Back to Journals » Infection and Drug Resistance » Volume 13
Whole Genome Sequencing of Ceftolozane-Tazobactam and Ceftazidime-Avibactam Resistant Pseudomonas aeruginosa Isolated from a Blood Stream Infection Reveals VEB and Chromosomal Metallo-Beta Lactamases as Genetic Determinants: A Case Report
Authors Alamri AM , Alfifi S, Aljehani Y, Alnimr A
Received 6 October 2020
Accepted for publication 4 November 2020
Published 23 November 2020 Volume 2020:13 Pages 4215—4222
DOI https://doi.org/10.2147/IDR.S285293
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Aisha M Alamri,1 Somayah Alfifi,2 Yasser Aljehani,3 Amani Alnimr4
1Department of Clinical Laboratory Sciences, College of Applied Medical Sciences Imam Abdulrahman Bin Faisal University, Dammam, Kingdom of Saudi Arabia; 2Department of Medical Laboratory Science, College of Applied Medical Sciences, Tabuk University, Tabuk, Kingdom of Saudi Arabia; 3Division of Thoracic Surgery, Department of Surgery. King Fahad Hospital of the University, College of Medicine, Imam Abdulrahman Bin Faisal University, Dammam, Kingdom of Saudi Arabia; 4Department of Microbiology, College of Medicine, Imam Abdulrahman Bin Faisal University, Dammam, Kingdom of Saudi Arabia
Correspondence: Aisha M Alamri; Amani Alnimr Tel +966538187263
; +966563181019
Email [email protected]; [email protected]
Abstract: Pseudomonas aeruginosa is a common gram-negative bacillus in nosocomial settings. Consideration of this organism is important due to its potential to acquire multi-drug resistance through various mechanisms causing severe infections, particularly in immunocompromised hosts. Here, we present a challenging case of a blood stream infection caused by a drug-resistant strain of P. aeruginosa in a debilitated young patient. A 31-year-old male patient with a complex history of multiple trauma following a vehicle accident that required several surgical interventions, is plagued by persistent bacteremia. An extensively drug-resistant strain of P. aeruginosa was repeatedly isolated that continued to grow in the patient’s blood cultures despite treatment with meropenem and colistin for an extended period. In addition to phenotypic characterization, the complete genome of the strain was sequenced and a genomic view was provided regarding its antimicrobial resistance (AMR) patterns, efflux pump genes, virulence determinants, phageomic signals, and genomic islands. The strain belongs to sequence type ST357 with dominant Class A (VEB), Class B, Class C (PDC-11) and D (OXA-10, OXA-50) β-lactamases, and injectosomes (type III secretion system) known to mediate high virulence. The pool of extended spectrum β-lactamases genes and the upregulated chromosomal efflux system are likely to account for the extended resistance pattern in this strain. In light of the global spread of ST357 isolates, it is essential to continue monitoring their resistance patterns and evaluate effective epidemiological tools to define the genetic determinants of emerging resistance. Intensified infection control measures are continuously required to stop dissemination of such strains in an institution where susceptible hosts are at risk of acquiring them.
