Back to Journals » Breast Cancer: Targets and Therapy » Volume 15
Unraveling the Pivotal Network of Ultrasound and Somatic Mutations in Triple-Negative and Non-Triple-Negative Breast Cancer
Authors Huang Y, Guo Y, Xiao Q , Liang S, Yu Q, Qian L, Zhou J, Le J, Pei Y, Wang L, Chang C, Chen S, Zhou S
Received 17 February 2023
Accepted for publication 1 July 2023
Published 11 July 2023 Volume 2023:15 Pages 461—472
DOI https://doi.org/10.2147/BCTT.S408997
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Harikrishna Nakshatri
Yunxia Huang,1,* Yi Guo,2,* Qin Xiao,3,* Shuyu Liang,2 Qiang Yu,4 Lang Qian,1 Jin Zhou,1 Jian Le,1 Yuchen Pei,5 Lei Wang,4 Cai Chang,1 Sheng Chen,4 Shichong Zhou1
1Department of Ultrasonography, Fudan University Shanghai Cancer Center, Shanghai Medical College, Fudan University, Shanghai, People’s Republic of China; 2Department of Radiology, Fudan University Shanghai Cancer Center, Fudan University, Shanghai, People’s Republic of China; 3Department of Electronic Engineering, Fudan University and Key Laboratory of Medical Imaging Computing and Computer Assisted Intervention of Shanghai, Shanghai, People’s Republic of China; 4Department of Breast Surgery, Fudan University Shanghai Cancer Center, Fudan University, Shanghai, People’s Republic of China; 5Precision Cancer Medical Center Affiliated to Fudan University Shanghai Cancer Center, Fudan University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shichong Zhou, Department of Ultrasonography, Fudan University Shanghai Cancer Center, Shanghai Medical College, Fudan University, Shanghai, 200032, People’s Republic of China, Tel +86 21-64175590, Email [email protected] Sheng Chen, Department of Breast Surgery, Fudan University Shanghai Cancer Center, Fudan University, Shanghai, 200032, People’s Republic of China, Tel +86 21-64175590, Email [email protected]
Purpose: The emergence of genomic targeted therapy has improved the prospects of treatment for breast cancer (BC). However, genetic testing relies on invasive and sophisticated procedures.
Patients and Methods: Here, we performed ultrasound (US) and target sequencing to unravel the possible association between US radiomics features and somatic mutations in TNBC (n=83) and non-TNBC (n=83) patients. Least absolute shrinkage and selection operator (Lasso) were utilized to perform radiomic feature selection. The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was utilized to identify the signaling pathways associated with radiomic features.
Results: Thirteen differently represented radiomic features were identified in TNBC and non-TNBC, including tumor shape, textual, and intensity features. The US radiomic–gene pairs were differently exhibited between TNBC and non-TNBC. Further investigation with KEGG verified radiomic–pathway (ie, JAK-STAT, MAPK, Ras, Wnt, microRNAs in cancer, PI3K-Akt) associations in TNBC and non-TNBC.
Conclusion: The pivotal network provided the connections of US radiogenomic signature and target sequencing for non-invasive genetic assessment of precise BC treatment.
Keywords: triple-negative breast cancer, radiomics, somatic mutation, signaling pathway
Introduction
Triple-negative breast cancer (TNBC) is an aggressive subtype of BC that does not express the estrogen receptor (ER), progesterone receptor (PR), or human epidermal growth factor receptor (HER2) amplification/overexpression, accounting for 10–20% of newly diagnosed BC.1 Studies have shown that the 5-year survival rate of patients with TNBC is only 77%, compared with 93% for other types.2 There is consensus that TNBC represents highly inter-and intra-tumor heterogeneity, and this may have implications for TNBC treatment choice. Advancement in genomics has fueled the efforts toward “precision oncology”, targeting cancers based on their genetic mutations, including TP53, BRCA, and PIK3CA.3–7 Genetic mutations are optimized to discover molecular biomarkers predicting prognosis and response to treatments, helping to foresee the emergence of unexplained drug resistance.8 Nevertheless, the comprehensive application of genomic profiles in clinical testing is limited by the low likelihood of identifying high responders using large panels of genes,9 and obtaining serial samples of tumor tissue is impractical and complicated by spatial heterogeneity and sampling bias. Therefore, there is an urgent need for a new approach to identify genetic information, which will facilitate the timely inclusion of patients with prognostic and predictive biomarkers into personalized treatment strategies.
