Back to Journals » OncoTargets and Therapy » Volume 12
Upregulation Of miR-153 Inhibits Triple-Negative Breast Cancer Progression By Targeting ZEB2-Mediated EMT And Contributes To Better Prognosis
Authors Shi D, Li Y, Fan L, Zhao Q, Tan B, Cui G
Received 17 July 2019
Accepted for publication 30 October 2019
Published 13 November 2019 Volume 2019:12 Pages 9611—9625
DOI https://doi.org/10.2147/OTT.S223598
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Dongliang Shi,1,2 Yong Li,1 Liqiao Fan,1 Qun Zhao,1 Bibo Tan,1 Guozhong Cui2
1The Third Department of General Surgery, The Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei, People’s Republic of China; 2The Second Department of Thyroid and Breast Surgery, Cangzhou Central Hospital, Cangzhou, Hebei, People’s Republic of China
Correspondence: Yong Li
The Third Department of General Surgery, The Fourth Hospital of Hebei Medical University, No.12 Jiankang Road, Shijiazhuang, Hebei 050019, People’s Republic of China
Email [email protected]
Background: Triple-negative breast cancer (TNBC) is the most malignant type of breast cancer. MicroRNAs (miRs) and their corresponding molecular targets are associated with the occurrence and development of various human malignancies. However, the roles of the microRNA-153 (miR-153) and zinc finger E-box-binding homeobox 2 (ZEB2)-induced epithelial–mesenchymal transition (EMT) in TNBC and predictive effect of miR-153 on the prognosis of TNBC have not been fully elucidated.
Materials and methods: Relative miR-153 expression level was examined by RT-qPCR assay in TNBC tissues of 60 patients and TNBC cell lines (SKBR3, BT-549 and MDA-MB-231). Cell proliferation ability, invasion ability and migration ability were measured by CCK8 assay, Transwell invasion assay and wound healing assay, respectively. Luciferase reporting experiment was used to confirm that there was a miR-153-binding site in ZEB2 3ʹ-UTR. The expression of ZEB2 in tissues and its relationship with miR-153 were analyzed with immunohistochemistry method. Relative ZEB2, E-cadherin, N-cadherin and Vimentin mRNA and protein expression levels were observed with RT-qPCR and Western blot, respectively. Based on risk factors, a prognostic model was established according to the Cox proportional risk model, and the prognostic risk factors of TNBC patients were predicted and analyzed.
Results: The expression of miR-153 in TNBC tissues and cells was declined (all P<0.01), and upregulation of miR-153 inhibited proliferation, invasion and migration of TNBC cells (all P<0.01). In addition, miR-153 regulated ZEB2/EMT link in TNBC, and ZEB2 overexpression reversed the tumor-suppressive effect of miR-153 in TNBC. Moreover, miR-153 was an independent predictive factor that was associated with excellent prognosis in TNBC patients.
Conclusion: miR-153 may inhibit TNBC proliferation, invasion and migration by regulating ZEB2/EMT link. Therefore, miR-153 is expected to be a molecular target and prognostic marker for TNBC.
Keywords: microRNA-153, zinc finger E-box-binding homeobox 2, epithelial-mesenchymal transition, proliferation, invasion, migration
Introduction
Breast cancer (BC) is one of the most common malignant tumors with the highest incidence in women, which attacks 2.1 million people worldwide each year.1 BC ranks first in the incidence of female malignant tumors in China,2 which has become a serious threat to women’s health. Triple-negative breast cancer (TNBC), a special subtype of BC, refers to one that lacks estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (Her-2) target expression, which accounts for 10–20% of BC.3 Unique molecular typing of TNBC determines its special clinicopathological characteristics, including higher histological grade, earlier onset age, earlier metastasis, higher invasiveness, shorter median survival time and higher mortality. Given that TNBC lacks ER, PR and Her-2 expression, excepting clinical chemotherapy, surgery and radiotherapy, no effective therapy method is available, which leads to early recurrence and metastasis. Therefore, it is necessary to better comprehend the molecular mechanism of occurrence and development of TNBC, to pinpoint prognostic molecular markers and to identify new prevention and treatment strategies.
MicroRNA (miRNA), a kind of endogenous non-coding single-stranded RNAs, consists of approximately 20–25 nucleotides, existing widely in eukaryotic organisms. It plays the role of gene silencing and post-transcriptional modification in regulating gene expression, mainly through partial pairing, especially between the 2 and 8 nucleotides at the mRNA 3ʹ-untranslated region and the miRNA 5ʹ-end.4 It is confirmed that miRNAs are related to a variety of human illnesses, including carcinoma.5 There is a growing body of evidence that miRNAs regulate the physiological activities of the cells, such as proliferation, tumorigenesis, invasion and migration.6,7 Accumulating studies show that miRNAs are involved in the pathogenesis of BC in which miR-153 was aberrantly expressed.8–10 Moreover, miR-153 was considered as a potential biomarker of TNBC.11 Modes of actions and prognostic value of miR-153 in TNBC, however, have not been fully illuminated.
