Back to Journals » Infection and Drug Resistance » Volume 15
Two Phenotypes of Klebsiella pneumoniae ST147 Outbreak from Neonatal Sepsis with a Slight Increase in Virulence
Authors Zou H, Shen Y, Li C, Li Q
Received 8 October 2021
Accepted for publication 9 December 2021
Published 4 January 2022 Volume 2022:15 Pages 1—12
DOI https://doi.org/10.2147/IDR.S343292
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Hua Zou,* Yan Shen,* Chunli Li, Qiuhong Li
Department of Laboratory Medicine, Chongqing Health Center for Women and Children, Chongqing, 400016, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chunli Li; Qiuhong Li Tel +8615823827032; +8613212377189
Email [email protected]; [email protected]
Purpose: Severe infection has been the leading causes of neonatal death, especially the emergency of multidrug-resistant bacteria such as carbapenem-resistant Enterobacteriaceae. This study aimed to investigate the outbreak of carbapenem-resistant Klebsiella pneumoniae (CR-KP) in neonatal wards and to explore the possible pathogenesis of Klebsiella pneumoniae.
Materials and Methods: CR-KP were collected from neonatal ward of Chongqing Health Center for Women and Children between 2017 and 2019. Broth microdilution method was used to evaluate the antimicrobial activities in vitro, at the same time, the virulence of the strain was evaluated by in vitro and in vivo experiments. At last, prokaryotic chain specific transcriptome sequencing was conducted to explore the possible pathogenesis of CR-KP.
Results: In this study, a total of 14 carbapenem-resistant-Klebsiella pneumoniae (CR-KP) strains were isolated from Chongqing Health Center for Women and Children, among which all CR-KP isolates were identified as NDM-1-producers. Molecular epidemiological studies revealed ST147 being the most common sequence type (ST). Moreover, we first found two phenotypes of K. pneumoniae with different virulence from the same specimen. Type I, which was a white and sticky colony had a slight increase in virulence with higher biofilm formation, serum resistance and virulence than Type II with gray colony. Compared with the Type II, 10 pathways were obviously changed in Type I especially amino acid metabolism, such as arginine and proline metabolism.
Conclusion: Our findings revealed a new potential threat of NDM-1-positive CR-KP with higher virulence in neonatal ICU ward. We found two phenotypes of K. pneumoniae with different virulent, which may be due to the difference expression of arginine and proline metabolism.
Keywords: neonates, carbapenem-resistant-Klebsiella pneumoniae, NDM-1, virulence
Introduction
Preterm birth, intrapartum-related complications (birth asphyxia) and neonatal sepsis/meningitis/pneumonia were the leading cause of neonatal death.1 A multi-country prospective cohort study in Asia found that about 40% of newborns die from severe neonatal infections.2 Appropriate empiric treatment of the infection is one of the effective means to reduce neonatal mortality. Nevertheless, as a multidrug-resistant pathogens continue to emerge, the treatment becomes increasingly complicated. A recent meta-analysis of Chinese literature (2009–2014) on neonatal sepsis revealed that more than 50% of E. coli and Klebsiella spp. were resistant to third-generation cephalosporin, and the resistance of ampicillin in E. coli was almost 80%.3 Carbapenems had become the last line of defense for neonatal treatment. However, severe neonatal infections caused by carbapenem-resistant bacteria have been reported from around the world since 2011. Surprisingly, studies have shown that the rates of Carbapenem-resistant Enterobacteriaceae (CRE) colonization in neonatal ICU units are not low from 5%~10%,4,5 and the outbreak of CRE in neonatal wards had also been reported.
Carbapenem-resistant -K. pneumoniae (CR-KP) has been reported from around the world causing outbreaks of neonatal infections.6 Ahmad7 and Bro8 had found the clonal outbreak of CR-KP, which emphasis need of proper surveillance and adequate infection control to limiting the spread of these organisms. The resistance mechanism in CR-KP is largely mediated by highly transmissible plasmid-encoded carbapenemase enzymes, including KPC, NDM, and OXA-48-like enzymes.6
However, a growing number of studies have found that some CR-KP possesses more invasive and highly pathogenic hypervirulent (hvKP) pathotype in the clinical context poses an additional challenge to the clinicians. Amir Mohammad firstly reported an outbreak of hypervirulent Klebsiella pneumoniae harbouring blaVIM-2 among mechanically ventilated drug-poisoning patients with high mortality rate in Iran in 2012.9 Then, Yen-Hua Huang had described a hybrid virulence plasmid, pVir (297,984 bp) resistant to carbapenem, tigecycline, and colistin from an ST11 strain in Taiwan. While, possessed pVir, TVGHCRE225 was less virulent than K1 and K2 strains, but was nevertheless more virulent than a carbapenem-resistant ST11.10 Yang Lu had also recovered a colistin-resistant strain carrying a 236 355bp hybrid virulence plasmid harbored the rmpA, rmpA2Δ, iucABCDiutA, iroBCDN, andyersiniabactin-encoding genes.11 This has occurred by two mechanisms. One was via hvKp strains gaining antimicrobial resistance genes, the other was CR-KP acquired a modified hvKp virulence plasmid. However, to date, information regarding the virulence of CR-KP in neonates remains elusive. Better understanding of these data may enable effective treatment of patients at high risk and potentially reduce the mortality rate of neonatal infection.
