three strains underwent numerous breaks and recombinations.
The plasmid pWYKP586-1 was 302kb in size and contained both virulence and antimicrobial resistance genes. Under antibiotic pressure, pWYKP586-1 was broken into two plasmids, pWYKP589-1 and pWYKP589-3, with size of 218kb and 84kb, respectively (Figures 3B, 4, 5A and 6A). Among them, pWYKP589-1 contained only virulence genes and pWYKP589-3 contained only drug resistance genes (Table S1, Figures 5A and 6A). After that, the two plasmids remained stable, and no breakage or recombination was observed in the strain WYKP594 (Figures 4 and 6A). The plasmid pWYKP589-2 was 110kb in size and contained antimicrobial resistance genes. Under the pressure of antibiotics, pWYKP589-2 was broken into two plasmids, pWYKP594-3 and pWYKP594-4, with size of 73kb and 51kb, respectively (Figure 6B). A portion of pWYKP594-3 was derived from the recombination of the plasmid pWYKP589-4 (Figure 5C). Plasmid mobility can be divided into conjugative, mobilizable, and non-mobilizable.38 Plasmids are considered to be conjugative in the presence of four essential modules (oriT, relaxase, T4CP, and T4SS). Plasmids are considered mobilizable when oriT is present, but relaxase, type IV coupling protein [T4CP], and T4SS are not present simultaneously. Plasmids are considered Non-mobilizable when oriT is not present. Plasmids pWYKP586-1, pWYKP589-3, pWYKP594-2, and pWYKP594-3 were predicted to carry a complete set of conjugative modules with all four essential modules (oriT, relaxase, type IV coupling protein [T4CP], and T4SS), suggesting that these four plasmids were conjugative (Table S1). Interestingly, pWYKP594-3 was predicted to be conjugative, arising from the breakage of pWYKP589-2 and the fusion of pWYKP589-4; however, pWYKP589-2 and pWYKP589-4 were non-mobilizable (Figures 4, 5C and 6B).
Aminoglycoside Resistance Emerges via Acquisition of the rmtB Gene
In addition to plasmids, other MGEs, such as insertion sequences (ISs) and transposons (Tns), surrounding resistance genes also play important roles in the horizontal transmission of acquired antimicrobial resistance (AMR) genes. Hence, we also analyzed the MGEs surrounding these four key resistant elements (blaKPC-2, blaSHV-12, rmtB and blaTEM-1B) to evaluate the evolution of these resistance genes in hv-CRKP. Moreover, two key resistance genes were focused on that played significant roles in the development of resistance to carbapenems (blaKPC-2) and aminoglycosides (rmtB). The plasmid pWYKP586-2 was 110kb in size with 55% GC content and contained two copies of blaKPC-2 and blaSHV-12 resistance genes located on two identical MGEs (Figure 5B). The segment was IS26–blaSHV-12–TnAs1–TnAs1–ISKpn6–blaKPC-2–ISKpn27–Tn3–IS26, which was flanked by IS26 (Figure 5B). Plasmid pWYKP589-2 was 110kb in size with 55% GC content and contained blaKPC-2, blaSHV-12, rmtB, and blaTEM-1B resistance genes. The resistance genes blaKPC-2 and blaSHV-12 were located on a MGEs, which was the same as the MGEs on the plasmid pWYKP586-2 (Figure 5B). The segment was IS26–blaSHV-12–TnAs1–TnAs1–ISKpn6–blaKPC-2–ISKpn27–Tn3–IS26, which was flanked by IS26. The resistance genes rmtB and blaTEM-1B were located on other MGEs with the structure of Tn3–IS26–IS903B–IS26–IS91–IS26–IS91–Tn3–blaTEM-1B–rmtB–IS15DI. The plasmids pWYKP586-2 and pWYKP589-2 were highly homologous in size and structure. The most important difference was that the MGEs containing the antimicrobial resistance genes blaSHV-12 and blaKPC-2 were replaced by MGEs containing the antimicrobial resistance genes rmtB and blaTEM-1B(Figure 5B). Notably, the rmtB gene encodes 16S rRNA methyltransferase (16S RMTase), which mediates high levels of resistance to aminoglycosides.