Zinc finger E-box-binding homeobox 2 (ZEB2), which is located on chromosome 2 of human, belongs to the E-box bound zinc finger protein family. ZEB2 is a protein molecule, being encoded by Zfhx1 B gene and consisting of 10 exons and 9 introns, which encode 1214 amino acids.12 Its intermediate structure is a variable region, with two independent and highly conservative zinc finger clusters on each side. Bilateral zinc finger clusters can independently bind to DNA containing CACCT (G) or CAGGTG (E-box) sequences to regulate the transcription of target genes.13,14 ZEB2 plays a variety of roles in tumorigenesis and development. The most important function of ZEB2 is to induce tumor cells to transform into mesenchymal phenotypes with high invasiveness and migration abilities, resulting in the acquisition of mesenchymal markers (N-cadherin and vimentin) and the loss of epithelial markers (E-cadherin), which is known as epithelial–mesenchymal transition (EMT). In this process, cellular structure, polarity, adhesion and migration change promote malignant progression of tumors, including TNBC.15
In our study, we not only analyzed the relationship between miR-153 and clinicopathological features, but also observed the effects of miR-153 on the proliferation, invasion and migration abilities of TNBC cells. We also focused on the relationship between miR-153 and ZEB2/EMT axis in TNBC and their effects on the occurrence and development of TNBC. The possible signal pathways and intrinsic regulatory mechanisms were also revealed. We further applied Cox proportional hazard model to analyze the risk factors of TNBC patients and confirmed the predictive value of miR-153 for the prognosis of TNBC patients. The results suggest that miR-153 may play an anticancer role by targeting the ZEB2/EMT axis, and miR-153 may be an underlying molecular target and prognostic marker for TNBC.
Materials And Methods
Clinical Tissue Samples
Sixty paired primary TNBCs and their corresponding paracancer tissues excised from patients were collected from the Department of Thyroid and Breast Surgery, Cangzhou Central Hospital (Cangzhou, China) between January 2012 and December 2013, with following-up as of 31 December 2018 (median follow-up of 69 months). All patients (with a median age of 51.5 years) had invasive ductal carcinoma. The tissue samples were quickly frozen after operation and stored at –80°C until specimens were tested. The research protocol was approved by the Institutional Ethics Committee of Cangzhou Central Hospital and written informed consent was obtained from all patients. The inclusion criteria in this study included modified radical mastectomy (complete mastectomy and systematic lymph node dissection of the axillary and thoracic intermuscular) for BC patients, histologically confirmed TNBC and complete clinical and follow-up data. The exclusion criteria included preoperative chemotherapy/radiotherapy or death in the perioperative period, lost to follow-up and IV stage of the first diagnosis. Tumor staging was based on the eighth edition American Joint Committee on Cancer (AJCC).
Cell Culture And Cell Transfection
TNBC cell lines (SKBR3, BT-549 and MDA-MB-231) and normal breast epithelial cell lines (MCF-10A) were purchased from the American Type Culture Collection (Invitrogen). All cell lines were cultured in RPMI-1640 medium (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 10% FBS (Invitrogen) in a humidified incubator with 5% CO2 at 37°C.
MicroRNA-153 (miR-153) mimics and their corresponding negative control (miR-NCs) as well as ZEB2 vector were acquired from the Genechem Company (Shanghai, China), and then verified by DNA alignment analysis. All cultured cells were transfected with the Lipofectamine 2000 reagent (Invitrogen) based on the commercial specifications. At 24 hr post-transfection, cells were gathered for in vitro assays.
RNA Extraction And Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
Total RNA was isolated from frozen tissue specimens and cultured cells with Trizol reagent (Invitrogen) in accordance with the manufacturer’s instructions. Reverse transcription was carried out using the TaqMan miRNA Reverse Transcript Kit (Applied Biosystems, Foster City, CA, USA). PCR reactions were conducted with TaqMan miRNA primers (Applied Biosystems) on an Applied Biosystems Prism7900 Fast Sequence Detection System. The expressions of miR-153 were quantified using U6 RNA. GAPDH, as endogenous control, was applied to normalize the expression level of ZEB2, E-cadherin, N-cadherin and vimentin. The 2−ΔΔCt method was used to quantify the relative expression levels of genes. The primer sequences are shown in Table 1.
 |
Table 1 Primer Sequences Of PCR |
Western Blot Analysis
RIPA Lysis buffer which was supplemented with phenylmethanesulfonyl fluoride (PMSF) was used to isolate the total protein from frozen tissue specimens and cultured cells. Loading buffer was added to cell lysates which were subjected to 10% SDS-PAGE, and then it was transferred onto polyvinylidene difluoride (PVDF) membranes. The 5% skimmed milk was used to block the membranes for 2 hrs. Subsequently, the membranes were incubated with antibodies against ZEB2, E-cadherin, N-cadherin and vimentin (Abcam, Cambridge, UK) at 4°C overnight, followed by three-time wash in TBST and incubation with the corresponding horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology, Danvers, MA, USA) for another 1 hr at 37°C. The proteins were visualized by an enhanced ECL Chemiluminescence kit (UltraSignal, Beijing, China) following the manufacturer’s instructions. The signals were exposed to X-ray films. β-actin was used as the loading control.