Keywords: whole genome sequencing, Pseudomonas aeruginosa, bacteremia, gram-negative bacilli, multidrug resistance, ceftolozane-tazobactam, ceftazidime-avibactam
Introduction
Pseudomonas aeruginosa is a common opportunistic pathogen that presents a therapeutic and infection control challenge in hospital settings due to its frequent resistance profile.1 Isolation of multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains of P. aeruginosa is increasing worldwide, with a subset of isolates exhibiting pan-resistance phenotypes.2,3 Complicated infections due to drug-resistant P. aeruginosa are often difficult to treat and associated with poor clinical outcomes.4 The key mechanisms involved in this organism’s resistance to carbapenems and various antimicrobial classes include the cumulative effects of AmpC β-lactamase expression, efficient efflux pump systems, and downregulation of the outer membrane proteins, of which OprD is well characterized.5 Other significant resistance determinants are the acquisition of resistance genes carried on mobile genetic elements such as plasmids and conjugative transposons.5
Ceftolozane–tazobactam is a novel intravenous combination of a third-generation cephalosporin with a β-lactamase inhibitor, tazobactam that was approved by the Food and Drug Administration (FDA) in 2014. In addition to being a poor substrate for AmpC hydrolysis, ceftolozane-tazobactam has demonstrated high in vitro activity against a range of MDR Gram negative bacteria including multidrug resistant P. aeruginosa (MDR-PA) mediated by mutations in the outer membrane proteins (OprD) and the efflux systems (MexY and MexAB).6 Similarly, ceftazidime-avibactam was FDA approved in 2015 and exhibited high potency against P. aeruginosa species, with high minimum inhibitory concentrations (MIC) to ceftazidime alone.7 Despite the growing evidence of successful treatment outcomes of MDR-PA using these two agents, we have previously shown that resistance to advanced cephalosporin-β-lactamase combinations was encountered among more than 65% of a MDR-PA cohort (n = 67) not previously exposed to the drug, of which >60% were found to have MIC > 256 µg/mL.8
Host factors that are linked to an increased risk of MDR-PA bacteremia include severe underlying primary illness, immunosuppression, bedridden status, presence of invasive devices, nosocomial origin, prior surgery, intensive care admission, previous use of broad-spectrum antimicrobials, and carriage of the organism as a colonizer at various sites.9–11 Herein, we describe a clinical case of sepsis caused by an XDR-PA strain (designated as PA179) that demonstrated an elevated resistance to a wide range of antimicrobial agents including ceftolozane-tazobactam and ceftazidime-avibactam. The phenotypic characteristics of the strain along with the key features of its complete genome sequence, such as resistance determinants, virulence genes, and phageomics, are presented.
Case Presentation
This is a case of a 31-year-old male who was a victim of motor vehicle accident in 2007. He sustained a severe head injury and cervical spine fracture, for which his spine was fixed with a plate and screw, and was intubated for several weeks. His recovery was slow overall, with gradual, mild improvement. Eventually, he became fully conscious and was discharged from hospitalization, although as a paraplegic. The patient sought medical advice over the years for recurrent chest infections, for which he was treated conservatively with antibiotics. Around mid-2016, he was referred to the cardiothoracic unit of our institution as his chest x-ray (CXR) showed a widened mediastinum (Figure 1). The initial laboratory investigations were normal apart from the following parameters: Hemoglobin 11.3 g/dL, hematocrit 35%, Mean corpuscular volume 67.7 fL, mean corpuscular hemoglobin 21.8 pg, albumin 2.7 g/L, creatinine 0.5 μmol/L, Na 135 mmol/L, and erythrocyte sedimentation rate 93 mm/h. Further radiological assessment, computed tomography (CT) of the neck, cervical spine, and chest revealed a superior mediastinal collection with signs of chronic esophageal perforation. The patient underwent a swallowing test and an upper gastroesophageal endoscopy that confirmed the presence of upper esophageal perforation due to erosion of the cervical spine plate and screw into the esophagus. Following a multi-disciplinary discussion represented by neurosurgeons, an otolaryngologist, a thoracic, and a general surgeon, the surgical approach was designed for this case. The patient underwent neck exploration through a cervical incision, the plate was removed, and the perforation was identified. The perforation was repaired using an interposition muscle flap from the strap muscle. A limited thoracotomy revealed extensive pleural adhesions, although no collection was identified. A feeding jejunostomy tube was created through mini-laparotomy, which the patient tolerated well. One week post-operatively, he developed a surgical site infection at the cervical wound. Exploration and drainage were performed along with antimicrobial therapy. This patient had a lengthy post-operative course with several admissions to the intensive care unit (ICU) due to repeated episodes of sepsis. Moreover, he required bronchoscopies and exchanging the tracheostomy. After a few weeks, he developed an acute surgical abdomen for which he underwent exploration and drainage of multiple intra-abdominal abscess collections, from which he recovered. However, he developed persistent status epilepticus over a few months that was difficult to manage. The patient eventually experienced cardiac arrest that could not be revived. The timeline of infections encountered in the patient is illustrated in Figure 2.