Mutations in rmpA and rmpA2 Genes Resulted in Decreased Hypermucoviscosity
The mutations were located in rmpA and rmpA2 associated with capsular polysaccharide synthesis. Whole-genome sequencing showed that the rmpA2 gene on plasmids pWYKP586-1, pWYKP589-1, and pWYKP594-1 was 300bp in size (Figures 7A and S3A). We found that rmpA2 gene on these three plasmids completely overlapped with rmpA2 on pK2044 at nucleotide position 393–692, and was truncated before nucleotide position 393 and after nucleotide position 692 (Figures 7A and S3A). Truncated rmpA2 may affect the transcription and expression of rmpA2, leading to a decrease in hypermucoviscosity.39 Whole-genome sequencing results showed that the size of rmpA gene on plasmids pWYKP586-1 and pWYKP589-1 was the same as that on pK2044, which was 633bp. However, the rmpA gene in pWYKP594-1 was split into two segments, 369bp and 300bp, respectively (Figures 7B, S3B and S3C). After alignment with the rmpA gene on plasmid pK2044, we found that the rmpA genes on pWYKP586-1 and pWYKP589-1 completely overlapped with rmpA on pK2044 (Figure S3B). There was a homologous region of 35bp between the two rmpA genes formed by fracture in the plasmid pWYKP594-1. The two rmpA genes on the plasmid pWYKP594-1 overlapped with the first and second halves of the rmpA gene on pK2044 (Figure S3C). Truncated rmpA may affect the transcription and expression of rmpA, leading to a further decrease in hypermucoviscosity of the strain.
 |
Figure 3 XbaI-PFGE, S1-PFGE and cgMLST of hv-CRKP WYKP586, WYKP587, WYKP589, and WYKP594. (A) The homology among hv-CRKP strains were detected by XbaI-PFGE. (B) Plasmid acquisition among hv-CRKP strains were detected by S1-PFGE. (C) Minimum-spanning tree of cgMLST profiles among hv-CRKP strains. The minimum-spanning tree was generated based on cgMLST analysis with 2358 conserved genome-wide genes. A cluster was defined at a distance of ≤15 alleles.28 |
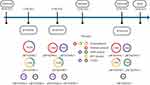 |
Figure 4 Timeline showing plasmid changes in 3 hv-CRKP isolates. Abbreviations: KPC, Klebsiella pneumoniae carbapenemase; MDR, multidrug resistance. |
Discussion
Zhou et al showed that ST11-KL47 hv-CRKP was replaced by ST11-KL64 hv-CRKP after 2016 in China and that ST11-KL64 hv-CRKP led to higher mortality in infected patients.40 Our previous study also showed that the most prevalent type of hv-CRKP is ST11-KL64.41 Owing to the hypervirulence and transmissibility of these strains, effective surveillance and strict infection control strategies are essential to prevent their further dissemination. Here, we report within-host resistance and virulence evolution of a fatal ST11-KL64 hv-CRKP strain under antibiotic pressure.