Cell Proliferation Assay
Cell proliferation ability was tested using a Cell Counting Kit-8 (CCK-8) (Beyotime Institute of Biotechnology, Shanghai, China), following the product instructions. In brief, the transfected cells (2×103 cells/well) were seeded into 96-well culture plates with complete medium to culture. At the appointed time, the CCK-8 assay solution (10 μL) and DMEM (100 μL) were added to every well, and then the cells were cultured for another 2 hrs. Auto-microplate reader was used to measure the absorbance at 450 nm.
Transwell Invasion Assay
Cell invasion ability was evaluated by Transwell invasion assay. Briefly, cells (2×105 cells) that were suspended in 200 μL of serum-free medium were seeded in the upper chamber of 24-well plates which were coated with Matrigel (BD Biosciences). The culture medium with 30% FBS was added to the lower chamber. The cells were incubated for 24 h at 37°C, fixed and then stained with 0.1% crystal violet (Sigma-Aldrich). Subsequently, the cells which had invaded through the matrigel membrane were quantified by microscopy.
Wound Healing Assay
For wound healing assay, the transfected cells were seeded in 60 mm dishes and cultured in DMEM. A scratch wound was made with a pipette tip in cell layers which were grown in serum-free DMEM for an additional 48 hrs. The wounded gaps were photographed with a light microscope, at 0 hr and 48 hrs, at 200×magnification, and the distances of migrating cells were measured.
Immunohistochemistry (IHC) And Result Evaluation
Immunohistochemical staining was performed according to standard biotin-streptomyces antibiotic protein-peroxidase (SP) immunohistochemical kit instructions. The 4-µm thickness paraffin tissue sections were dewaxed in xylene, followed by rehydration in graded ethanol. The antigens were thermally repaired with 0.01 mol/L sodium citrate buffer (pH 6.0) and cooled at 37°C for 40 mins. The endogenous peroxidase activity was blocked by incubation with 3% hydrogen peroxide ion water at 37°C for 15 mins and nonspecific protein blocking solution 1% bovine serum albumin in PBS at 37°C for 45 mins, successively. The slices were incubated with anti-ZEB2 antibody (rabbit polyclonal; ab138222; Abcam) overnight at 4°C, washed, incubated with biotin labelled anti-II working fluid at 37°C for 20 mins and then incubated with streptomyces ovalbumin working solution labeled with horseradish peroxidase at 37°C for 20 mins. Finally, the slices were stained with diaminobenzidine (DAB), slightly re-stained with hematoxylin, dehydrated with alcohol gradient, transparentized with xylene and sealed with neutral gum. The primary antibody was replaced by blocking solution as negative controls.
The immunostaining results were evaluated by two independent pathologists by randomly selecting five high-power fields (10×40) for observation. The summation of cell color intensity score and percentage score of chromogenic cells was used as a semi-quantitative evaluation standard. The intensity of staining was evaluated semiquantitatively as follows: 0, no staining; 1, weak; 2, moderate; 3, strong. Percentage of staining cells was evaluated semiquantitatively as follows: 0, <5%; 1,5–25%; 2, 25––50%; 3, >50% of the cells in the high-power fields. The results (ranging from 0 to 6) of summing the above scores were split into negative (final scores, <1) and positive (final scores, ≥1).
Luciferase Reporter Assays
The ZEB2 3′UTR luciferase reporter gene plasmid was synthesized with the plasmid pMIR. These wild-type (WT) or mutant (MUT) 3′UTRs which were, respectively, defined as ZEB2 WT or ZEB2 MUT were cloned into pMIR plasmid at the downstream site of the luciferase vector. Luciferase reporter gene assay was conducted in 96-well plates with the Dual-Luciferase Assay System (Promega, Madison, WI, USA). ZEB2 WT or ZEB2 MUT and miR-153 mimics were co-transfected into cells with Lipofectamine 2000 (Invitrogen). After 48 hrs, Dual-Luciferase Reporter Assay Kit (Promega) was applied to measure the luciferase activity following the manufacturer’s instructions. The detection result was analyzed by Spectra Max M5 instrument software (Molecular Devices, San Francisco, CA, USA).
Statistical Analysis
Statistical software SPSS25.0 (IBM, Chicago, IL, USA) was applied to analyze the experimental results. All quantitative data were expressed as mean ± SE. Student’s t-test or one-way analysis of variance (ANOVA) was used to compare two or multiple groups, respectively. The Mann–Whitney U-test was performed to test the differences of miR-153 expression between TNBCs and their corresponding adjacent noncancerous tissues. Spearman correlation analysis was carried out to assess the relationship between miR-153 and ZEB2. Disease-free survival (DFS) curves and overall survival (OS) curves were generated using the Kaplan–Meier method, and the differences in survival rate were calculated using the log-rank test. Prognostic factors were assessed by univariate and multivariate analyses (Cox’s proportional hazard regression model). A P-value <0.05 was regarded as having statistical differences.