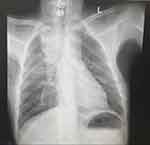 |
Figure 1 Standard PA chest x-ray demonstrating widened superior mediastinum with homogenous opacity seen in the right upper lung zone. |
 |
Figure 2 Microbiological summary timeline. |
An XDR-P. aeruginosa strain (PA179) was repeatedly isolated from the urinary tract of the patient 16 months after his admission, followed by an episode of sepsis during which the same strain was grown from multiple blood culture samples. The identification of suspected P. aeruginosa colonies was confirmed using VITEK MS (BioMérieux, Craponne, France) based on matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) technology with automated broth microdilution (BMD) susceptibility testing using VITEK 2 Software Version 7.01 and AST Card-GN99 (BioMérieux, Craponne, France). Additionally, E tests (AB Biodisk, Solna, Sweden) were used to measure the MIC of two carbapenems, imipenem and meropenem, two β-lactam/β-lactamase inhibitor combinations, ceftazidime-avibactam and ceftolozane-tazobactam, and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) 2017 guidelines for all antimicrobials tested. The susceptibility pattern of the blood isolate revealed an extensive drug resistance pattern, as shown in Table 1.
 |
Table 1 Phenotypic Antimicrobial Susceptibility Pattern of the Strain PA179 |
Whole Genome Sequencing (WGS) and Bioinformatics Tools and Servers
A range of bioinformatics online tools were used to analyze the sequenced genome including: Soap denovo: de novo draft assembly for the human-sized genomes: (http://soap.genomics.org.cn/soapdenovo.html), Reference-Assisted Genome Ordering Utility (Ragout): (https://github.com/fenderglass/Ragout), Multi-locus sequence typing (MLST): in silico multilocus sequence typing website (http://cge.cbs.dtu.dk/services/), Prokaryotic Genome Annotation Pipeline annotation system (PGAAP): (https://github.com/ncbi/pgap), Rapid Annotation using Subsystem Technology (RAST): (https://rast.nmpdr.org/), Virulence factor database (VFDB) and Virulence factor analyser (VFanalyser): (http://www.mgc.ac.cn/VFs/), PHAge Search Tool (PHAST): (http://phast.wishartlab.com/) and clustered regularly interspaced short palindromic repeats finder (CRISPR): (https://crispr.i2bc.paris-saclay.fr/Server/).
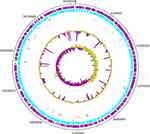
The isolate was subjected to whole genome sequencing as follows: total genomic DNA was prepared using DNeasy blood and tissue kits (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations, followed by sequencing using an Illumina HiSeq 4000 system (Illumina, San Diego, CA, USA) at the Beijing Genomics Institute (BGI, Shenzhen, China). Genomic DNA was sheared randomly to construct three read libraries with lengths of 6580692 bp using a Bioruptor ultrasonicator (Diagenode, Denville, NJ, USA) and physicochemical methods. The paired-end fragment libraries were sequenced according to the Illumina HiSeq 4000 system protocol. Low-quality raw reads from paired-end sequencing; those with consecutive bases covered by fewer than five reads were discarded. The sequenced reads were assembled using SOAPdenovo v1.05 software. The complete annotated genome sequences of strain PA179 were deposited in the National Centre for Biotechnology Information (NCBI) and published under the accession number CP058257 (Table 2/Supplementary Table S1). The general genomic features of strain PA179 are summarized in Table 2. Initial assembly using Soap denovo produced 171 scaffolds with 1,88,134 a N50 value and a total sequence length of 69,09,802 bp and coverage of 99.97 x (Supplementary Table S1). Thereafter, it was rearranged to form the complete genome using Ragout (Reference-Assisted Genome Ordering Utility) tool, which resulted in a single contig of size 6580692 bp (Figure 3). In silico analysis of Multi-locus sequence typing (MLST) classified PA179 under the sequence type ST357 (Supplementary Table S1). PGAAP annotation deciphering the PA179 strain revealed that the chromosome comprised 6083 genes, 6023 coding sequences (CDS), 56 tRNA genes, 20 rRNA genes, and 4 non-coding RNAs (Supplementary Table S1).