Radiomics is an attractive approach mining high-throughput quantitative image features associated with different variables of interest to create decision support models.10–13 Ultrasound (US) imaging is commonly applied in BC detection and diagnosis because it can assess the morphology, orientation, internal structure, and margins of the entire tumor.14 US radiomic features and algorithms have been recently explored for their role in differentiating TNBC.15–17 Moreover, a change in the radiogenomics landscape, delta-radiomics,18 during follow-up may provide a non-invasive predictive marker for such patients because numerous biopsies and repeated genomic testing are often impossible. However, the disconnection between the predictor model and biological meaning has limited the widespread clinical translation of such tools. Although efforts to reintroduce biological meaning into radiomics have attracted much attention in the field of genomic correlates, most studies have focused on a few genes or have relied on observer-dependent semiquantitative features that make replication difficult.10,15 Only a few studies have investigated the associations between tumor imaging phenotype and the underlying molecular landscape.19,20
Therefore, by performing a pathway analysis in TNBC and non-TNBC, we aim to discover the associations between US phenotypes of breast cancer and their underlying molecular biology derived from gene mutation data.
Methods
Study Design and Participants
The study participants design was illustrated in Figure S1. A total of 443 suspected BC patients were recruited between January 1, 2020 and June 30, 2021. In this prospective study, we excluded participants whose tumors were not present or not complete in the subtracted image on US (N=21) and who underwent biopsy before US examination (n = 35). Then, we excluded patients with benign lesions (n = 37). Eighty-three TNBC and 267 non-TNBC BC patients were included. To resolve the class imbalance problem, 184 non-TNBC patients were randomly excluded. Finally, 83 TNBC and 83 non-TNBC BC patients were included. This study was approved by the institutional review board of Fudan University Shanghai Cancer Center. Informed consent has been signed by each participant and the study was conducted in accordance with the Declaration of Helsinki.
US Radiomic Features Extraction
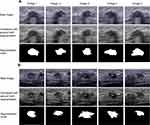
Aixplorer US system (Aixplorer, Supersonic Imagine, Aix-en-Provence, France) equipped with a 4–15 MHz linear array transducer was used to conduct US examination. Standardized breast US examination was performed by a certified radiologist (J Zhou.) with 8 years of experience, as previously described.21 The regions of interest (ROI) of US images were delineated using a trained U-NET network22 (Figure 1). Segmentation revision was performed by an experienced radiologist (L Qian, with 5 years of diagnosis experience in breast medical images) where necessary (where contour of lesion was not precisely drawn). An open-source package PyRadiomics (version 3.0) in Python was used to determine radiomics features on the US image for each ROI.23 A total of 555 radiomic features were included in the analysis. The radiomic features were divided into five groups: (I) First-order statistics (eg, mean, standard deviation, skewness); (II) Shape and size-based features (eg, sphericity, maximal diameter, elongation); (III) Textual features (eg, entropy, gray-level variances: ngtdm [neighborhood grey-tone difference matrix], glszm [size-zone matrix], glcm [co-occurrence matrix], glrlm [run-length matrix], gldm [grey-level difference method]); (IV) Wavelet-based features (eg, Wavelet energy, Wavelet median); (V) Laplace of Gaussian (LoG) features (eg, LoG Kurtosis, LoG Gray-level non uniformity). First-order statistics are based on the histogram of voxel intensity values of the image. Shape and size-based features are based on the 2D representation of a tumor. Textual features measure textural structures. Wavelet-based features are calculated by first applying Wavelet decomposition to the image before computing groups I and III features. Laplace of Gaussian (LoG) features are calculated by applying a LoG filter to the image first, which results in highlighted edges, after which I and III features are computed.
 |
Figure 1 Radiomics pipeline. |
Gene Mutation and Pathway Analysis
Both the tumor samples and matched blood DNA were sequenced using a custom-designed genetic panel of 511 breast cancer-specific genes to detect somatic mutation, as previously reported.24 The panel was designed based on the integration of The Cancer Genome Atlas (TCGA),25 Memorial Sloan Kettering Cancer Center (MSKCC)26 and FUSCC-TNBC datasets.27 DNA fragments captured by probes were pooled and sequenced, 1000× median coverage for tumor genomic DNA and 400× median coverage for bold genomic DNA, thereby eliminating germline variant interference and ensuring somatic mutations. A mutation was included only if it was shift deletion and insertion, in-frame deletion and insertion, missense, nonsense, and splice site modifications.