Results
Relative miR-153 Expression Was Decreased In TNBC Tissues And Cells
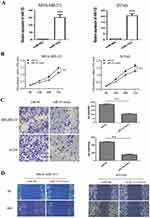
To explore the biological role of miR-153 in TNBC, we measured the relative expression of miR-153 in TNBC tissues with RT-qPCR. The results revealed that the relative expression of miR-153 in TNBC tissues was obviously lower than that in corresponding adjacent noncancerous tissues (P< 0.01, Figure 1A). To further discuss the clinical significance of miR-153 in TNBC, the relationship between expression levels of miR-153 and clinicopathological feature was analyzed. On the basis of the cutoff value, which was defined as the median of the cohort, the relative miR-153 expression in TNBC tissues was supposed as either low (n=30) or high (n=30). It was noted that the expression of miR-153 was downregulated in TNBC tissues with larger tumor size (P=0.035), lymph node metastasis (P=0.008) and advanced TNM stages (P=0.031) (Table 2). Furthermore, we compared the relative expression of miR-153 in TNBC cells (SKBR3, BT-549 and MDA-MB-231) and normal breast epithelial cells (MCF-10A) with RT-qPCR. The results showed that the relative expression of miR-153 in SKBR3, BT-549 and MDA-MB-231 cells was remarkably lower than that in MCF-10A cells (P<0.01, Figure 1B). These data suggested that miR-153 might play an important role against TNBC.
 |
Table 2 Correlation Between miR-153 Expression And Clinical Features (n=60) |
Upregulation Of miR-153 Diminished Proliferation, Invasion And Migration Of TNBC Cells
To further analyze the molecular characteristics of miR-153 in TNBC, gain-of-function assays were carried out with miR-153 mimics. The expression of miR-153 was remarkably upregulated in the miR-153 mimic group compared to the miR-NC group (P<0.001, Figure 2A). CCK8 assay was performed to detect the effect of miR-153 on the proliferation ability of TNBC cells. The results suggested that ectogenic miR-153 significantly repressed the proliferation ability of TNBC cells (P< 0.01, Figure 2B). Meanwhile, the effects of miR-153 on the invasion ability of TNBC cells were likewise examined. Exogenic miR-153 obviously weakened the invasion ability of MDA-MB-231 and BT-549 cells, indicating that miR-153 attenuated the invasion ability of TNBC cells (P< 0.01, Figure 2C). In addition, wound healing assay revealed that overexpression of miR-153 diminished the migration ability of TNBC cells (Figure 2D). All these results verified that miR-153 could effectively depress the proliferation, invasion and migration of TNBC cells.
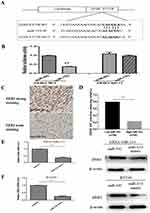
ZEB2 Was Predicted As A Direct Target Of miR-153 In TNBC
Because of the large number of downstream molecular targets of miR-153, it was unrealistic to study each molecular target. We could only look for a downstream target that might play an important role in the process of EMT of TNBC as well as the occurrence and development of TNBC through the TargetScan (http://www.targetscan.org/vert_71/), PicTar (https://pictar.mdc-berlin.de) and mi-Randa (http://www.microrna.org/microrna/getGeneForm.do) databases. It is found that ZEB2 was a target gene, and there was a miR-153-binding site in ZEB23′-untranslated region (3ʹ-UTR, Figure 3A). The most important function of ZEB2 is to induce tumor cells to transform into mesenchymal phenotypes from epithelial phenotypes. To further investigate whether miR-153 could target ZEB2 and the underlying molecular mechanism, luciferase activity assay was conducted on cells co-transfected with miR-NC or miR-153 plasmids and wild-type (WT) or mutant (MUT) ZEB2 3′UTR vectors. As predicted, miR-153 mimics obviously decreased the relative firefly luciferase activity when the predicted sites were not mutated (P< 0.01, Figure 3B).
To figure out the correlation between miR-153 and ZEB2 in TNBC tissues, IHC assay was first performed. The positive expression of ZEB2 was mainly located in the cytoplasm of TNBC cells. The expression of ZEB2 with low miR-153 expression in TNBC tissues was remarkably higher than that with high miR-153 expression (Figure 3C). Moreover, Spearman correlation analysis implied that miR-153 expression was negatively correlated with ZEB2 expression in TNBC (r=−0.567, P< 0.001, Figure 3D). In order to further explore the relationship between miR-153 and ZEB2 in TNBC cells, RT-qPCR assay and Western blot assay also were performed, respectively. The results indicated that ZEB2 mRNA and protein were both significantly reduced in miR-153 upregulated TNBC cells (Figure 3E and F).
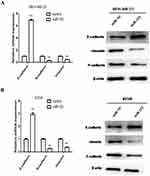
miR-153 Suppressed ZEB2-Induced EMT In TNBC
The relationship between miR-153 and EMT markers induced by ZEB2 was further examined by RT-qPCR assay and Western blot assay, respectively. The expression of mRNA and protein of epithelial marker (E-cadherin) was remarkably increased in miR-153 upregulated TNBC cells, while the expression of mRNA and protein of mesenchymal markers (N-cadherin and Vimentin) was reduced in TNBC cells transfected with miR-153 mimic (Figure 4A and B). These results displayed that miR-153 might inhibit the progression of TNBC cells by depressing ZEB2-induced EMT.