 |
Table 2 Summary of Genome Features of the Strain PA179 |
With regard to the genomic view of antimicrobial effects, resistance to β-lactam agents was observed by the presence of blaOXA-10 (J03427), blaOXA-50 (AY306130), blaVEB-9, blaPAO, and the class C beta-lactamase PDC-11. Notably, four Metallo-β-lactamases (MBL) fold metallo-hydrolase (belonging to Metallo-beta-lactamase super family proteins) were found among the chromosomal gene cluster (WP_003091024.1). Aminoglycoside resistant determinants, namely, aph(3’)-IIb, aac(6’)-Il (aac(6’)-Il_U13880), ant(2’ ‘‘)-Ia (ant(2’’)-Ia_X04555), aac(6’)-Il (aac(6’)-Il_U13880), ant(2’’)-Ia (ant(2’’)-Ia_X04555), aac(6’)-Il (aac(6’)-Il_U13880), ant(2’’)-Ia (ant(2’’‘)-Ia_X04555), and aph(3’)-IIb_CP006832. The fluoroquinolone-related resistance genes crpP (crpP_HM560971) also existed. Phenicol resistance gene catB7 (AF036933), and tetracycline resistance genes tet(A) (tet(A) (AY196695)) were present (Supplementary Table S2). Additionally, manual examination of RAST output predicted that P. aeruginosa PA179 may contain major efflux pump proteins comprising the RND family of MexCD-OprJ, cobalt efflux protein CorC, multidrug efflux proteins from the MATE family, macrolide-specific efflux protein MacA, and potassium efflux system KefA. Multi-drug resistant efflux pumps of MexX, MexA, and MexE were also detected. (Supplementary Table S1 and S2).
The virulence factor identification VFDB-organized VFanalyser was used to predict the presence of over 222 different virulence factors, including bacteriocins and invasions (Supplementary Table S3). The most remarkable feature of the PA179 genome is the presence of multiple type secretion system proteins. Type 1 (six genes), Type 2 (15 genes), Type 3 (36 genes), Type 4 (20 genes), Type 5 (two genes), and Type 7 (five genes) were identified using both RAST and prokaryotic genome annotation pipeline (PGAAP) annotation system. (Supplementary Table S3). A phage genome with a cluster of genetic rearrangements was observed using PHAST. A total of three phages in the PA197 genome was predicted, from one intact phage, one consisted of an 18.4 kb Pseudomonas phage, NC_030929, with a 62.25% GC content, and 25 CDS were detected. The second phage consisted of a 6.6 kb Pseudomonas phage Pf1 (NC_001331), 62.47% GC content, and eight protein-coding sequences. Furthermore, a partial genome of Pseudomonas phage B3 NC_006548 (19.4 kb region length) was also observed in PA179 with 22 CDS. Prophage visualization and details can be seen in Supplementary Figures 1–4. A total of three CRISPR genes, clustered regularly interspaced short palindromic repeats, were predicted using the CRISPR finder online tool and consisted of sequences ranging from 447 bp to 1047 bp CRISPR size with a 28 bp direct repeat (DR) length. These CRISPR-Cas systems confer flexible, sequence-specific immunity against bacteriophages and mobile genetic elements. (Supplementary Table S4, Supplementary Figures 1–4).