Materials and Methods
Bacterial Strains
This retrospective study was performed in Chongqing Health Center for Women and Children in Southwest China.14 non-repetitive nosocomial CR-KP strains were collected from September, 2017 to August, 2018 from 14 different patients. All the isolates were identified at the species level by the VITEK MS (bioMerieux, Hazelwood, MO, United States) automated system, and routine antimicrobial susceptibility testing was performed by using the BD Phoenix™ system. According to the breakpoint recommendations by Clinical and Laboratory Standards Institute 2020, isolates which were non susceptible to at least one of the carbapenems by the broth microdilution method, with the criteria of minimum inhibitory concentrations (MICs) of ≥2 mg/mL for ertapenem, ≥4 mg/mL for imipenem, or ≥4 mg/mL for meropenem, were included in the study.
Antibiotics and in vitro Susceptibility Testing
According to the recommended guidelines of Clinical and Laboratory Standards Institute (CLSI, M100-S27),12 ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), ertapenem (ETP), imipenem (IPM), meropenem (MEM), piperacillin tazobactam (TZP), tigecycline (TGC), colistin (COL), fosfomycin (FOS), levofloxacin (LEV), ciprofloxacin (CIP), amikacin (AMK) and gentamicin (GEN) were determined using the broth microdilution method. At the same time, K. pneumoniae ATCC 700603 was selected as the standard strains for MIC detection. All standard strains were purchased from American Type Culture Collection and all antibiotics were purchased from Meilunbio in China.
DNA Amplification and Analysis
Total DNA was extracted by boiling. Briefly, single colonies were picked from overnight culture of each CR-KP, resuspended in 200 ul of sterile distilled water, and boiled at 100 °C for 10 minutes. After centrifugation at 15000g for 15 minutes, supernatants were collected and stored at −20 °C. Polymerase chain reaction (PCR) was used to detect the potential presence of carbapenemase genes, including blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA−48, and sequencing was used to confirm the variants of these carbapenemase genes. Moreover, ESBLs, AmpC, OmpF and OmpC genes were also determined by using primers as described previously.13 Nine representative virulence-associated genes including the pLVPK-like plasmid genetic loci such asenterobactin(entB), mucoviscosity-associated gene A (magA), regulator of mucoid phenotype gene A(rmpA)and aerobactin(iutA)were amplified by PCR as previously described.14 Capsular serotype-specific genes (K1, K2, K5, K20, K54, and K57) were amplified by PCR as previously described.14 If negative, wzi loci were amplified and sequenced to determine capsular serotype-specific genes.15 All the PCR products were gel purified and sequenced.
Multilocus Sequence Typing (MLST) and Pulsed-Field Gel Electrophoresis (PFGE)
Multilocus sequence typing (MLST) was performed by the amplifications of the internal fragments of seven housekeeping genes of CR-KP isolates (rpaB, gapA, mdh, pgi, phoE, infB and tonB) according to the database (https://pubmlst.org/organisms). The clonal relationships of the isolates were further determined by pulsed-field gel electrophoresis (PFGE). Briefly, genomic DNAs of the CR-KP isolates were prepared in agarose blocks and digested with restriction enzyme XbaI. DNA fragments were separated using a CHEF II D-Mapper XA PFGE system with running conditions as described previously. The dendrogram of the PFGE profiles was clustered by the Dice coefficient and the unweighted pair group method (UPGAMA) on the basis of the dice similarity by the Quantity One software package 4.6.16 DNA patterns sharing ≥95% similarity (SA) was defined as Clusters.17
String Test
A single CR-KP colony was inoculated on 5% sheep blood agar plate, 37°C, overnight. The colony was gently pulled up with the inoculation loop and repeated for 3 times. If length was longer than 5mm, it was considered positive for the “string test”, and the strain was hypermucoviscous phenotype.
Serum Bactericidal Assay and Virulence Behavior in the Galleria mellonella Infection Model
Serum bactericidal activity was analyzed according to what was previously described. Briefly, 25 μL of a bacterial inoculum of 1×106 CFU/mL was mixed with 75 μL of fresh mixed human serum. Samples were taken at h, 1h, 2h and 3h, and bacterial cell count was performed in tryptone soy agar (TSA) plates. Hypervirulent K. pneumoniae isolate was used as positive control. All experiments were performed in triplicate. Bacterial survival of <1% after 3 h of incubation with serum was considered as susceptible. On the other hand, survival percentages of 1–90% or >90% were considered as intermediate and resistant, respectively. The G. mellonella infection model was utilized to evaluate the virulence of the strains. Fresh single colonies were selected and cultured in LB broth (2mL) at 37°C for logarithmic growth phase. Bacterial inoculum of 1×108 CFU/mL, 1×106 CFU/mL and 1×104 CFU/mL were injected into the G. mellonella. Larvae survival was analyzed during 96 h, and Kaplan-Meier killing curves of G. mellonella were generated using the log rank test with p < 0.05. Each assay was performed in triplicate.