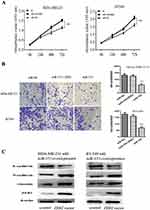
Overexpression Of ZEB2 Attenuated The Inhibitory Effect Of miR-153 On Tumor Proliferation, Invasion And EMT In TNBC
To further investigate the influence of ZEB2 on the inhibitory effect of TNBC cells mediated by miR-153, we co-transfected the ZEB2 overexpression vector (without 3′-UTR) and miR-153 overexpression plasmid into TNBC cells. We observed that the upregulation of ZEB2 partially reversed the miR-153-mediated suppressive effect on tumor proliferation and invasion in TNBC cells (all P< 0.01, Figure 5A and B). In addition, in TNBC cells with the ZEB2 overexpression vector, EMT abilities and ZEB2 expression which were found to be decreased by the miR-153 upregulated were restored, as compared with the control group (Figure 5C). These results showed that miR-153 repressed TNBC cell malignant evolution by downregulating the ZEB2/EMT axis.
The Relationship Between The Expression Of miR-153 And The Prognosis Of TNBC Patients
We performed Cox regression analysis of univariate and multivariate of potential risk factors that might affect the prognosis of patients with TNBC. Univariate analysis revealed that tumor size, lymph node metastasis, TNM stage, tumor thrombus, and miR-153 affected DFS and OS of TNBC patients (Tables 3 and 4). Multivariate analysis confirmed that tumor thrombus and miR-153 were significantly associated with DFS and OS of TNBC patients (Tables 3 and 4). Moreover, based on the expression of miR-153, TNBC patients with complete clinical survival information were analyzed by Kaplan–Meier estimates (Figure 6A and B). Results from the statistical analysis indicated that the upregulation of miR-153 contributed to better DFS and OS in TNBC patients. Therefore, miR-153 might serve as an independent prognostic factor in TNBC patients.
 |
Table 3 Univariate And Multivariate Analysis Of Clinical Characteristics In Relation To Disease-Free Survival (DFS) |
 |
Table 4 Univariate And Multivariate Analysis Of Clinical Characteristics In Relation To Overall Survival (OS) |
Discussion
In the field of molecular biology of cancer, miRNAs have attracted increasing attention, due to their involvement in a series of vital life processes, including biological behavior of malignant tumors, such as growth, cloning, invasion and migration.6,7 Especially in TNBC, an increasing number of miRNAs have been proved to play a significant role in tumorigenesis and development. However, emerging studies have confirmed that different miRNAs play diverse roles in TNBC, some as tumorigenic agents, and some as tumor inhibitors. For example, as tumorigenic agents, miR-25,16 miR-20,17 miR-224,18 miR-13519 and miR-30120 promote the occurrence and development of TNBC, whereas miR-124,15 miR-4417,21 miR-4306,22 miR-199,23 miR-128724 and miR-317825 as tumor inhibitors inhibit the oncogenesis and development of TNBC. In this study, relative miR-153 expression in TNBC tissues and cells was remarkably lower than that in corresponding adjacent noncancerous tissues and normal breast epithelial cells. These data showed that miR-153 might act as an inhibitor to suppress the neoplasia and development of TNBC.
miR-153, as one of the conservative miRNAs, was initially found in seven miRNAs specifically expressed in human and mouse brain tissues.26 Current studies have confirmed that miR-153 is involved in the development and progression of a variety of malignancies,27–32 such as prostate carcinoma, colorectal cancer, hepatocellular malignancy, non-small-cell lung carcinoma, gastric malignant tumor, glioma and BC. However, its role in various tumors is not exactly identical. In prostate cancer, miR-153 has been reported as an oncogene for the first time, and it has been verified that it can promote the proliferation of carcinoma cells by inhibiting the expression of PTEN.27 miR-153 promotes the malignant evolution of colorectal carcinoma by regulating MMP-9 to increase the invasiveness of cancer cells and directly regulating FOXO3a to enhance platinum-based chemotherapy resistance.28 In addition, miR-153 promotes the oncogenesis and development of hepatocellular carcinoma via activating the Wnt/β-catenin signal pathway.29 However, it also has been reported that miR-153 plays a role similar to anti-oncogene in a number of tumors. miR-153 suppresses the progression of non-small-cell lung carcinoma via regulating ADAM19.30 In gastric cancer, miR-153 depresses the oncogenesis and progression of tumor cells by regulating KLF5.31 miR-153 inhibits glioma progression by directly regulating SNAI1 and downregulation of miR-153 is related to poor prognosis in glioma sufferers.32 Therefore, these evidence suggest that the biological behavior of miR-153 seems to have cell and tissue specificity, playing distinct roles in different tumors. With regard to the study of miR-153 in BC, Li et al have reported that miR-153 suppresses EMT of BC via targeting MTDH.8 In addition, miR-153 also plays a critical role in the apoptosis of BC cells.9 Moreover, Wang et al pointed out that miR-153 could regulate the TGF–β signaling pathway to repress the malignant evolution of BC cells.10 Fkih M’hamed et al have found that the expression levels of miR-153 in TNBC and non-TNBC were similar.33 Nevertheless, another study suggested that the expression of miR-153 was different in diverse molecular types. Compared with Luminal A, Luminal B and HER-2 positive breast cancer, the expression of miR-153 was lower in TNBC.8 However, the biological role of miR-153 in TNBC, especially the specific molecular mechanism of miR-153 in the occurrence and development of TNBC remains elusive. In our study, we analyzed the potential function of miR-153 in TNBC through a series of cell experiments. We found that the upregulation of miR-153 repressed the proliferation, invasion and migration of TNBC cells. These findings provide a new perspective for understanding the underlying mechanism of miR-153 in inhibiting the malignant evolution of TNBC.