Multiple strains isolated from the same patient at different times have highly conserved core genomes. Our longitudinal data suggests that the use of distinct antimicrobial drugs may drive the acquisition of resistance. This study showed that under the pressure of antibiotics, the acquisition of antimicrobial resistance was accompanied by a decrease in the hypermucoviscosity and virulence of strains. Whole-genome sequencing analysis revealed that the plasmids of the strains underwent a large number of breaks and recombinations, and the number of plasmids changed, but the total size of plasmids in each strain remained relatively stable. Jin et al studied the evolution of tigecycline and polymyxin resistance in ST11 hypervirulent carbapenem-resistant Klebsiella pneumoniae during treatment with tigecycline and polymyxin. During the evolution process, the plasmids did not undergo a large number of breaks and recombinations, and the number of plasmids was three.29 Dong et al tracked microevolutionary events among ST11 CR-hvKP outbreak strains. Their study found that each of the three ST11 CR-hvKP isolates (CR-HvKP1, CR-HvKP4 and CR-HvKP5) harbored five plasmids with sizes of 178, 177.5, 99.7, 11.9, and 5.6 Kb, respectively.42 Chen et al studied the within-patient microevolution of carbapenem-resistant, hypervirulent K. pneumoniae isolates. During the evolutionary process, the number of plasmids in the strains and the total size of the plasmids showed dynamic changes. Treatment with gentamicin and ciprofloxacin correlated with the appearance of an MDR plasmid that encoded resistance genes to both classes of antimicrobial drugs. The 165kb MDR plasmid was obtained and subsequently downsize to 100kb resistant plasmids. Throughout the evolutionary process, the K. pneumonia virulence plasmid and pKPC2 were stable and showed few changes over the course of a year.43 In this study, plasmids pWYKP586-2 and pWYKP589-2 were highly homologous in size and structure. The most important difference was that the MGEs containing the drug resistance genes blaSHV-12 and blaKPC-2 was replaced by MGEs containing the drug resistance genes rmtB and blaTEM-1B. Notably, rmtB gene encodes 16S rRNA methyltransferase (16S RMTase), which mediates high levels of resistance to aminoglycosides. Studies have shown that rmtB is one of the most widespread 16S rRNA methylase genes.20,44
Classic pLVPK-like and pK2044-like virulence plasmids are generally regarded as nonconjugative because they lack complete conjugative elements, which may limit their self-transmissibility.5,45 Notably, the strain WYKP586 initially isolated in this study harbored the superplasmid pWYKP586-1, which had the following characteristics: (i) a plasmid that harbors virulence and antimicrobial resistance genes; (ii) harboring a complete set of conjugative machinery with all four essential modules (oriT, relaxase, type IV coupling protein [T4CP], and T4SS) that guarantees self-transmissibility. The presence of such superplasmids increases the risk of simultaneous transfer of virulence and antimicrobial resistance genes. With the popularization of whole-genome sequencing technology, superplasmids have been discovered. Zhou et al found that antimicrobial resistance genes and the virulence factor iuc operon co-occur on the conjugative plasmid p1864-1.46 Xia et al identified a rare conjugative plasmid, pCY814036-iucA, carrying a virulence-associated iuc operon (iucABCDiutA) coding for aerobactin and determinants of multidrug resistance (MDR).47 Jia et al reported a superplasmid (pSZS128-Hv-MDR) coharboring hypervirulence and MDR genes, and possessing complete conjugative regions.48 Previous studies36,49 have shown that some virulence plasmids, such as p15WZ-82 Vir and pK2606, have complete conjugative elements and may promote rapid dissemination of virulence-encoding elements among gram-negative bacterial pathogens. In this study, although the plasmid pWYKP586-1 remained relatively stable during serial passages in vitro, it split into a virulence plasmid (pWYKP589-1) and a resistance plasmid (pWYKP589-3) in the host. The resistance plasmid (pWYKP589-3) was a conjugative plasmid harboring complete conjugative elements, and the virulence plasmid (pWYKP589-1) was a mobilizable plasmid lacking complete conjugative elements. Perhaps the superplasmid pWYKP586-1 is relatively stable in vitro, but is not stable in the host, which is the fitness cost of acquiring resistance to antimicrobial agents.
Capsular polysaccharides are acidic lipopolysaccharides that coat the surface of Klebsiella pneumoniae to form a protective barrier. It has the ability of anti-neutrophils, macrophages, and dendritic cells, inhibiting early inflammatory responses and resisting bacitracin, so that KP can escape the killing of the host immune system.50 The capsular polysaccharide of the hvKP strain is positively regulated by the mucoid phenotype regulator gene, rmpA/rmpA2.51 Long et al introduced the blaNDM-5 plasmid into the hvKP strain using a transformation test.52 After introduction of the blaNDM-5 plasmid into hvKP, the expression levels of virulence genes related to capsular polysaccharide synthesis decreased, leading to a significantly reduced mucoid phenotype and capsular polysaccharide content. Some studies have reported that the acquisition of polymyxin resistance in hvKP is accompanied by fitness costs such as reduced capsular polysaccharide production and virulence.53,54 Fursova et al55 showed that under the selective pressure of 100 mg /l ampicillin or 10 mg /l ceftriaxone, the expression levels of drug resistance and virulence genes in the cKp, hvKp, and CR-hvKP strains were altered. These studies only studied the transcription and expression levels of virulence genes after the acquisition of drug resistance but did not study the change in virulence genes at the molecular level, leading to a reduction in expression levels. Pu et al showed that the rmpA genes on all 11 ST11-KL64 CRKP were interrupted by ISKpn26, resulting in a decreased hypermucoviscosity of the strain and a negative string test.56 In this study, we found for the first time that as aminoglycoside resistance emerged via acquisition of the rmtB gene, the hypermucoviscosity and virulence of the ST11-KL64 hv-CRKP strains decreased due to internal mutations in the rmpA and rmpA2 genes.