The genomic islands representing mobile elements in the PA179 genome were predicted using the online server “Island Viewer”. This was identified in 47 islands in the genome, with several embedded mobile genetic elements. The islands were visualized using Island Viewer software (Figure 4 and Supplementary data Table S5).
 |
Figure 4 Genomic Islands in P. aeruginosa PA179. |
Discussion
Complicated infections caused by MDR and XDR P. aeruginosa represent a significant clinical threat with limited therapeutic options, particularly in cases of bacteremia that carry a poor prognosis and high mortality.12,13 In this study, we present a case of refractory sepsis in a bedridden patient with complex underlying morbidities causing persistent bacteremia by a virulent XDR-PA strain, which exhibited high-level resistance profiles to antimicrobials including cephalosporin-β-lactamase inhibitors, ceftolozane-tazobactam, and ceftazidime-avibactam. Despite prolonged combination antimicrobial therapy using colistin and meropenem, eradication of the infection was not achieved. Such difficult to treat pathogens warrant further investigation at the molecular level to elucidate the underlying determinants of resistance. In institutions, these cases have clinical impacts such as selection of empirical antimicrobial regimens in high-risk groups, and intensifying infection control measures in the affected units.
In the increasingly high-throughput genome sequencing era, a large collection of sequence data has become available in multiple databases. We utilized a range of software and online servers to analyze the general genome annotations, virulence factors, and resistant determinants of PA179 relative to the well-characterized, reference strain PAO1. Sequence analysis of strain PA179 revealed its belonging to the international, high-risk XDR clone ST357, outbreaks of which has been reported in many countries, including Central and Western Europe, Korea, Lebanon, and Gulf Co-operation Council states (GCC).14–17 A pool of β-lactamase genes, such as Class A (VEB), C (PDC-11), D (OXA-10, OXA-50), and resistance genes targeting the other antimicrobial classes of tetracycline, aminoglycosides, and chloramphenicol were identified. The majority of these genes were found to be carried on mobile genetic elements such as integrons and conjugative transposons, which aid their dissemination to other susceptible strains in clinical and community settings, leading to serious threats. Similarly, the acquisition of such resistant genes was evident in many P. aeruginosa strains reported globally.18,19
Chromosomal, non-mobile MBL are usually linked to other genera conferring intrinsic resistance, eg, some Bacillus spp., Stenotrophomonas maltophilia, Chryseobacterium indologenes and non-clinical environmental genera like Aeromonas.20 The contribution of this genetic element to the phenotypic expression of high level, broad-spectrum β-lactam resistance is obscure and requires further investigation.
Despite the wide dissemination of mobile MBL genes in P. aeruginosa, there have been reports of cohorts of carbapenem-resistant strains that did not harbor any of the known acquired MBL genes such as IMP, VIM, and NDM.16,21 Other uncommon genes linked to resistance to ceftolozane-tazobactam and ceftazidime-avibactam are GES and VEB; the latter was detected in the current strain PA179, which can explain the reduced susceptibility to the two agents.22 VEB-9 is a rare integron-bourne broad-spectrum β-lactamase encoding gene that was reported earlier in the GCC.23,24 The VEB-like family has also been reported in Europe, Thailand, Iran, China, and the United States.25 The high transmissibility of these mobile genetic elements encoding broad-spectrum resistance mechanisms reduces the efficacy of newly developed therapeutic agents.