It is well known that in the occurrence and development of cancer, miRNAs can play a carcinogenic or anticarcinogenic role only in combination with their regulated target genes. Up to now, it has been proved that miR-153 has a large number of target genes, such as PTEN,27 FOXO3a,28 ADAM19,30 KLF531 and MTDH.8 To elucidate the potential mechanism of miR-153 in TNBC, we used biological software to predict the target gene of miR-153. ZEB2 has been proved to be a candidate target gene of miR-153, mainly based on the biological role of ZEB2 in tumor progression.34 The earliest report about ZEB2 was that ZEB2 mRNA was found in the embryo of xenopus.35 Later, it was found that ZEB2 was associated with the regulation of cell differentiation, growth, apoptosis, embryo development, inflammation and other life activities. One of the main functions of ZEB2 is to promote tumorigenesis and development by regulating EMT. Previous studies have confirmed that ZEB2 is related to infiltration and metastasis of multiple types of carcinomas, such as gastric malignancy,36 liver malignant tumor37 and non-small cell lung carcinoma.38 In BC, Ang et al pointed out that the positive expression rates of ZEB2 mRNA and protein in ER-positive and ER-negative BC were different, but there was no significant statistical difference.39 Another study found that the positive expression rate of ZEB2 in TNBC was significantly higher than that in ER-positive BC (Figure S1).40 As a repressor of EMT, ZEB2 is involved in the regulation of various biological functions of BC cells through complex signaling pathways. Si et al reported that there was a feedback inhibition process between ZEB2 and GATA3, two main inhibitors of EMT, and the damage of the feedback loop was an important molecular mechanism for BC progression.41 In addition, He et al reported that ZEB2 could form functional protein complexes with CdGAP in a GAP-independent manner and then regulate E-cadherin expression, thereby promoting the proliferation and metastasis of BC cells.42 Both miR-204/ZEB2 axis43 and miR-29b/TET1/ZEB2 axis44 were involved in EMT and malignant evolution of BC cells. Zou et al showed that FAT10 could bind to ZEB2 directly, reducing its ubiquitination and enhancing its protein stability in BC cells, which contributed to the progress of BC.45 Recently, a study revealed that miR-124 targeted ZEB2-mediated EMT and promoted the invasion of TNBC.15 There may be more ZEB2 involved signaling pathways that promote TNBC progression, which, however, have not yet been revealed. In the present study, we used luciferase activity assay to prove that ZEB2 was a direct target downstream of miR-153. Immunohistochemical method was used to confirm that ZEB2 was negatively regulated by miR-153. RT-qPCR and Western blot further verified this conclusion and confirmed that miR-153 inhibited ZEB2-mediated EMT. Therefore, miR-153 might inhibit the progression of TNBC by regulating the ZEB2/EMT signaling pathway. Furthermore, we also found that the upregulation of ZEB2 partially attenuated the inhibitory effect of miR-153 on TNBC cells. These data revealed that miR-153 inhibited the malignant evolution of TNBC by targeting the ZEB2/EMT axis. In addition, Cox’s proportional hazard regression model of risk factors that might affect the prognosis of patients with TNBC confirmed that the expression of miR-153 was related to DFS and OS. Upregulating the expression of miR-153 is helpful to improve DFS and OS in TNBC patients.
There are also some limitations to our study. Firstly, more clinical tissue samples and cell lines are needed to examine the biological functions of miR-153 in TNBC tissues and cells. Second, TNBC is a kind of disease in which multiple genes and multiple steps are involved. More miRs and their molecular targets, as well as the corresponding signaling pathways and potential molecular mechanisms, need to be further confirmed. In addition, we have only carried out studies at the tissue and molecular levels, lacking further experiments in vivo.
In conclusion, our study confirms that miR-153 may repress the malignant progression of TNBC by regulating the ZEB2/EMT axis, and the overexpression of ZEB2 can partially weaken the inhibitory function mediated by miR-153. These results provide a novel thinking for us to explore the occurrence and development of TNBC. In addition, upregulation of the expression of miR-153 may contribute to better prognosis of TNBC patients. Therefore, the miR-153/ZEB2/EMT axis is expected to serve as a new target pathway for the prevention and treatment of TNBC and miR-153 as an independent prognostic marker of TNBC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492.