CRKP is mainly derived from nosocomial infection,57 while hvKP is mainly derived from community-acquired infection.58 When the patient was not admitted to the hospital, antibiotics were not administered, and the main threats to KP survival came from host immune serum and neutrophil killing. Capsular polysaccharides are coated on the surface of hvKP to form a protective barrier, which has the ability of anti-neutrophils, macrophages, and dendritic cells, inhibiting the early inflammatory response and resisting bacitracin, so that KP can escape the killing of the host immune system. After the patient was admitted to the hospital and treated with antibiotics, the main threat for KP was changed to antibiotics. Wyres et al showed that hvKP may be subject to some sort of constraint for horizontal antibiotic resistance gene transfer and shows more conserved pan-genomic diversity than MDR clones.59 In this study, we found that internal mutations in the mucoid phenotype regulatory genes rmpA and rmpA2 may reduced the surface capsular polysaccharide content of the strain, which was conducive to the acquisition of drug resistance genes through horizontal transmission to avoid being killed by antibiotics. Clinicians should not only pay attention to changes in bacterial resistance, but also to changes in bacterial virulence when treating patients with infection.
Conclusion
In summary, we report a case of the complex evolution of ST11-KL64 hv-CRKP in the host. It was found that under the pressure of antibiotics, the antimicrobial resistance of the strains increased, and the virulence decreased. Whole-genome sequencing analysis showed that the hv-CRKP plasmids underwent a large number of breaks and recombinations under antibiotic pressure. During this process, a new antibiotic resistance gene, rmtB, is acquired through the transfer of MGEs, and aminoglycoside resistance is obtained. Mutations in the mucoid phenotype regulator genes rmpA and rmpA2 led to a decrease in mucilage and virulence of the strain. Our research suggests that rapid changes in antimicrobial resistance and virulence of ST11 hv-CRKP under antibiotic pressure require wide attention.
Data Sharing Statement
The genome data of the four hv-CRKP strains were submitted to the National Center for Biotechnology Information (NCBI) under BioProject accession number PRJNA962850.
Ethics Approval and Informed Consent
This study was approved by the Medical Ethics Council of Shanghai Fifth People’s Hospital (Approval No. 11, 2022), and written informed consent has been provided by the patient’s son to have the case details published.
Acknowledgments
We thank the authority of NTUH-K2044 by Jin-Town Wang from National Taiwan University Hospital.
We extend our thanks to Personal Biotechnology Co., Ltd. Shanghai, China, for the support of whole-genome sequencing and the prediction of coding DNA sequences in the genome.
Funding
This study was supported by the Outstanding Young Medical Technical Talents and Pharmaceutical Talents Training Program supported by Health Commission of Minhang District, Shanghai (No. MWYJYX07), Natural Science Research Funds of Minhang District, Shanghai(No. 2023MHZ042), Medical Specialty Construction Project of Minhang District, Shanghai (No. 2020MWTZB04), Key Medical Specialty funded by Shanghai Fifth People’s Hospital, Fudan University (No. 2020WYZDZK08).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. Fems Microbiol Rev. 2017;41(3):252–275. doi:10.1093/femsre/fux013
2. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of st11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/S1473-3099(17)30489-9
3. Tian D, Wang M, Zhou Y, Hu D, Jiang X. Genetic diversity and evolution of the virulence plasmids encoding aerobactin and salmochelin in Klebsiella pneumoniae. Virulence. 2021;12(1):1323–1333. doi:10.1080/21505594.2021.1924019
4. Lan P, Jiang Y, Zhou J, Yu Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;25:26–34. doi:10.1016/j.jgar.2021.02.020
5. Yang X, Dong N, Zhang R, Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 2021;29(1):65–83. doi:10.1016/j.tim.2020.04.012
6. Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327–336. doi:10.1093/jac/dkz446
7. Xie Y, Tian L, Li G, et al. Emergence of the third-generation cephalosporin-resistant hypervirulent Klebsiella pneumoniae due to the acquisition of a self-transferable bladha-1-carrying plasmid by an st23 strain. Virulence. 2018;9(1):838–844.