Pearson correlation was used to examine whether a radiomic feature had a significant association with mutant genes for each genotype. This process was repeated for each deep learning feature and all the 511 genes. Differential mutation of each gene was evaluated with mutational information using Fisher’s exact test (p < 0.05 as statistically significant). Least absolute shrinkage and selection operator (Lasso) regression was used to develop radiomic feature selection for discriminating TNBC and non-TNBC. First, N features significantly different between TNBC and non-TNBC were selected as the independent variable using t-test. A pre-selected feature set was formed by removing the features with high absolute value correlation (>c) with other features. Lasso regression was then used to identify the radiomic features significantly different between TNBC and non-TNBC. The biologic processes associated with US radiomic factor were identified using KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analyses of gene mutation data. Specifically, a single US radiomic feature could determine an association with a gene set. Only the pathway with more than two gene hits and p < 0.05 (Benjamini–Hochberg Procedure) was classified as mutated pathway related to the US radiomic feature for a given US factor-gene set pair.
Statistical Analysis
Python version 3.7 and R version 4.2.0 was used for all statistical analyses. The radiomic signature was established using an elastic net regression model via Lasso. Features with high similarity were removed to reduce multicollinearity of the features in the model. The regularization parameters α were turned using a grid search under 10-fold cross-validation to reduce model overfitting. The correlation between mutated genes and radiomic features was identified via Spearman correlation. All statistical results were considered significant at p<0.05.
Results
The Characteristics of Radiomic Features in TNBC and Non-TNBC
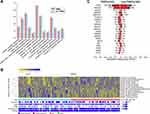
The overall study design and flow chart of patient inclusion is shown in Figures 1 and S1. The clinicopathological analysis showed that there were no significant differences between the TNBC and non-TNBC groups in age (p = 0.526), menopausal status (p = 0.534), T stage (p = 0.895), N stage (p = 0.102), and histologic category (p = 0.663), but there was significant difference in pathology grade (p = 0.017). An integrated radiomic-genomic analysis of TNBC and non-TNBC was conducted to evaluate the relationship between radiomic phenotypes and molecular pathways (Table S1). Thirteen robust and non-redundant radiomic features (Figure 2A) were identified to quantify a panel of phenotypic characteristics, such as intratumor homogeneity and heterogeneity. Unsupervised clustering revealed clusters of radiomic expression patterns between TNBC and non-TNBC (Figure 2B). The first-order (minimum- and median-) and entropy (join-, run-, and dependence-) radiomics features were overrepresented in TNBC compared with those in non-TNBC. First-order statistics describe the distribution of voxel intensities within the image region defined by the mask through commonly used and basic metrics. Joint entropy measures the randomness or variability in the neighbourhood intensity values. Run entropy measures the uncertainty or randomness in the distribution of run lengths and gray levels. A higher entropy value indicates that the texture patterns are more heterogeneous.
Shape (perimeter), first-order (energy), inverse variance, maximum probability, busyness, small area emphasis (SAE), and gray-level non-uniformity normalized (GLNN) were overrepresented in non-TNBC compared with those in TNBC. Shape features describe tumor aggressiveness. Tumors are described as spiculated or having “ill-defined borders”, indicating their potential to spread to contiguous structures and association with advanced stages.16,28–30 Energy measures the magnitude of voxel values in an image. SAE measures the distribution of small size zones. A greater SAE value indicates more smaller size zones and more fine textures. GLNN measures the similarity of gray-level intensity values in the image. A lower GLNN value indicates a greater similarity in intensity values while a larger value indicates a greater sum of the squares of these values. These textural features are sensitive to tumor radiographic heterogeneity. Inverse variance measures the variations in the intensity of voxels close to each other and thus quantifies another aspect of homogeneity. Maximum probability is defined as the occurrence of the most predominant pair of neighbouring intensity values. Busyness measures the change from a pixel to its neighbour. A high busyness value indicates a “busy” image, with rapid changes of intensity between pixels and its neighborhood. Busyness and inverse variance emphasize voxel pattern from close range intensity. These textural features are related to radiographic homogeneity.