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338.
3. Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi:10.1038/35021093.
4. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi:10.1016/j.cell.2009.01.035
5. Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer–a brief overview. Adv Biol Regul. 2015;57:1–9. doi:10.1016/j.jbior.2014.09.013.
6. Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9(11):831–842. doi:10.1038/nrg2455.
7. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002.
8. Li W, Zhai L, Zhao C, Lv S. MiR-153 inhibits epithelial-mesenchymal transition by targeting metadherin in human breast cancer. Breast Cancer Res Treat. 2015;150(3):501–509. doi:10.1007/s10549-015-3346-y.
9. Wu X, Li L, Li Y, Liu Z. MiR-153 promotes breast cancer cell apoptosis by targeting HECTD3. Am J Cancer Res. 2016;6(7):1563–1571.
10. Wang J, Liang S, Duan X. Molecular mechanism of miR-153 inhibiting migration, invasion and epithelial-mesenchymal transition of breast cancer by regulating transforming growth factor beta (TGF-beta) signaling pathway. J Cell Biochem. 2019;120(6):9539–9546. doi:10.1002/jcb.28230.
11. Fkih M’hamed I, Privat M, Ponelle F, Penault-Llorca F, Kenani A, Bignon YJ. Identification of miR-10b, miR-26a, miR-146a and miR-153as potential triple-negative breast cancer biomarkers. Cell Oncol (Dordr). 2015;38(6):433–442. doi:10.1007/s13402-015-0239-3.
12. Nelles L, Van de Putte T, van Grunsven L, Huylebroeck D, Verschueren K. Organization of the mouse Zfhx1b gene encoding the two-handed zinc finger repressor smad-interacting protein-1. Genomics. 2003;82(4):460–469. doi:10.1016/S0888-7543(03)00169-1
13. Verschueren K, Remacle JE, Collart C, et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with smad proteins and binds to 5ʹ-CACCT sequences in candidate target genes. J Biol Chem. 1999;274(29):20489–20498. doi:10.1074/jbc.274.29.20489.
14. Remacle JE, Kraft H, Lerchner W, et al. New mode of DNA binding of multi-zinc finger transcription factors: δEF1 family members bind with two hands to two target sites. EMBO J. 1999;18(18):5073–5084. doi:10.1093/emboj/18.18.5073.
15. Ji H, Sang M, Liu F, Ai N, Geng C. MiR-124 regulates EMT based on ZEB2 target to inhibit invasion and metastasis in triple-negative breast cancer. Pathol Res Pract. 2019;215(4):697–704. doi:10.1016/j.prp.2018.12.039.
16. Chen H, Pan H, Qian Y, Zhou W, Liu X. MiR-25-3p promotes the proliferation of triple negative breast cancer by targeting BTG2. Mol Cancer. 2008;17(1):4. doi:10.1186/s12943-017-0754-0.
17. Bai X, Han G, Liu Y, Jiang H, He Q. MiRNA-20a-5p promotes the growth of triple-negative breast cancer cells through targeting RUNX3. Biomed Pharmacother. 2018;103:1482–1489. doi:10.1016/j.biopha.2018.04.165.
18. Zhang L, Zhang X, Wang X, He M, Qiao S. MicroRNA-224 promotes tumorigenesis through downregulation of caspase-9 in triple-negative breast cancer. Dis Markers. 2019;2019:7378967. doi:10.1155/2019/7378967.
19. Lv ZD, Xin HN, Yang ZC, et al. miR-135b promotes proliferation and metastasis by targeting APC in triple-negative breast cancer. J Cell Physiol. 2019;234(7):10819–10826. doi:10.1002/jcp.27906.
20. Song H, Li D, Wu T, et al. MicroRNA-301b promotes cell proliferation and apoptosis resistance in triple-negative breast cancer by targeting CYLD. BMB Rep. 2018;51(11):602–607. doi:10.5483/BMBRep.2018.51.11.168
21. Wong CK, Gromisch C, Ozturk S, et al. MicroRNA-4417 is a tumor suppressor and prognostic biomarker for triple-negative breast cancer. Cancer Biol Ther. 2019;20(8):1113–1120. doi:10.1080/15384047.2019.1595285.
22. Zhao Z, Li L, Du P, et al. Transcriptional downregulation of mir-4306 serves as a new therapeutic target for triple negative breast cancer. Theranostics. 2019;9(5):1401–1416. doi:10.7150/thno.30701.
23. Wu A, Chen Y, Liu Y, Lai Y, Liu D. MiR-199b-5p inhibits triple negative breast cancer cell proliferation, migration and invasion by targeting DDR1. Oncol Lett. 2018;16(4):4889–4896. doi:10.3892/ol.2018.9255.
24. Schwarzenbacher D, Klec C, Pasculli B. MiR-1287-5p inhibits triple negative breast cancer growth by interaction with phosphoinositide 3-kinase CB, thereby sensitizing cells for PI3Kinase inhibitors. Breast Cancer Res. 2019;21(1):20. doi:10.1186/s13058-019-1104-5.
