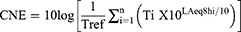Back to Journals » Journal of Multidisciplinary Healthcare » Volume 17
Variations in the Cadherin 23 Gene Associated With Noise-Induced Hearing Loss
Authors Jiao J, Yu S, Gu G, Chen G, Zhang H, Zheng Y
Received 5 December 2023
Accepted for publication 11 March 2024
Published 6 April 2024 Volume 2024:17 Pages 1473—1482
DOI https://doi.org/10.2147/JMDH.S453417
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Jie Jiao,1 Shanfa Yu,2 Guizhen Gu,1 Guoshun Chen,3 Huanling Zhang,3 Yuxin Zheng4
1The Third People’s Hospital of Henan Province (Henan Hospital for Occupational Diseases), Zhengzhou, Henan, People’s Republic of China; 2Henan Medical College, Zhengzhou, Henan, People’s Republic of China; 3Wugang Institute for Occupational Health, Wugang, Henan, People’s Republic of China; 4School of Public Health, Qingdao University, Qingdao, Shandong, People’s Republic of China
Correspondence: Shanfa Yu, Henan Medical College, No. 100, Science Avenue, Zhengzhou, Henan, 451191, People’s Republic of China, Tel\Fax +86-0371-6257-6891, Email [email protected]
Background: The relationship between CDH23 gene variants and NIHL is unclear. This study investigates the association between cadherin 23 (CDH23) gene variants and noise-induced hearing loss (NIHL).
Methods: This is a case-control study. Workers who were exposed to noise from a steel factory in North China were recruited and divided into two groups: the case group (both ears’ high-frequency threshold average [BHFTA] ≥ 40dB) and the control group (BHFTA ≤ 25 dB). This study used the generalised multifactor dimensionality reduction method to analyse the association among 18 single-nucleotide polymorphisms (SNPs) in CDH23 and NIHL. Logistic regression was performed to investigate the main effects of SNPs and the interactions between cumulative noise exposure (CNE) and SNPs. Furthermore, CNE was adjusted for age, gender, smoking, drinking, physical exercise and hypertension.
Results: This study recruited 1,117 participants. The results showed that for rs11592462, participants who carried the GG genotype showed an association with NIHL greater than that of those who carried the CC genotype. Accordingly, genetic variation in the CDH23 gene could play an essential role in determining individual susceptibility to NIHL.
Conclusion: Genetic variations in the CDH23 gene may play an important role in determining individual susceptibility to NIHL. These results provide new insight into the pathogenesis and early prevention of NIHL.
Keywords: cadherin 23 gene variants, noise-induced hearing loss risk, Chinese population, single-nucleotide polymorphisms, cumulative noise exposure
Introduction
Noise-induced hearing loss (NIHL) is sensory deafness caused by the long-term exposure of the auditory system to a noisy environment.1 Furthermore, NIHL is a complex disease resulting from environmental and intrinsic factors. Studies on environmental factors include noise, organic solvents, heat, vibrations, smoking and drinking.1–3 However, when exposed to similar levels of noise and environmental factors, the morbidity of NIHL varies widely; some workers develop NIHL, and others do not.4 Genetic variations could explain some of the inconsistent results.