Association with Somatic Mutation
A list of the top highly somatic mutated genes is presented in Figure 2C. The US radiomic–gene pair were differently exhibited between TNBC and non-TNBC (Figure 3). A total of 27 genes were significantly associated with the 12 radiomic features in the TNBC group and 29 genes were significantly associated with 13 radiomic features in the non-TNBC group, but only one radiomic–gene pair (Wavelet-HL ngtdm busyness and BRCA1) and five genes (MAP3K1, RYR2, TYK2, BRCA1, JAK1) were shared in both TNBC and non-TNBC (Figure 3A and B). The complete lists of all the associations between somatic mutation and US radiomics in TNBC and non-TNBC are presented in Tables S2 and S3 (p value <0.05).
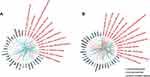 |
Figure 3 The association between mutated genes and radiomic features ((A), TNBC; (B), non-TNBC). |
US radiomics differently presented among the gene-mutated TNBC and non-TNBC (Table 1). Original glcm Joint entropy, original shape2D perimeter, and Wavelet-LH first-order energy were underrepresented in BRCA1-mutated TNBC while Wavelet-HL ngtdm busyness, Wavelet-HL glcm inverse variance, and Wavelet-HL glcm maximum were underrepresented in BRCA1-mutated non-TNBC. Moreover, Wavelet-HL gldm dependence entropy and Wavelet-HH glrlm run entropy were overrepresented in both BRCA1-mutated TNBC and non-TNBC (Figure 3A and B). The representation of these features indicates that BRCA1-mutated TNBC is more likely to be homogeneous and smaller, while BRCA1-mutated non-TNBC is more likely to be heterogeneous. FOXA1 and ATM were differently mutated in TNBC and non-TNBC groups. FOXA1-mutated non-TNBC was associated with overrepresentation of original glcm joint entropy, while ATM-mutated TNBC was associated with underrepresentation of Wavelet-L first-order minimum. Moreover, TP53-mutated non-TNBC was associated with overrepresentation of Wavelet-HL ngtdm busyness, original shape2D perimeter, Wavelet-LH first-order energy, and underrepresentation of Wavelet-LH first-order minimum. These results indicate that TP53-mutated non-TNBC is more likely to be heterogeneous.
 |
Table 1 Radiomic-Genomic Association Within TNBC and Non-TNBC |
Association with KEGG Pathway Analysis
The associations of somatic gene mutations at the pathway level were examined through KEGG analysis (Figure 4). A total of 55 and 106 radiomic pathway associations were identified in TNBC and non-TNBC groups, respectively (Tables S4 and S5). The distinct radiomic features were associated with distinct biological processes in TNBC and non-TNBC groups. For example, Wavelet-based intensity features were mainly associated with category of human diseases pathway in both TNBC and non-TNBC. Textural features were mainly associated with category of human diseases pathway in TNBC, while Wavelet-based textural features were mainly associated with category of human diseases and environmental information processing pathway in non-TNBC. Shape features were mainly associated with category of organismal systems pathway in non-TNBC.
Further examples of radiomic–pathway in BC are illustrated in Figure 5. Eleven signaling pathways were associated with 5 radiomic features in TNBC, and 13 signaling pathways were associated with 9 radiomic features in non-TNBC, respectively. Radiomic features were associated with 8 signal pathways (EGFR tyrosine kinase inhibitor resistance, JAK-STAT signaling pathway, MAPK signaling pathway, microRNAs in cancer, necroptosis, PI3K-Akt signaling pathway, NOD-like receptor signaling pathway, Th17 cell differentiation) in both TNBC and non-TNBC groups. However, radiomic features were associated with three signal pathways (HIF-1 signaling pathway, chemokine signaling pathway, and Toll-like receptor signaling pathway) only in TNBC. Besides, radiomic features were associated with five signal pathways (cell cycle, choline metabolism in cancer, PD-L1 expression and PD-1 checkpoint pathway in cancer, Ras signaling pathway, and Wnt signaling pathway) only in non-TNBC. Notably, radiomic features were differently correlated to the pathways in TNBC and non-TNBC groups, with only three features (3/11) shared between the TNBC and non-TNBC groups (original shape2D perimeter, Wavelet HL ngtdm busyness, and original glcm joint entropy). The Wavelet-based intensity and textural features were correlated with non-TNBC. These features included Wavelet HH glrlm run entropy, Wavelet HL gldm dependence entropy, Wavelet HH glszm SAE, Wavelet LH first-order energy, which are sensitive to complex patterns of high variation, as well as Wavelet HL glcm inverse variance, Wavelet HL glcm maximum probability, which emphasize voxel pattern from close range intensity. The textural and Wavelet-based intensity features were also correlated with TNBC. These features included original glcm joint entropy, original glrlm GLNN, which are sensitive to complex patterns of high variation, as well as Wavelet LH first-order minimum, Wavelet LL first-order minimum, which emphasize voxel pattern from close range intensity.