25. Kong P, Chen L, Yu M, et al. MiR-3178 inhibits cell proliferation and metastasis by targeting Notch1 in triple-negative breast cancer. Cell Death Dis. 2018;9(11):1059. doi:10.1038/s41419-018-1091-y.
26. Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi:10.1186/gb-2004-5-3-r13.
27. Wu Z, He B, He J, Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate. 2013;73(6):596–604. doi:10.1002/pros.22600.
28. Zhang L, Pickard K, Jenei V, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73(21):6435–6447. doi:10.1158/0008-5472.CAN-12-3308.
29. Hua HW, Jiang F, Huang Q, Liao Z, Ding G. MicroRNA-153 promotes Wnt/β-catenin activation in hepatocellular carcinoma through suppression of WWOX. Oncotarget. 2015;6(6):3840–3847. doi:10.18632/oncotarget.2927.
30. Shan N, Shen L, Wang J, He D, Duan C. MiR-153 inhibits migration and invasion of human non-small-cell lung cancer by targeting ADAM19. Biochem Biophys Res Commun. 2015;456(1):385–391. doi:10.1016/j.bbrc.2014.11.093.
31. Ouyang Y, Yuan W, Qiu S. MicroRNA-153 functions as a tumor suppressor in gastric cancer via targeting Kruppel-like factor 5. Exp Ther Med. 2018;16(2):473–482. doi:10.3892/etm.2018.6226.
32. Zhao W, Yin CY, Jiang J, Kong W, Xu H, Zhang H. MicroRNA-153 suppresses cell invasion by targeting SNAI1 and predicts patient prognosis in glioma. Oncol Lett. 2019;17(1):1189–1195. doi:10.3892/ol.2018.9706.
33. Fkih M’hamed I, Privat M, Trimeche M, Penault-Llorca F, Bignon YJ, Kenani A. miR-10b, miR-26a, miR-146a and miR-153 expression in triple negative vs non triple negative breast cancer: potential biomarkers. Pathol Oncol Res. 2017;23(4):815–827. doi:10.1007/s12253-017-0188-4.
34. Zhou J, Xie M, Shi Y, et al. MicroRNA-153 functions as a tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer cells. Oncol Rep. 2015;34(1):111–120. doi:10.3892/or.2015.3952.
35. Murray D, Precht P, Balakir R, Horton WE
36. Wang GJ, Jiao BP, Liu YJ, Li YR, Deng BB. Reactivation of microRNA‐506 inhibits gastric carcinoma cell metastasis through ZEB2. Aging (Albany NY). 2019;11(6):1821–1831. doi:10.18632/aging.101877.
37. Zhang X, Xu X, Ge G, et al. MiR498 inhibits the growth and metastasis of liver cancer by targeting ZEB2. Oncol Rep. 2019;41(3):1638–1648. doi:10.3892/or.2018.6948.
38. Tong X, Su P, Yang H, et al. MicroRNA-598 inhibits the proliferation and invasion of non-small cell lung cancer cells by directly targeting ZEB2. Exp Ther Med. 2018;16(6):5417–5423. doi:10.3892/etm.2018.6825.
39. Ang L, Zheng L, Wang J, et al. Expression of and correlation between BCL6 and ZEB family members in patients with breast cancer. Exp Ther Med. 2017;14(5):3985–3992. doi:10.3892/etm.2017.5101.
40. Karihtala P, Auvinen P, Kauppila S, Haapasaari KM, Jukkola-Vuorinen A, Soini Y. Vimentin, zeb1 and Sip1 are up-regulated in triple-negative and basal-like breast cancers: association with an aggressive tumour phenotype. Breast Cancer Res Treat. 2013;138(1):81–90. doi:10.1007/s10549-013-2442-0.
41. Si W, Huang W, Zheng Y, et al. Dysfunction of the reciprocal feedback loop between GATA3- and ZEB2-nucleated repression programs contributes to breast cancer metastasis. Cancer Cell. 2015;27(6):822–836. doi:10.1016/j.ccell.2015.04.011.
42. He Y, Northey JJ, Pelletier A, et al. The Cdc42/Rac1 regulator CdGAP is a novel E-cadherin transcriptional co-repressor with Zeb2 in breast cancer. Oncogene. 2017;36(24):3490–3503. doi:10.1038/onc.2016.492.
43. Wang Y, Zhou Y, Yang Z, et al. MiR-204/ZEB2 axis functions as key mediator for MALAT1-induced epithelial-mesenchymal transition in breast cancer. Tumour Biol. 2017;39(7):1010428317690998. doi:10.1177/1010428317690998.
44. Wang H, An X, Yu H, et al. MiR-29b/TET1/ZEB2 signaling axis regulates metastatic properties and epithelial-mesenchymal transition in breast cancer cells. Oncotarget. 2017;8(60):102119–102133. doi:10.18632/oncotarget.22183.
45. Zou Y, Ouyang Q, Wei W, Yang S, Zhang Y, Yang W. FAT10 promotes the invasion and migration of breast cancer cell through stabilization of ZEB2. Biochem Biophys Res Commun. 2018;506(3):563–570. doi:10.1016/j.bbrc.2018.10.109.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.