Cadherin 23 (CDH23) is located on chromosome 10 and is a component of the stereocilia tip links, which are thought to gate the mechanotransduction channel in hair cells.5 Moreover, CDH23, also known as otocadherin, is part of the cadherin superfamily of calcium-dependent cell-surface adhesion proteins and plays a crucial role in the lateral and stereocilia tip-links of the inner ear’s sensory hair cells, which control the hearing process.6 Tip-links are extracellular filaments that are the ‘gate cables’ for opening mechanotransduction channels, which transduce mechanical forces arising from sound waves and head movement, allowing hearing and balance to be maintained.5 In animals, CDH23 is the first and only gene linked with a predisposition to NIHL in waltzer mice (the CDH23v mutation).7 Previous studies revealed that genetic variations in CDH23 are a key determinant of age-related hearing loss and early-onset progressive hearing loss. Mice carrying this allele (CDH23v, CDH23 [753A] and CDH23c.753A/G) are more susceptible noise damage.8–10 Mutations in CDH23 in mice cause stereocilia disorganisation, leading to deafness and vestibular disorders. In humans, mutations in CDH23 lead to both non-syndromic and syndromic hearing loss.9–11 Evidence from epidemiological studies suggests an inconsistent association between genetic variations and NIHL.4 One study reported that the associations between 63 polymorphisms in CDH23 and NIHL were negative in the Polish and Swedish populations.11 Other studies12,13 described a positive association between CDH23 polymorphisms and NIHL. We hypothesize that genetic variation in the CDH23 gene may play an important role in individual susceptibility to NIHL. However, the relationship between CDH23 gene variants and NIHL is unclear and is the focus of this paper. This study investigates the association between CDH23 gene variants and NIHL.
Materials and Methods
Participants
This is a case-control study in which workers in a steel factory in North China who were exposed to noise were recruited and divided into two groups: the case group (both ears’ high-frequency threshold average [BHFTA] ≥40 dB) and the control group (BHFTA ≤25 dB). The case group was defined as the average binaural hearing level (HL) of a high frequency >40 dB. The control group included a binaural HL of any frequency <25 dB. The details are described in a previous trial.14 This study was conducted following the Declaration of Helsinki and approved by the hospital’s ethics committee. All participants provided informed consent.
Inclusion and Exclusion Criteria
The inclusion criteria were as follows: (1) exposure to occupational noise >80 dB, with a cumulative time of noise exposure ≥3 years; (2) older than 18 years; and (3) provided informed consent. The exclusion criteria were as follows: (1) served in the air force or artillery, had a history of head trauma, explosion deafness or familial deafness or had a history of contagious diseases (mumps, measles or rubella) or treatment with an ototoxic drug (aminoglycoside); (2) had tympanitis, Meniere’s syndrome, conductive hearing loss, exaggerated hearing loss, feigned deafness, sudden deafness, toxic deafness, deafness by contagious disease, tumours and autoimmunological/other diseases that affect hearing; (3) the pure-tone audiogram showed horizontal lines or near-horizontal lines; and (4) had impairment of hearing loss in the speech spectrum that was more severe than the hearing loss in high frequencies.
Following previous studies, the participants were selected and divided into case and control groups.14 The case group was defined as reflecting the average binaural HL of a high frequency ≥40 dB. The individually matched control group was defined as indicating the binaural HL of any frequency <25 dB. The control group was individually matched with a case group of the same gender, age (±5 years), type of work and duration of noise exposure (±2 years) to control environmental confounders.
Data Collection
All of the study participants worked in a steel factory in North China. This study selected 392 participants in the case group and 725 participants in the control group for CDH23 SNP analysis. Participation was voluntary, and written informed consent was obtained from all the participants. They were interviewed by trained study nurses at a convenient location using a standardised and structured questionnaire. Information on the name, sex, date of birth, height, weight, noise exposure history (including air force and artillery), tenure, history of past diseases (head trauma, measles, mumps, rubella, tympanitis, Meniere’s syndrome, explosion deafness and familial deafness), smoking history, drinking history, medication history, physical exercise habits and other diseases that could induce hearing loss were collected through in-person interviews. Data on hypertension and the results of pure-tone audiometry were acquired through a professional test. A morphological otology examination and otoscopy were requested, including bilateral auricle malformation, external auditory canal malformation and stenosis, tympanic membrane perforation and adhesion and calcification.