 |
Figure 5 The association between key signaling pathways of breast cancer and radiomic features in TNBC and non-TNBC. |
Discussion
We evaluated the associations between breast cancer US imaging and genomic sequencing. Our results found 13 US radiomic features differently underrepresented between TNBC and non-TNBC. Furthermore, these radiomic features were associated with a large number of gene mutation and signaling pathways, with different patterns in TNBC and non-TNBC. TNBC exhibited textural and Wavelet-based intensity features associated with human diseases, while non-TNBC exhibited Wavelet-based intensity and Wavelet-based textural features associated with human diseases.
Radiologic imaging has presented characteristic features in TNBC.31,32 TNBC frequently presented with a mass (62–100%)33–35 and less frequently associated with focal asymmetric density (9–11%) and microcalcification (6-%12%)33,35 on mammography. The distinctive US features of TNBC included a well-circumscribed margin in 21–27% of the lesions with posterior acoustic enhancement (24–41%)36 and absence of echogenic halo (85%).36 MRI exhibited a higher presence of rim enhancement (76–88%) in TNBC.37,38 However, these studies are based on semantic features leading to subjective discrepancies. Advanced handcrafted radiomic features, the mathematical result of image pixel values within ROI, can quantitatively and automatically identify and interpret cancer imaging. Wu et al reported that the margin of TNBC is sharper, with a more regular shape than non-TNBC by using nine quantitative US features.16 In this study, we extracted US radiomic features based on automatic segmentation using u-net convolutional neural network (CNN) (Figure 6), of which a number of radiomic features were differently represented between TNBC and non-TNBC. For TNBC, the first-order (minimum- and median-) and entropy (join-, run-, and dependence-) radiomics features were overrepresented in this study, indicated that TNBC are more likely to be heterogeneous. The imaging phenotypes like heterogeneity may capture and convey pathologic characteristics like lymphocyte infiltration,39 and molecular pathways such as tumor metabolism.40 It is feasible that entropy may be a radiomic signature of the biologic activity in TNBC.
US radiomic features can reflect the pathophysiologic processes of cell proliferation, apoptosis, and metastasis in breast cancer.41,42 Previous radiogenomics studies have demonstrated that specific imaging phenotypes are associated with specific genomic mutations.10,19 For example, imaging features are associated with TP53/PIK3CA mutation based on MRI43 and US21 in breast cancer but with limited imaging features. Our study found the radiogenomic features were distinct between TNBC and non-TNBC, with few shared radiomic–gene and radiomic–pathway pairs. The underrepresentation of original glcm joint entropy, original shape2D perimeter, and Wavelet-LH first-order energy in TNBC indicated BRCA1 mutation. Similarly, previous studies showed that BRCA-associated BC exhibits benign morphologic features, with circumscribed margin and rim enhancement in MRI and acoustic enhancement in US.44 Cancers with BRCA mutation are associated with a deficiency in homologous recombination repair of DNA double-strand breaks, resulting genomic instability cytogenetic changes45,46 and poor prognosis.47,48 Recent trials have proved that olaparib, a targeted drug for BRAC1 mutation in breast cancer, increases objective response rate of patients with somatic BRCA1/2 mutation.49 ATM, a prognostic and predictive indicator for TNBC,50 is associated with the underrepresentation of Wavelet-LL first-order minimum. FGFR3 mutation, a therapeutic option for TNBC,51 is associated with the original shape2D perimeter. These findings suggest that homogenous radiomic features in TNBC may be associated with gene mutation, and thus could be a predictor of molecular targets in TNBC precision oncology.
The association between radiomic phenotypes and molecular pathways in TNBC and non-TNBC showed different landscapes. Recent studies have shown that targeting both vertical and horizontal pathways is a promising strategy for BC treatment. For example, the PI3K-Akt signaling pathway is a promising therapeutic target for TNBC.52 Therefore, a tailored anti-PI3K-Akt therapeutic cocktail can be suggested for a patient with high probability of a PI3K-Akt signaling pathway. Thus, evaluating the parallel signaling may guide the development of combination therapies. Previous study has shown that TNBC patients with residual disease after NAC harbored alterations of cell cycle progression, PI3K/Akt/mTOR, and EGFR tyrosine kinase inhibitor-resistance pathways.53 Compared to the associations at the genomic level, we found more statistically significant associations for somatic mutations and signaling pathway. Our findings exhibited the pivotal network of TNBC and non-TNBC, which may provide a non-invasive method to evaluate the efficacy of treatment.