P. aeruginosa is known to retain an array of virulence factors implicated in invasive infections. The PA179 strain was found to possess a range of secretion systems, T1SS–T7SS. Studies have shown that type protein secretion system mutants have contributed to lower survival rates in a Caenorhabditis elegans laboratory model.26 This was also shown in a retrospective analysis of 85 cases of P. aeruginosa bacteremia, where mortality was six-fold greater than infection with a strain that did not express type III secreted toxins, although host factors largely contributed to the prognosis.27 Furthermore, these secretion systems appear to play a major role in biofilm-specific antibiotic resistance, as evidenced by diminished biofilm activity among T6SS-deficient P. aeruginosa mutants.28 Such systems represent a major virulence determinant that has been reported in a number of non-related bacterial species, including Legionella pneumophila and Helicobacter pylori, highlighting the ability to spread easily.29 WGS studies have also described similar protein secretion systems in the P. aeruginosa VRFPA04 genome and the global clone ST235, which has been linked to hospital outbreaks and poor clinical outcomes.30 These secretion systems are currently under investigation as potential vaccine targets for immunotherapy.31
Alternatively, bacteriophages exist as components in many bacterial genomes, including P. aeruginosa, where certain prophages are essential to the biofilm life cycle, contributing to bacterial persistence on surfaces and medical devices.32 Strain PA179 was found to contain three phages of different sizes, a common feature described in many genome sequencing studies of the high-risk P. aeruginosa clones, ST235 and ST111, reported worldwide.18,30,33 Currently, phage therapy remains under investigation for P. aeruginosa blood steam infections.34,35
Overall, the current report reflects the importance of whole genome sequencing as a powerful, lab-based tool that can support AMR surveillance systems by pinpointing resistant foci and linking phenotypic data to the genomic basis of resistance. Moreover, virulence and potential clone dissemination can be predicted by the genomic power of analyzing the mobile genetic elements that serve as AMR vehicles. To the best of our knowledge, this is the first WGS study performed on an invasive P. aeruginosa strain in Saudi Arabia. A limitation of this study is the inability to define the strain as pandrug-resistant since colistin BMD, the method endorsed by the CLSI, was not performed despite the preliminary resistance suggested by the automated system (VITEK 2). The lack of availability of BMD assays in many diagnostic laboratories further complicates the challenges in treating such XDR pathogens, and necessitates the development of user-friendly colistin susceptibility testing tools with clinically impactful, rapid turn-around times. Future potential treatment agents of PDR-PA include novel cephalosporins such as a combination of cefiderocol and carbapenem-beta-lactamase, eg imipenem-cilastatin-relebactam. However, further microbiological studies are required to infer the mechanism of potential resistance to these agents and assess their cross-resistance with existing drugs in clinical use.36 This is based on the case in which the strain isolated in 2017 was highly resistant to two novel agents prior to introducing them to our hospital. Thus, intense preventive measures are consistently required in healthcare facilities with debilitated patients who are at an increased risk of infections by virulent, drug-resistant strains.
Abbreviations
MDR, multi-drug resistant; XDR, extensively-drug resistant; MBL, metallo-β-lactamases; OXA, oxacillinases; PA, Pseudomonas aeruginosa; OprD, outer membrane proteins; FDA, Food Drug Agency; MIC, minimum inhibitory concentration; MLST, multi-locus sequence typing; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; BMD, broth microdilution.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author (Aisha Alamri) upon reasonable request.
Ethics Approval and Informed Consent
The material presented is original, unpublished, and has not been submitted elsewhere. This study was a part of a project approved by the ethical committee of the Institutional Review Board at Imam Abdulrahman Bin Faisal University (IRB 2020-03-163). Written informed consent was obtained from the patient for publication of this case report and any accompanying images.
Acknowledgment
The authors thank the Deanship of Scientific Research (DSR) at Imam Abdulrahman Bin Faisal University (IAU) for funding this work (2019-016-CAMS-NF).