Pure-Tone Audiometry Detection
The audiometry was carried out using the AS216 audiometer (Interacoustics A/S Company, Denmark). A trained occupational health physician performed all of the audiometric tests on 6,297 workers using standard procedures in quiet test rooms with a background noise level of <25 dB(A). The data of pure-tone air conduction hearing threshold tests were recorded at frequencies of 500, 1,000, 2,000, 3,000, 4,000 and 6,000 Hz after participants had not been exposed to noise for at least 48 h. The averages of 3,000, 4,000 and 6,000 Hz were calculated as the threshold levels at a high frequency for each ear. The hearing thresholds at speech frequency were calculated by the average of 500, 1,000 and 2,000 Hz for each ear. The raw audiometric data were refined by the confounding effects of age and gender based on the Diagnostic Criteria of Occupational NIHL (National Health and Family Planning Commission of the People’s Republic of China. Diagnosis of occupational noise-induced deafness [GBZ49-2014]. Available online: http://www.nhc.gov.cn/zwgkzt/pyl/201410/12e4ec65af8e46248bb45d366a0d5021.shtml [accessed 29 October 2014]).15 In addition, the participants’ ears were also inspected according to this standard.
Cumulative Noise Exposure Calculation
The cumulative noise exposure (CNE) calculation was based on the Occupational Health Standard of the People’s Republic of China: Measurement of Noise in the Workplace (National Health and Family Planning Commission of the People’s Republic of China. Measurement of noise in the workplace [GBZ/T189.8–2007]. Available online: http://www.nhc.gov.cn/zwgkzt/pyl/201410/1a150c9e20f846b8a651d2fd69c6bdb0.shtml [accessed 30 October 2014]);16 noise exposure levels were assessed from 8 am to 4 pm during the participants’ time of work at the representational sites of each type of work using noise dosimeters (NoisePro series, Quest Technologies, Austin, Minnesota, USA). Noise exposure was evaluated with equivalent continuous dB(A)-weighted sound pressure levels (LAeq,8h). Furthermore, previously recorded data about the noise exposure levels of the factory were collected. The CNE was calculated to determine the actual noise exposure for each participant based on every phase of occupational history, which was defined as follows:17
where LAeq,8hi is the equivalent continuous A-weighted noise exposure level in decibels normalised to an 8-hour workday, occurring over a time interval (Ti) in years, with a total of n different noise level exposure phases (ie years spent working in different noise environments/completing noise tasks) and Tref: is 1 year, which represents the noise exposure received over 1 year.
Single-Nucleotide Polymorphism Selection and Genotyping
A total of 18 SNPs in the CDH23 genes were selected for genotyping based on the following criteria: minor allele frequencies of more than 5%; laboratory evidence of function or prior association with human disease studies. Detailed information on the 15 SNPs is presented in Table 1. Genomic DNA was extracted from blood samples using a DNA extraction kit (Shanghai Lifefeng Biotech Co., Ltd., Shanghai, China). The SNP genotyping work was performed using the SNPscan method.18 Additionally, PCR products were sequenced using an ABI3730XL DNA analyser (Applied Biosystems, Foster City, CA, USA) and the results were analysed using the GeneMapper 4.1 software (Applied Biosystems). The entire analysis was performed as a blinded process. The concordance rates for the quality control samples were 99–100% for all assays. All of the SNPs in the controls were in the Hardy–Weinberg equilibrium (HWE) (p > 0.05), except one SNP group (rs1227049) with minor allele frequencies of <10%, which were excluded from the final analysis. A total of 14 SNPs in CDH23 (rs3802720, rs7087735, rs10999947, rs3752752, rs3752751, rs10999978, rs3747867, rs17712523, rs10762480, rs3802711, rs11592462, rs10466026, rs4747194 and rs4747195) were included in the final analysis.