8. Xie M, Yang X, Xu Q, et al. Clinical evolution of st11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun Biol. 2021;4(1):650. doi:10.1038/s42003-021-02148-4
9. Zhang R, Lin D, Gu D, Chen G, Chen S. Emergence of carbapenem-resistant serotype k1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2015;60(1):709–711. doi:10.1128/AAC.02173-15
10. Dong N, Lin D, Zhang R, Chan EW, Chen S. Carriage of blakpc-2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73(12):3317–3321. doi:10.1093/jac/dky358
11. Huang YH, Chou SH, Liang SW, et al. Emergence of an xdr and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018;73(8):2039–2046. doi:10.1093/jac/dky164
12. Lam MMC, Wyres KL, Wick RR, et al. Convergence of virulence and mdr in a single plasmid vector in mdr Klebsiella pneumoniae st15. J Antimicrob Chemother. 2019;74(5):1218–1222. doi:10.1093/jac/dkz028
13. Turton J, Davies F, Turton J, Perry C, Payne Z, Pike R. Hybrid resistance and virulence plasmids in “high-risk” clones of Klebsiella pneumoniae, including those carrying blandm-5. Microorganisms. 2019;7(9):1–11. doi:10.3390/microorganisms7090326
14. Zhao Y, Zhang X, Torres VVL, et al. An outbreak of carbapenem-resistant and hypervirulent Klebsiella pneumoniae in an intensive care unit of a major teaching hospital in Wenzhou, China. Front Public Health. 2019;7:229. doi:10.3389/fpubh.2019.00229
15. Serio AW, Keepers T, Andrews L, Krause KM. Aminoglycoside revival: review of a historically important class of antimicrobials undergoing rejuvenation. Ecosal Plus. 2018;8(1):1–20. doi:10.1128/ecosalplus.ESP-0002-2018
16. Raut A, Sharma D, Suvarna V. A status update on pharmaceutical analytical methods of aminoglycoside antibiotic: amikacin. Crit Rev Anal Chem. 2022;52(2):375–391. doi:10.1080/10408347.2020.1803042
17. Yang W, Hu F. Research updates of plasmid-mediated aminoglycoside resistance 16s rRNA methyltransferase. Antibiotics. 2022;11(7). doi:10.3390/antibiotics11070906
18. Wachino JI, Doi Y, Arakawa Y. Aminoglycoside resistance: updates with a focus on acquired 16s ribosomal RNA methyltransferases. Infect Dis Clin North Am. 2020;34(4):887–902. doi:10.1016/j.idc.2020.06.002
19. Kawai A, Suzuki M, Tsukamoto K, Minato Y, Doi Y. Functional and structural characterization of acquired 16s rRNA methyltransferase npmb1 conferring pan-aminoglycoside resistance. Antimicrob Agents Chemother. 2021;65(10):e100921. doi:10.1128/AAC.01009-21
20. Camelena F, Morel F, Merimeche M, et al. Genomic characterization of 16s rRNA methyltransferase-producing Escherichia coli isolates from the Parisian area, France. J Antimicrob Chemother. 2020;75(7):1726–1735. doi:10.1093/jac/dkaa105
21. Uchida H, Tada T, Tohya M, et al. Emergence in Japan of an isolate of Klebsiella pneumoniae co-harbouring bla(kpc-2) and rmtb. J Glob Antimicrob Resist. 2019;17:157–159. doi:10.1016/j.jgar.2018.11.026
22. Amin M, Mehdipour G, Navidifar T. High distribution of 16s rRNA methylase genes rmtb and arma among Enterobacter cloacae strains isolated from an ahvaz teaching hospital, Iran. Acta Microbiol Immunol Hung. 2019;66(3):337–348. doi:10.1556/030.66.2019.009
23. Nagasawa M, Kaku M, Kamachi K, et al. Loop-mediated isothermal amplification assay for 16s rRNA methylase genes in gram-negative bacteria. J Infect Chemother. 2014;20(10):635–638. doi:10.1016/j.jiac.2014.08.013