Limitations of the Study
Several limitations still need to be addressed in our study. First, the relatively small number of patients from a single institution limited the statistical power of the results. Therefore, our results must be validated in larger prospective studies with multicenter research. Second, downstream biomarkers were not studied at the mRNA and protein levels. However, the radiogenomic investigations of breast cancer on the somatic mutation level may provide the basis for examining in-depth US image-to-molecule and gene feature associations. Furthermore, animal models should be used to validate the intrinsic biological meaning of radiomic features.
Conclusion
In conclusion, ultrasound radiomic features can help to characterize TNBC and non-TNBC noninvasively and are associated with subtype-related gene mutation and signaling pathway, which offer potential guidance for targeted therapy.
Acknowledgments
We thank Zhenping Zhang for the continuous support for this study.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82071945, 81830058, 91959207) and Joint Fund Project by Shanghai Anticancer Association Fudan University Shanghai Cancer Center (YJQN202109).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Shen M, Pan H, Chen Y, Xu YH, Yang W, Wu Z. A review of current progress in triple-negative breast cancer therapy. Open Med. 2020;15(1):1143–1149. doi:10.1515/med-2020-0138
2. Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109(9):1721–1728. doi:10.1002/cncr.22618
3. Ghosh M, Saha S, Bettke J, et al. Mutant p53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell. 2021;39(4):494–508 e495. doi:10.1016/j.ccell.2021.01.003
4. Wellenstein MD, de Visser KE. Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity. 2018;48(3):399–416. doi:10.1016/j.immuni.2018.03.004
5. Hurvitz SA, Goncalves A, Rugo HS, et al. Talazoparib in patients with a germline BRCA-mutated advanced breast cancer: detailed safety analyses from the Phase III EMBRACA trial. Oncologist. 2020;25(3):e439–e450. doi:10.1634/theoncologist.2019-0493
6. Eikesdal HP, Yndestad S, Elzawahry A, et al. Olaparib monotherapy as primary treatment in unselected triple negative breast cancer. Ann Oncol. 2021;32(2):240–249. doi:10.1016/j.annonc.2020.11.009
7. Hu H, Zhu J, Zhong Y, et al. PIK3CA mutation confers resistance to chemotherapy in triple-negative breast cancer by inhibiting apoptosis and activating the PI3K/AKT/mTOR signaling pathway. Ann Transl Med. 2021;9(5):410. doi:10.21037/atm-21-698
8. Jiang YZ, Liu Y, Xiao Y, et al. Molecular subtyping and genomic profiling expand precision medicine in refractory metastatic triple-negative breast cancer: the FUTURE trial. Cell Res. 2021;31(2):178–186. doi:10.1038/s41422-020-0375-9
9. Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31(11):1491–1505. doi:10.1016/j.annonc.2020.07.014
10. Park AY, Han MR, Park KH, et al. Radiogenomic analysis of breast cancer by using B-mode and vascular US and RNA sequencing. Radiology. 2020;295(1):24–34. doi:10.1148/radiol.2020191368
11. Guo Y, Hu Y, Qiao M, et al. Radiomics analysis on ultrasound for prediction of biologic behavior in breast invasive ductal carcinoma. Clin Breast Cancer. 2018;18(3):e335–e344. doi:10.1016/j.clbc.2017.08.002
12. Xiong L, Chen H, Tang X, et al. Ultrasound-based radiomics analysis for predicting disease-free survival of invasive breast cancer. Front Oncol. 2021;11:621993. doi:10.3389/fonc.2021.621993
13. Jiang M, Li CL, Luo XM, et al. Ultrasound-based deep learning radiomics in the assessment of pathological complete response to neoadjuvant chemotherapy in locally advanced breast cancer. Eur J Cancer. 2021;147:95–105. doi:10.1016/j.ejca.2021.01.028