 |
Table 1 The Results of Hardy-Weinberg Test and MAF |
Statistical Analysis
This study used the SPSS 22.0 (IBM, Chicago, USA) software program to conduct statistical analysis. Before the analysis, the HWE test was checked for each SNP among the control participants using a chi-square test. All SNPs in the controls were in the HWE (p > 0.05). Continuous variables were expressed as mean ± standard deviation, while categorical variables were expressed as frequencies (%). An independent series samples t-test was used for two comparisons when each datum conformed to a normal distribution. In contrast, the non-normally distributed continuous data were compared using non-parametric tests. The count data were tested using the chi-square test. Four genetic models were used: additive, dominant, recessive and co-dominant inheritance. The unconditional logistic regression model was used to estimate the odds ratios (ORs) and 95% confidence intervals (CIs) for associations between SNPs, NIHL, different CNE strata (CNE <97 dB[A]/year and CNE ≥97 dB[A]/year) and different noise levels (<85 dB and >85 dB) in different genotype strata. This was completed after adjusting for potential confounding factors, such as smoking, alcohol consumption, hypertension, physical exercise and CNE. The Bonferroni correction was performed to control multiple tests. In the process of Bonferroni correction, significant associations with SNPs (p < 0.05) were detected. The significance threshold was determined by applying a Bonferroni correction, calculated as 0.05/n, where n (n = 18) represents the number of SNPs analysed either genome-wide or chromosome-wide. Consequently, the adjusted 5% threshold for chromosome-wide significance was set as 0.00625. The Haploview 4.2 software package (MIT, The Broad Institute of MIT and Harvard, Cambridge, Mass, USA) was used to investigate the disequilibrium links between the SNPs. The generalised multifactor dimensionality reduction tool (v.0.9) was applied to detect the best multilocus model associated with NIHL after adjustment for covariates, including smoking, drinking, blood pressure, physical exercise and CNE. Finally, 1,000 permutations were performed to gain a permutated value of these models.
Results
General Characteristics
This study included 1,117 participants. The detailed characteristics are presented in Table 2. The BHFTA in the case group was significantly greater than in the control group (51.3 ± 9.0 vs 18.6 ± 8.2, respectively, p < 0.05). The results showed that there were no statistically significant differences between the case and control individuals in the distribution of age, tenure exposure to noise, CNE, sex, drinking status or blood pressure (p > 0.05). However, the physical exercise results reflected the opposite (p < 0.05). The participants who were smokers showed a greater association with NIHL compared with participants who did not smoke (p < 0.05). The variables (smoking, drinking and physical exercise) were further adjusted in the unconditional logistic regression.
 |
Table 2 Selected Characteristics of the Participants in the Study |
The Association Between Cadherin 23 Single-Nucleotide Polymorphisms and Noise-Induced Hearing Loss
The CDH23 SNPs are shown in Table 3 (presented only for SNPs with significant results; the full results of the associations between CDH23 SNPs and NIHL are detailed in Table S1). In Table 3, it is demonstrated that after adjusting for age, tenure, exposure to noise, CNE, sex, smoking, drinking, physical exercise, and blood pressure, participants carrying the CC genotype (adjusted OR: 2.40; 95% CI: 1.26–4.56) and CG + CC genotypes (adjusted OR: 2.47; 95% CI: 1.31–4.66) exhibited the indicated associations. However, no significant differences were detected in the genotype of the other 13 SNPs between the case and control participants (p > 0.05). None of the associations remained statistically significant following the Bonferroni adjustment.
 |
Table 3 The Significant Associations Between the CDH23 SNPs and NIHL |
Stratification Analysis of Cadherin 23 by Noise Exposure Levels and Cumulative Noise Exposure
The associations between the CDH23 SNPs and NIHL stratified by noise exposure levels and CNE are listed in Tables 4 and 5 (presented only for SNPs with significant results; the full results of the stratification analysis of CDH23 by noise exposure levels and CNE are detailed in Table S1). For rs10999947, when the noise exposure levels were >85 dB(A), compared with the GG genotype, an association with NIHL was observed for workers carrying the GA (adjusted OR: 1.43; 95% CI: 1.01–2.01) and GA + AA (adjusted OR: 1.40; 95% CI: 1.01–1.95) genotypes.