24. CLSI. Performance Standards for Antimicrobial Susceptibility Testing.
25. Hua Y, Wang J, Huang M, et al. Outer membrane vesicles-transmitted virulence genes mediate the emergence of new antimicrobial-resistant hypervirulent Klebsiella pneumoniae. Emerg Microbes Infect. 2022;11(1):1281–1292. doi:10.1080/22221751.2022.2065935
26. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi:10.1371/journal.pcbi.1005595
27. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi:10.1093/bioinformatics/btu153
28. Wang M, Goh YX, Tai C, Wang H, Deng Z, Ou HY. Vrprofile2: detection of antibiotic resistance-associated mobilome in bacterial pathogens. Nucleic Acids Res. 2022;50(W1):W768–773. doi:10.1093/nar/gkac321
29. Jin X, Chen Q, Shen F, et al. Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae st11 during treatment with tigecycline and polymyxin. Emerg Microbes Infect. 2021;10(1):1129–1136. doi:10.1080/22221751.2021.1937327
30. Alikhan NF, Petty NK, Ben ZN, Beatson SA. Blast ring image generator (brig): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi:10.1186/1471-2164-12-402
31. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. doi:10.1093/bioinformatics/btr039
32. Wang X, Chen G, Wu X, et al. Increased prevalence of carbapenem resistant Enterobacteriaceae in hospital setting due to cross-species transmission of the bla ndm-1 element and clonal spread of progenitor resistant strains. Front Microbiol. 2015;6:595. doi:10.3389/fmicb.2015.00595
33. Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi:10.1128/jcm.33.9.2233-2239.1995
34. Mukherjee S, Bhadury P, Mitra S, et al. Hypervirulent Klebsiella pneumoniae causing neonatal bloodstream infections: emergence of ndm-1-producing hypervirulent st11-k2 and st15-k54 strains possessing plvpk-associated markers. Microbiol Spectr. 2023;11(2):e412122. doi:10.1128/spectrum.04121-22
35. Semenec L, Cain AK, Dawson CJ, et al. Cross-protection and cross-feeding between Klebsiella pneumoniae and Acinetobacter baumannii promotes their co-existence. Nat Commun. 2023;14(1):702. doi:10.1038/s41467-023-36252-2
36. Tian D, Wang W, Li M, et al. Acquisition of the conjugative virulence plasmid from a cg23 hypervirulent Klebsiella pneumoniae strain enhances bacterial virulence. Front Cell Infect Microbiol. 2021;11:752011. doi:10.3389/fcimb.2021.752011
37. Wang L, Zhang Y, Liu Y, et al. Effects of chlorogenic acid on antimicrobial, antivirulence, and anti-quorum sensing of carbapenem-resistant Klebsiella pneumoniae. Front Microbiol. 2022;13:997310. doi:10.3389/fmicb.2022.997310
38. Loyola IH, Brito IL. Characterizing conjugative plasmids from an antibiotic-resistant dataset for use as broad-host delivery vectors. Front Microbiol. 2023;14:1199640. doi:10.3389/fmicb.2023.1199640
39. Lai YC, Peng HL, Chang HY. Rmpa2, an activator of capsule biosynthesis in Klebsiella pneumoniae cg43, regulates k2 cps gene expression at the transcriptional level. J Bacteriol. 2003;185(3):788–800. doi:10.1128/JB.185.3.788-800.2003
40. Zhou K, Xiao T, David S, et al. Novel subclone of carbapenem-resistant Klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis. 2020;26(2):289–297. doi:10.3201/eid2602.190594
41. Zhou C, Wu Q, He L, et al. Clinical and molecular characteristics of carbapenem-resistant hypervirulent Klebsiella pneumoniae isolates in a tertiary hospital in shanghai, China. Infect Drug Resist. 2021;14:2697–2706. doi:10.2147/IDR.S321704