Author Contributions
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content, agreed to submit to the current journal, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kanj SS, Kanafani ZA. Current concepts in antimicrobial therapy against resistant gram-negative organisms: extended-spectrum beta-lactamase-producing Enterobacteriaceae, carbapenem-resistant Enterobacteriaceae, and multidrug-resistant Pseudomonas aeruginosa. Mayo Clin Proc. 2011;86(3):250–259. doi:10.4065/mcp.2010.0674
2. Tuon FF, Gortz LW, Rocha JL. Risk factors for pan-resistant Pseudomonas aeruginosa bacteremia and the adequacy of antibiotic therapy. Braz J Infect Dis. 2012;16(4):351–356. doi:10.1016/j.bjid.2012.06.009
3. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
4. Thaden JT, Park LP, Maskarinec SA, Ruffin F, Fowler VG, van Duin D. Results from a 13-year prospective cohort study show increased mortality associated with bloodstream infections caused by pseudomonas aeruginosa compared to other bacteria. Antimicrob Agents Chemother. 2017;61(6):e02671–16. doi:10.1128/AAC.02671-16
5. Botelho J, Grosso F, Peixe L. Antibiotic resistance in Pseudomonas aeruginosa - mechanisms, epidemiology and evolution. Drug Resist Updat. 2019;44:100640. doi:10.1016/j.drup.2019.07.002
6. Rodríguez-Martínez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(11):4783–4788. doi:10.1128/AAC.00574-09
7. Chalhoub H, Tunney M, Elborn JS, et al. Avibactam confers susceptibility to a large proportion of ceftazidime-resistant Pseudomonas aeruginosa isolates recovered from cystic fibrosis patients. J Antimicrob Chemother. 2015;70(5):1596–1598. doi:10.1093/jac/dku551
8. Alnimr AM, Alamri AM. Antimicrobial activity of cephalosporin-beta-lactamase inhibitor combinations against drug-susceptible and drug-resistant. J Taibah Univ Med Sci. 2020;15(3):203–210. doi:10.1016/j.jtumed.2020.04.004
9. Mascitti H, Duran C, Nemo EM, et al. Factors associated with bacteremia due to multidrug-resistant organisms among bacteraemic patients with multidrug-resistant organism carriage: a case control study. Antimicrob Resist Infect Control. 2018;7:116. doi:10.1186/s13756-018-0412-3
10. Tartof SY, Kuntz JL, Chen LH, et al. Development and assessment of risk scores for carbapenem and extensive β-lactam resistance among adult hospitalized patients with pseudomonas aeruginosa infection. JAMA Netw Open. 2018;1(6):e183927. doi:10.1001/jamanetworkopen.2018.3927
11. Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother. 2006;50(1):43–48. doi:10.1128/AAC.50.1.43-48.2006
12. Bisbe J, Gatell JM, Puig J, et al. Pseudomonas aeruginosa bacteremia: univariate and multivariate analyses of factors influencing the prognosis in 133 episodes. Rev Infect Dis. 1988;10(3):629–635. doi:10.1093/clinids/10.3.629
13. Zhang Y, Chen XL, Huang AW, et al. Mortality attributable to carbapenem-resistant Pseudomonas aeruginosa bacteremia: a meta-analysis of cohort studies. Emerg Microbes Infect. 2016;5:e27. doi:10.1038/emi.2016.22
14. Sung JY, Koo SH, Cho HH, Kwon KC. Nosocomial infection by sequence type 357 multidrug-resistant Acinetobacter baumannii isolates in a neonatal intensive care unit in Daejeon, Korea. Ann Lab Med. 2013;33(4):279–282. doi:10.3343/alm.2013.33.4.279
15. Zowawi HM, Syrmis MW, Kidd TJ, et al. Identification of carbapenem-resistant Pseudomonas aeruginosa in selected hospitals of the Gulf Cooperation Council States: dominance of high-risk clones in the region. J Med Microbiol. 2018;67(6):846–853. doi:10.1099/jmm.0.000730
16. Mihara T, Kimura T, Momiyama K, et al. Secondary in-hospital epidemiological investigation after an outbreak of Pseudomonas aeruginosa ST357. J Infect Chemother. 2020;26(3):257–265. doi:10.1016/j.jiac.2019.09.014
17. Pelegrin AC, Saharman YR, Griffon A, et al. High-risk international clones of carbapenem-nonsusceptible pseudomonas aeruginosa endemic to indonesian intensive care units: impact of a multifaceted infection control intervention analyzed at the genomic level. mBio. 2019;10(6). doi:10.1128/mBio.02384-19