 |
Table 4 The Association Between SNPs of CDH23 and NIHL Risk Stratified Analysis by Noise Exposure Level |
 |
Table 5 The Association Between SNPs of CDH23 and NIHL Risk Stratified Analysis by CNE |
As shown in Tables 4 and 5, for rs3802711, for noise exposure levels ≥85 dB(A) and CNE ≥97 dB(A)/year, compared with workers carrying the GG genotype, an association with NIHL was observed for GA (noise exposure levels ≥85 dB[A]: adjusted OR: 1.51; 95% CI: 1.06–2.15; CNE ≥97 dB[A]/year: adjusted OR: 1.50; 95% CI: 1.04–2.15) and GA + AA (noise exposure levels ≥85 dB[A]: adjusted OR: 1.53; 95% CI: 1.09–2.15; CNE ≥97 dB[A]/year: adjusted OR: 1.53; 95% CI: 1.07–2.18) genotypes.
For rs11592462 (noise exposure levels ≥85 dB[A] and CNE ≥97 dB[A]/year), compared with workers carrying the CC genotype, an association with NIHL was observed for the GG (noise exposure levels ≥85 dB[A]: adjusted OR: 2.53; 95% CI: 1.16–5.52; CNE ≥97 dB[A]/year: adjusted OR: 2.46; 95% CI: 1.09–5.55) genotype. A similar pattern was also observed when comparing workers carrying the GG genotype.
This study made a post hoc correction using the Bonferroni adjustment, which adjusts the statistical significance level to reduce the probability of committing a type I error (rejecting the true null hypothesis). However, none of the associations remained statistically significant following the Bonferroni adjustment.
Association of Cadherin 23 Haplotypes with Noise-Induced Hearing Loss
Haplotypes were inferred based on the observed genotypes using the Haploview software. The 10 SNPs, that is, rs1227049, rs3752752, rs10999947, rs3752751, rs10762480, rs3802711, rs11592462, rs4747195, rs4747194 and rs10466026 constructed the haplotypes. The results suggest a significant association between the 10 SNP haplotypes and NIHL. After applying the Bonferroni correction, none of the associations remained statistically significant (Table S2).
Evaluation of the Interaction Effect Between Cadherin 23 Single-Nucleotide Polymorphisms
Generalised multifactor dimensionality reduction was performed to reveal interactions between the SNPs. None of the significant SNP–SNP interactions were found in this study (p > 0.05; see Table S3 for details). Specific information about SNPs on chromosomes is shown in Table S4.
Discussion
Noise-induced hearing loss is caused by prolonged exposure to high levels of noise in the workplace and is classified as a serious occupational disease. There are two types of hearing loss: temporary and permanent threshold shifts.19 Studies have shown that oxidative stress plays an important role in hearing loss. High levels of noise can lead to free radical damage. Moreover, reactive oxygen species (ROS) and lipid peroxides increase during and after noise exposure, leading to hearing loss.20 At the hair cell level, noise can lead to ischaemia/reperfusion effects in cochlear blood sources, resulting in an increase in ROS and damage to DNA synthesis and cell membranes and can serve as a starting factor for apoptosis. Moreover, the combination of hair cell damage and apoptosis leads to hearing loss.21 Antioxidants, such as vitamin B12, folic acid and N-acetylcysteine can have a positive impact on patients with noise overexposure and reflect a promising new treatment strategy for preventing the effects of noise on hearing.19
This study is the first comprehensive analysis of associations between genetic polymorphisms in CDH23 genes and NIHL. Using SNPs (14 in total) located on CDH23 genes, significant associations were observed for rs10999947, rs3802711, rs11592462, rs10762480, rs3752751, rs3752752 and rs3747867 concerning NIHL overall and/or various CNE strata and noise exposure levels.