42. Dong N, Yang X, Zhang R, Chan EW, Chen S. Tracking microevolution events among st11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg Microbes Infect. 2018;7(1):146. doi:10.1038/s41426-018-0146-6
43. Chen Y, Marimuthu K, Teo J, et al. Acquisition of plasmid with carbapenem-resistance gene blakpc2 in hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis. 2020;26(3):549–559. doi:10.3201/eid2603.191230
44. Yu FY, Yao D, Pan JY, et al. High prevalence of plasmid-mediated 16s rRNA methylase gene rmtb among Escherichia coli clinical isolates from a Chinese teaching hospital. BMC Infect Dis. 2010;10:184. doi:10.1186/1471-2334-10-184
45. Chen Y, Chang H, Lai Y, Pan C, Tsai S, Peng H. Sequencing and analysis of the large virulence plasmid plvpk of Klebsiella pneumoniae cg43. Gene. 2004;337:189–198. doi:10.1016/j.gene.2004.05.008
46. Zhou Y, Ai W, Guo Y, et al. Co-occurrence of rare arma-, rmtb-, and kpc-2-encoding multidrug-resistant plasmids and hypervirulence iuc operon in st11-kl47 Klebsiella pneumoniae. Microbiol Spectr. 2022;10(2):e237121. doi:10.1128/spectrum.02371-21
47. Xia P, Yi M, Yuan Y, et al. Coexistence of multidrug resistance and virulence in a single conjugative plasmid from a hypervirulent Klebsiella pneumoniae isolate of sequence type 25. Msphere. 2022;7(6):e47722. doi:10.1128/msphere.00477-22
48. Jia X, Zhu Y, Jia P, et al. Emergence of a superplasmid coharboring hypervirulence and multidrug resistance genes in Klebsiella pneumoniae poses new challenges to public health. Microbiol Spectr. 2022;10(6):e263422. doi:10.1128/spectrum.02634-22
49. Yang X, Wai-Chi CE, Zhang R, Chen S. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol. 2019;4(12):2039–2043. doi:10.1038/s41564-019-0566-7
50. Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi:10.4161/viru.22718
51. Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J Intern Med. 2020;287(3):283–300. doi:10.1111/joim.13007
52. Long D, Zhu L, Du F, et al. Phenotypical profile and global transcriptomic profile of hypervirulent Klebsiella pneumoniae due to carbapenemase-encoding plasmid acquisition. BMC Genomics. 2019;20(1):480. doi:10.1186/s12864-019-5705-2
53. Choi MJ, Ko KS. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother. 2015;59(11):6763–6773. doi:10.1128/AAC.00952-15
54. Zhang Y, Wang X, Wang S, et al. Emergence of colistin resistance in carbapenem-resistant hypervirulent Klebsiella pneumoniae under the pressure of tigecycline. Front Microbiol. 2021;12:756580. doi:10.3389/fmicb.2021.756580
55. Fursova AD, Fursov MV, Astashkin EI, et al. Early response of antimicrobial resistance and virulence genes expression in classical, hypervirulent, and hybrid hvkp-mdr Klebsiella pneumoniae on antimicrobial stress. Antibiotics. 2022;11(1):7. doi:10.3390/antibiotics11010007
56. Pu D, Zhao J, Lu B, et al. Within-host resistance evolution of a fatal st11 hypervirulent carbapenem-resistant Klebsiella pneumoniae. Int J Antimicrob Agents. 2023;61(4):106747. doi:10.1016/j.ijantimicag.2023.106747
57. Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (cre) strains in China. Ebiomedicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
58. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):e00001–19. doi:10.1128/CMR.00001-19
59. Wyres KL, Wick RR, Judd LM, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15(4):e1008114. doi:10.1371/journal.pgen.1008114
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