18. Taiaroa G, Samuelsen Ø, Kristensen T, Økstad OAL, Heikal A. Complete genome sequence of pseudomonas aeruginosa K34-7, a carbapenem-resistant isolate of the high-risk sequence type 233. Microbiol Resour Announc. 2018;7(4). doi:10.1128/MRA.00886-18
19. Witney AA, Gould KA, Pope CF, et al. Genome sequencing and characterization of an extensively drug-resistant sequence type 111 serotype O12 hospital outbreak strain of Pseudomonas aeruginosa. Clin Microbiol Infect. 2014;20(10):O609–18. doi:10.1111/1469-0691.12528
20. Palzkill T. Metallo-β-lactamase structure and function. Ann N Y Acad Sci. 2013;1277:91–104. doi:10.1111/j.1749-6632.2012.06796.x
21. Turton JF, Wright L, Underwood A, et al. High-resolution analysis by whole-genome sequencing of an international lineage (Sequence Type 111) of pseudomonas aeruginosa associated with metallo-carbapenemases in the United Kingdom. J Clin Microbiol. 2015;53(8):2622–2631. doi:10.1128/JCM.00505-15
22. Hawkey PM, Warren RE, Livermore DM, et al. Treatment of infections caused by multidrug-resistant Gram-negative bacteria: report of the British Society for Antimicrobial Chemotherapy/Healthcare Infection Society/British Infection Association Joint Working Party. J Antimicrob Chemother. 2018;73(suppl_3):iii2–iii78. doi:10.1093/jac/dky027
23. Poirel L, Mugnier PD, Toleman MA, et al. ISCR2, another vehicle for bla(VEB) gene acquisition. Antimicrob Agents Chemother. 2009;53(11):4940–4943. doi:10.1128/AAC.00414-09
24. Poirel L, Rotimi VO, Mokaddas EM, Karim A, Nordmann P. VEB-1-like extended-spectrum beta-lactamases in Pseudomonas aeruginosa, Kuwait. Emerg Infect Dis. 2001;7(3):468–470. doi:10.3201/eid0703.010322
25. Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45(6):568–585. doi:10.1016/j.ijantimicag.2015.03.001
26. Sana TG, Hachani A, Bucior I, et al. The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem. 2012;287(32):27095–27105. doi:10.1074/jbc.M112.376368
27. El-Solh AA, Hattemer A, Hauser AR, Alhajhusain A, Vora H. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit Care Med. 2012;40(4):1157–1163. doi:10.1097/CCM.0b013e3182377906
28. Zhang L, Hinz AJ, Nadeau JP, Mah TF. Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance. J Bacteriol. 2011;193(19):5510–5513. doi:10.1128/JB.00268-11
29. Nagai H, Kubori T. Type IVB secretion systems of legionella and other gram-negative bacteria. Front Microbiol. 2011;2:136. doi:10.3389/fmicb.2011.00136
30. Brüggemann H, Migliorini LB, Sales RO, et al. Comparative genomics of nonoutbreak pseudomonas aeruginosa strains underlines genome plasticity and geographic relatedness of the global clone ST235. Genome Biol Evol. 2018;10(7):1852–1857. doi:10.1093/gbe/evy139
31. Frank DW, Vallis A, Wiener-Kronish JP, et al. Generation and characterization of a protective monoclonal antibody to Pseudomonas aeruginosa PcrV. J Infect Dis. 2002;186(1):64–73. doi:10.1086/341069
32. Rice SA, Tan CH, Mikkelsen PJ, et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009;3(3):271–282. doi:10.1038/ismej.2008.109
33. Guzvinec M, Izdebski R, Butic I, et al. Sequence types 235, 111, and 132 predominate among multidrug-resistant pseudomonas aeruginosa clinical isolates in Croatia. Antimicrob Agents Chemother. 2014;58(10):6277–6283. doi:10.1128/AAC.03116-14
34. Oechslin F, Piccardi P, Mancini S, et al. Synergistic interaction between phage therapy and antibiotics clears pseudomonas aeruginosa infection in endocarditis and reduces virulence. J Infect Dis. 2017;215(5):703–712. doi:10.1093/infdis/jiw632
35. Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health. 2018;2018(1):60–66. doi:10.1093/emph/eoy005
36. Portsmouth S, van Veenhuyzen D, Echols R, et al. Cefiderocol versus imipenem-cilastatin for the treatment of complicated urinary tract infections caused by Gram-negative uropathogens: a Phase 2, randomised, double-blind, non-inferiority trial. Lancet Infect Dis. 2018;18(12):1319–1328. doi:10.1016/S1473-3099(18)30554-1
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.