In this study, the tested associations did not remain statistically significant after the Bonferroni adjustment. In two recent studies on associations between NIHL and the hereditary spastic paraplegia gene, Yang et al did not detect a significant association with NIHL using SNP analysis. Furthermore, they found only significant associations between two haplotypes (GGC and GGT) and NIHL; however, Annelies et al detected significant results after a single SNP analysis.22,23 One possible reason for this may be the stabilisation of sequencing or the methods used for establishing groups, which employed different ages, sex and smoking or drinking statuses. Qian et al24 confirmed alcohol as a risk factor for hearing loss. Furthermore, in another study on the association between NIHL and gene analysis, the researchers found an association between NIHL and the rs41281334 of CDH23, providing more evidence on the CDH23 gene and NIHL.25
Overall, this study suggests that the CDH23 polymorphism (rs11592462) may have a significant association with NIHL, which is consistent with a Polish population study.26 Furthermore, the CDH23 polymorphism (rs3802711) may have an increased NIHL risk, as was found in a previous study.23
In terms of stratified analysis by noise exposure level and/or CNE, when noise exposure levels were >85 dB(A) or when CNE was >97 dB(A)/year, this study suggests that the CDH23 polymorphism (rs11592462) may exhibit a significant association with NIHL, as well as the CDH23 polymorphisms (rs3802711) and (rs10762480).
In summary, this study found that CDH23 polymorphisms (rs11592462, rs3802711 and rs10762480) are significantly associated with NIHL, which is consistent with an existing study.27,28 This suggests that CDH23 gene polymorphisms play an important role in the development of NIHL, which was proven by analysing the association between CDH23 haplotypes and NIHL.
Cadherin 23 plays an essential long-term role in the normal structure of the ciliary bundle of the cochlea.29–31 Cadherin 23 mutations were first associated with susceptibility to NIHL in the population, and this study recommends that CDH23 could be an early indicator of hearing loss in routine screening.11 Although some studies did not reveal the association between gene variations in CDH23 and NIHL,11,32 a recent study suggests that CDH23 gene variations show a significant association with NIHL,13 which is consistent with this study’s results.
This study has several strengths. First, it is an NIHL cohort population-based case-control study and, accordingly, has a more comprehensive dataset of noise exposure than previous studies. Second, the cases included in this study were selected using the diagnosis of occupational noise-induced deafness (standard GBZ 49–2014) and the controls were diagnosed using the same standard as the cases. Hence, the results differ from those of previous studies11,14 in which the population was selected based on susceptibility to noise and, to some degree, hearing loss. Furthermore, the participants in the case group were diagnosed with NIHL specifically, as opposed to more generic hearing loss, which provided a better dataset for studying NIHL. Third, the power size of the study in the case of hearing loss was bigger than in previous studies.
This study also has limitations that should be considered. First, when the Bonferroni correction was applied, the significance of the findings was not detected. However, replicating findings in independent populations is more important than obtaining highly significant p-values. Furthermore, it is necessary to test other correction methods to support these findings. Second, workers can be exposed to noise in other places such as the community, but this was too complex to consider in the present study. One recent review,33 based on different high-noise exposure groups, suggests that the noise intensity of daily life has little effect on the results for high-intensity noise exposure studies. In addition, in a previous study, relevant analysis was conducted34 in which the participants of two studies were sourced from the noise exposure cohort established by the research group at an earlier stage. The previous study selected 286 participants in the case and control groups for CDH23 SNP analysis.34 This paper, based on the analysis results, expanded the number of cases and controls to verify previous results. The current research focused on a specific pathway; additional SNPs should be tested in the future.
Conclusion
The genetic variations in the CDH23 gene may play an important role in determining individual susceptibility to NIHL. These results provide new insight into the pathogenesis and early prevention of NIHL.
Data Sharing Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
This study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of the Henan Medical College. Signed informed consent was obtained from all participants.
Funding
National Natural Science Foundation of China (81372940, 81872574); National Science and technology support program (2014BAI12B03).
Disclosure
All of the authors had no any personal, financial, commercial, or academic conflicts of interest separately for this work.