14. Guo R, Lu G, Qin B, Fei B. Ultrasound imaging technologies for breast cancer detection and management: a review. Ultrasound Med Biol. 2018;44(1):37–70. doi:10.1016/j.ultrasmedbio.2017.09.012
15. Chen Q, Xia J, Zhang J. Identify the triple-negative and non-triple-negative breast cancer by using texture features of medical ultrasonic image: a STROBE-compliant study. Medicine. 2021;100(22):e25878. doi:10.1097/MD.0000000000025878
16. Wu T, Sultan LR, Tian J, Cary TW, Sehgal CM. Machine learning for diagnostic ultrasound of triple-negative breast cancer. Breast Cancer Res Treat. 2019;173(2):365–373. doi:10.1007/s10549-018-4984-7
17. Ko ES, Lee BH, Kim HA, Noh WC, Kim MS, Lee SA. Triple-negative breast cancer: correlation between imaging and pathological findings. Eur Radiol. 2010;20(5):1111–1117. doi:10.1007/s00330-009-1656-3
18. Nardone V, Reginelli A, Grassi R, et al. Delta radiomics: a systematic review. Radiol Med. 2021;126(12):1571–1583. doi:10.1007/s11547-021-01436-7
19. Zhu Y, Li H, Guo W, et al. Deciphering genomic underpinnings of quantitative MRI-based radiomic phenotypes of invasive breast carcinoma. Sci Rep. 2015;5:17787. doi:10.1038/srep17787
20. Rios Velazquez E, Parmar C, Liu Y, et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 2017;77(14):3922–3930. doi:10.1158/0008-5472.CAN-17-0122
21. Huang Y, Qiang Y, Jian L, et al. Ultrasonic features and molecular subtype predict somatic mutations in TP53 and PIK3CA genes in breast cancer. Acad Radiol. 2022;29(12):e261–e270. doi:10.1016/j.acra.2022.02.021
22. Ronneberger OFP, Brox T. U-net: convolutional networks for biomedical image segmentation.
23. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi:10.1158/0008-5472.CAN-17-0339
24. Lang GT, Jiang YZ, Shi JX, et al. Characterization of the genomic landscape and actionable mutations in Chinese breast cancers by clinical sequencing. Nat Commun. 2020;11(1):5679. doi:10.1038/s41467-020-19342-3
25. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi:10.1038/nature11412
26. Razavi P, Chang MT, Xu G, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34(3):427–438 e426. doi:10.1016/j.ccell.2018.08.008
27. Jiang YZ, Ma D, Suo C, et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell. 2019;35(3):428–440 e425. doi:10.1016/j.ccell.2019.02.001
28. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi:10.1245/s10434-010-0985-4
29. Zhang Y, Liu F, Zhang H, et al. Diagnostic value of radiomics analysis in contrast-enhanced spectral mammography for identifying triple-negative breast cancer. Front Oncol. 2021;11:773196. doi:10.3389/fonc.2021.773196
30. Ye H, Hang J, Zhang M, et al. Automatic identification of triple negative breast cancer in ultrasonography using a deep convolutional neural network. Sci Rep. 2021;11(1):20474. doi:10.1038/s41598-021-00018-x
31. Zhang HX, Sun ZQ, Cheng YG, Mao GQ. A pilot study of radiomics technology based on X-ray mammography in patients with triple-negative breast cancer. J Xray Sci Technol. 2019;27(3):485–492. doi:10.3233/XST-180488
32. Youk JH, Son EJ, Chung J, Kim JA, Kim EK. Triple-negative invasive breast cancer on dynamic contrast-enhanced and diffusion-weighted MR imaging: comparison with other breast cancer subtypes. Eur Radiol. 2012;22(8):1724–1734. doi:10.1007/s00330-012-2425-2
33. Kojima Y, Tsunoda H. Mammography and ultrasound features of triple-negative breast cancer. Breast Cancer. 2011;18(3):146–151. doi:10.1007/s12282-010-0223-8
34. Yang WT, Dryden M, Broglio K, et al. Mammographic features of triple receptor-negative primary breast cancers in young premenopausal women. Breast Cancer Res Treat. 2008;111(3):405–410. doi:10.1007/s10549-007-9810-6
35. Gao B, Zhang H, Zhang SD, et al. Mammographic and clinicopathological features of triple-negative breast cancer. Br J Radiol. 2014;87(1039):20130496. doi:10.1259/bjr.20130496
36. Zheng FY, Lu Q, Huang BJ, et al. Imaging features of automated breast volume scanner: correlation with molecular subtypes of breast cancer. Eur J Radiol. 2017;86:267–275. doi:10.1016/j.ejrad.2016.11.032
37. Matsuda M, Tsuda T, Ebihara R, et al. Triple-negative breast cancer on contrast-enhanced MRI and synthetic MRI: a comparison with non-triple-negative breast carcinoma. Eur J Radiol. 2021;142:109838. doi:10.1016/j.ejrad.2021.109838
38. Dogan BE, Gonzalez-Angulo AM, Gilcrease M, Dryden MJ, Yang WT. Multimodality imaging of triple receptor-negative tumors with mammography, ultrasound, and MRI. AJR Am J Roentgenol. 2010;194(4):1160–1166. doi:10.2214/AJR.09.2355
39. Ko ES, Kim JH, Lim Y, Han BK, Cho EY, Nam SJ. Assessment of invasive breast cancer heterogeneity using whole-tumor magnetic resonance imaging texture analysis: correlations with detailed pathological findings. Medicine. 2016;95(3):e2453. doi:10.1097/MD.0000000000002453
40. Ganeshan B, Skogen K, Pressney I, Coutroubis D, Miles K. Tumour heterogeneity in oesophageal cancer assessed by CT texture analysis: preliminary evidence of an association with tumour metabolism, stage, and survival. Clin Radiol. 2012;67(2):157–164. doi:10.1016/j.crad.2011.08.012
41. Zhang H, Sui X, Zhou S, Hu L, Huang X. Correlation of conventional ultrasound characteristics of breast tumors with axillary lymph node metastasis and Ki-67 expression in patients with breast cancer. J Ultrasound Med. 2019;38(7):1833–1840. doi:10.1002/jum.14879
42. Khairalseed M, Javed K, Jashkaran G, Kim JW, Parker KJ, Hoyt K. Monitoring early breast cancer response to neoadjuvant therapy using H-scan ultrasound imaging: preliminary preclinical results. J Ultrasound Med. 2019;38(5):1259–1268. doi:10.1002/jum.14806
43. Moon WK, Chen HH, Shin SU, Han W, Chang RF. Evaluation of TP53/PIK3CA mutations using texture and morphology analysis on breast MRI. Magn Reson Imaging. 2019;63:60–69. doi:10.1016/j.mri.2019.08.026
44. Ha SM, Chae EY, Cha JH, Kim HH, Shin HJ, Choi WJ. Association of BRCA mutation types, imaging features, and pathologic findings in patients with breast cancer with BRCA1 and BRCA2 mutations. AJR Am J Roentgenol. 2017;209(4):920–928. doi:10.2214/AJR.16.16957
45. Olivier M, Taniere P. Somatic mutations in cancer prognosis and prediction: lessons from TP53 and EGFR genes. Curr Opin Oncol. 2011;23(1):88–92. doi:10.1097/CCO.0b013e3283412dfa
46. Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7(4):a016600. doi:10.1101/cshperspect.a016600
47. Pop LA, Cojocneanu-Petric RM, Pileczki V, et al. Genetic alterations in sporadic triple negative breast cancer. Breast. 2018;38:30–38. doi:10.1016/j.breast.2017.11.006
48. Sporikova Z, Koudelakova V, Trojanec R, Hajduch M. Genetic markers in triple-negative breast cancer. Clin Breast Cancer. 2018;18(5):e841–e850. doi:10.1016/j.clbc.2018.07.023
49. Tung NM, Robson ME, Ventz S, et al. TBCRC 048: Phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J Clin Oncol. 2020;38(36):4274–4282. doi:10.1200/JCO.20.02151
50. Geenen JJJ, Linn SC, Beijnen JH, Schellens JHM. PARP inhibitors in the treatment of triple-negative breast cancer. Clin Pharmacokinet. 2018;57(4):427–437. doi:10.1007/s40262-017-0587-4
51. Chew NJ, Nguyen EV, Su SP, et al. FGFR3 signaling and function in triple negative breast cancer. Cell Commun Signal. 2020;18(1):13. doi:10.1186/s12964-019-0486-4
52. Pascual J, Turner NC. Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann Oncol. 2019;30(7):1051–1060. doi:10.1093/annonc/mdz133
53. Di Cosimo S, Appierto V, Silvestri M, et al. Targeted-gene sequencing to catch triple negative breast cancer heterogeneity before and after neoadjuvant chemotherapy. Cancers. 2019;11(11):1753. doi:10.3390/cancers11111753
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.