References
1. Mehrparvar AH, Mirmohammadi SJ, Hashemi SH, et al. Concurrent effect of noise exposure and smoking on extended high-frequency pure-tone thresholds. Int J Audiol. 2015;54(5):301–307. doi:10.3109/14992027.2014.978906
2. Upile T, Sipaul F, Jerjes W, et al. The acute effects of alcohol on auditory thresholds. BMC Ear Nose Throat Disord. 2007;7:4. doi:10.1186/1472-6815-7-4
3. Taylor W, Pearson J, Mair A, Burns W. Study of noise and hearing in jute weaving. J Acoust Soc Am. 1965;38:113–120. doi:10.1002/ajhb.20744
4. Sliwinska-Kowalska M, Pawelczyk M. Contribution of genetic factors to noise-induced hearing loss: a human studies review. Mutat Res. 2013;752(1):61–65. doi:10.1016/j.mrrev.2012.11.001
5. Siemens J, Lillo C, Dumont RA, et al. Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature. 2004;428(6986):950–955. doi:10.1038/nature02483
6. Bouzid A, Smeti I, Chakroun A, et al. CDH23 methylation status and presbycusis risk in elderly women. Front Aging Neurosci. 2018;10:241. doi:10.3389/fnagi.2018.00241
7. Holme RH, Steel KP. Progressive hearing loss and increased susceptibility to noise-induced hearing loss in mice carrying a Cdh23 but not a Myo7a mutation. J Assoc Res Otolaryngol. 2004;5(1):66–79. doi:10.1007/s10162-003-4021-2
8. Noben-Trauth K, Zheng QY, Johnson KR. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat Genet. 2003;35(1):21–23. doi:10.1038/ng1226
9. Davis RR, Newlander JK, Ling X, Cortopassi GA, Krieg EF, Erway LC. Genetic basis for susceptibility to noise-induced hearing loss in mice. Hear Res. 2001;155(1–2):82–90. doi:10.1016/s0378-5955(01)00250-7
10. Miyasaka Y, Shitara H, Suzuki S, et al. Heterozygous mutation of Ush1g/Sans in mice causes early-onset progressive hearing loss, which is recovered by reconstituting the strain-specific mutation in Cdh23. Hum Mol Genet. 2016;25(10):2045–2059. doi:10.1093/hmg/ddw078
11. Konings A, Van Laer L, Wiktorek-Smagur A, et al. Candidate gene association study for noise-induced hearing loss in two independent noise-exposed populations. Ann Hum Genet. 2009;73(2):215–224. doi:10.1111/j.1469-1809.2008.00499.x
12. Yang M, Tan H, Zheng JR, et al. Association of cadherin CDH23 gene polymorphisms with noise induced hearing loss in Chinese workers. Wei Sheng Yan Jiu. 2006;35(1):19–22.
13. Kowalski TJ, Pawelczyk M, Rajkowska E, Dudarewicz A, Sliwinska-Kowalska M. Genetic variants of CDH23 associated with noise-induced hearing loss. Otol Neurotol. 2014;35(2):358–365. doi:10.1097/MAO.0b013e3182a00332
14. Xu X, Yang Q, Jiao J, et al. Genetic variation in POU4F3 and GRHL2 associated with noise-induced hearing loss in Chinese population: a case-control study. Int J Environ Res Public Health. 2016;13(6):561. doi:10.3390/ijerph13060561
15. National Health and Family Planning Commission of the PRC. Diagnosis of occupational noise deafness (instead of GBZ 49-2007). Available fromhttp://www.nhc.gov.cn/zwgkzt/pyl/201410/12e4ec65af8e46248bb45d366a0d5021.shtml.
16. National Health and Family Planning Commission of the PRC. Measurement of physical factors in the workplace -Part 8: noise. Available fromhttp://www.nhc.gov.cn/zwgkzt/pyl/201410/1a150c9e20f846b8a651d2fd69c6bdb0.shtml
17. Xie HW, Qiu W, Heyer NJ, et al. The use of the kurtosis-adjusted cumulative noise exposure metric in evaluating the hearing loss risk for complex noise. Ear Hear. 2016;37(3):312–323. doi:10.1097/AUD.0000000000000251
18. Ting JC, Ye Y, Thomas GH, Ruczinski I, Pevsner J. Analysis and visualization of chromosomal abnormalities in SNP data with SNPscan. BMC Bioinf. 2006;7:25. doi:10.1186/1471-2105-7-25
19. Abbasi M, Pourrajab B, Tokhi MO. Protective effects of vitamins/antioxidants on occupational noise-induced hearing loss: a systematic review. J Occup Health. 2021;63(1):e12217. doi:10.1002/1348-9585.12217
20. Henderson D, Bielefeld EC, Harris KC, Hu BH. The role of oxidative stress in noise‐induced hearing loss. Ear Hear. 2006;27(1):1–19. doi:10.1097/01.aud.0000191942.36672.f3
21. Quaranta N, Dicorato A, Matera V, D’Elia A, Quaranta A. The effect of alpha‐lipoic acid on temporary threshold shift in humans: a preliminary study. Acta Otorhinolaryngol Ital. 2012;32:380–385.
22. Yang M, Tan H, Yang Q, et al. Association of hsp70 polymorphisms with risk of noise-induced hearing loss in Chinese automobile workers. Cell Stress Chaperones. 2006;11(3):233–239. doi:10.1111/j.1469-1809.2008.00499.x
23. Konings A, Van Laer L, Michel S, et al. Variations in HSP70 genes associated with noise-induced hearing loss in two independent populations. Eur J Hum Genet. 2009;17(3):329–335. doi:10.1038/ejhg.2008.172
24. Qian PY, Zhao ZX, Liu SY, et al. Alcohol as a risk factor for hearing loss: a systematic review and meta-analysis. PLoS One. 2023;18(1):e0280641. doi:10.1371/journal.pone.0280641
25. Jiang Z, Fa B, Zhang X, et al. Identifying genetic risk variants associated with noise-induced hearing loss using a novel strategy for evaluating individual susceptibility. Hear Res. 2021;407:108281. doi:10.1016/j.heares.2021.108281
26. Sliwinska-Kowalska M, Noben-Trauth K, Pawelczyk M, Kowalski TJ. Single nucleotide polymorphisms in the cadherin 23 (CDH23) gene in polish workers exposed to industrial noise. Am J Hum Biol. 2008;20(4):481–483. doi:10.1002/ajhb.20744
27. Jiang CM. Single nucleotide polymerase in CDH23, PMCA2 gene in workers exposed to industrial noise. Master’s Deg Thesis Guangdong Pharmaceutical University. 2012.
28. Zheng QY, Yan D, Ouyang XM, et al. Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum Mol Genet. 2005;14(1):103–111. doi:10.1093/hmg/ddi010
29. Kazmierczak P, Sakaguchi H, Tokita J, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449(7158):87–91. doi:10.1038/nature06091
30. Muller U. Cadherins and mechanotransduction by hair cells. Curr Opin Cell Biol. 2008;20(5):557–566. doi:10.1016/j.ceb.2008.06.004
31. Sakaguchi H, Tokita J, Müller U, Kachar B. Tip links in hair cells: molecular composition and role in hearing loss. Curr Opin Otolaryngol Head Neck Surg. 2009;17(5):388–393. doi:10.1097/MOO.0b013e3283303472
32. Wang SL, Yu LG, Liu RP, et al. Gene-gene interaction of GJB2, SOD2, and CAT on occupational noise-induced hearing loss in Chinese han population. Biomed Environ Sci. 2014;27(12):965–968. doi:10.3967/bes2014.131
33. Lie A, Skogstad M, Johannessen HA, et al. Occupational noise exposure and hearing: a systematic review. Int Arch Occup Environ Health. 2016;89(3):351–372. doi:10.1007/s00420-015-1083-5
34. Jiao J, Gu GZ, Chen GS, et al. Relationship research among CDH23 gene and the risk of noise-induced hearing loss. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2020;38(2):84–90. doi:10.3760/cma.j.issn.1001-9391.2020.02.002
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.