Biofilm Assay
Biofilms-quantification was performed as previously described. The strain was placed in LB liquid medium and shaken overnight at 37°C. From this culture, 10 μL of a bacterial suspension contained 1×106 CFU/mL was used to inoculate 96-well polystyrene plates containing 90 μL of LB liquid, 37°C for 24 h. Subsequently, the medium was removed from the plates and each well was washed three times with phosphate buffer. After dyeing with 1% crystal violet for 15min, the dyeing solution was sucked out and cleaned with distilled water until the sucked liquid was colorless and dried. Finally, Crystal violet was fully dissolved with 20ul anhydrous ethanol and the absorbance value was measured at 570nm. The yield of biofilm formation of the strains was interpreted as follows: OD > 0.6 as strong-producing, 0.4 < OD ≤0.6 as moderate-producing and OD < 0.4 as weak-producing.
Standard Strains
E. coli ATCC25922 was selected as the standard strains for MIC detection. K. pneumoniae ATCC70063 was selected as the standard strains for biofilm assay, serum bactericidal assay and virulence behavior in the Galleria mellonella infection model. All standard strains were purchased from American Type Culture Collection.
Prokaryotic Chain Specific Transcriptome Sequencing
Total RNA was extracted from the tissue using TRIzol® Reagent according the manufacturer’s instructions (Invitrogen) and genomic DNA was removed using DNase I (TaKara). RNA-seq transcriptome library was prepared following TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA) using 2μg of total RNA. Shortly, ribosomal RNA (rRNA) depletion instead of poly(A) purification is performed by Ribo-Zero Magnetic kit (epicenter) and then all mRNAs were broken into short (200nt) fragments by adding fragmentation buffer firstly. Secondly double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, CA) with random hexamer primers (Illumina). When the second strand cDNA was synthesized, dUTP was incorporated in place of dTTP. Then the synthesized cDNA was subjected to end-repair, phosphorylation and “A” base addition according to Illumina’s library construction protocol. The second strand cDNA with dUTP was recognized and degraded by UNG enzyme. Libraries were size selected for cDNA target fragments of 200bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by TBS380, paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq×TEN (2 × 150bp read length). The processing of original images to sequences, base-calling, and quality value calculations were performed using the Illumina GA Pipeline (version 1.6), in which 150bp paired-end reads were obtained. A Perl program was written to select clean reads by removing low-quality sequences, reads with more than 5% of N bases (unknown bases) and reads containing adaptor sequences.
Data generated from Illumina platform were used for bioinformatics analysis. All of the analyses were performed using the free online platform of Majorbio Cloud Platform (www.majorbio.com) from Shanghai Majorbio Bio-pharm Technology Co.
Ethical Considerations
The data and samples analyzed in the present study were obtained in accordance with the standards and approved by Chongqing Health Center for Women and Children the Institutional Review Board and Biomedical Ethics Committee. For this study, samples were collected at the microbiology laboratory of our hospital, with no contact with the patient. This study was retrospective and there was no patient identification performed during data collection. Therefore, the ethics committee determined that informed consent was not required.
Results
General Characteristics and Antimicrobial Susceptibility of CR-KP Isolates
A total of 14 non-repetitive CR-KPstrains were isolated in our hospitals. These non-duplicated isolates were mainly isolated from blood (n = 5), respiratory tract samples (n = 4), catheter (n = 2), secretion (n = 2) and urine (n = 1). For the antimicrobial susceptibility of these isolates, our study showed that all CR-KP resistant to β-lactam antibiotics, including cephalosporin, β-lactam/β-lactamase inhibitors and carbapenems. More notably, all the CR-KP were not susceptible to tigecycline considered as a last-resort drug to treat these serious CRE infections. Fortunately, these CR-KP isolates were mostly susceptible to amikacin and gentamicin, with 100.00% and 92.85% susceptibility rates (Table 1).
 |
Table 1 General Characteristics and Antimicrobial Susceptibility of CR-KP Isolates |
Molecular Characteristics of CR-KP Isolates
Three STs were identified among 14 CR-KP isolates, which included 12 isolates for ST147 and one isolate for ST3444, another for ST17. PFGE showed there were three clusters. Cluster A had 12 isolates of ST147 (85.71%), which represented the largest group of STs. Within this group, each isolate had a similar PFGE pattern that exceeded SA 0.95, thus indicating that most of the isolates shared a clonal relationship. ST3444 belonged to Cluster B was similar to cluster A with SA 0.93. ST17 divided into Cluster C showed different PFGE patterns with SA 0.71, suggesting that they had a polyclonal origin (Figure 1).
Genotypic Distribution of the CR-KP Isolates
We observed that all the 14 CR-KP strains were positive for blaNDM-1 with 92.86% co-expressed blaCTXM-14 and blaSHV-2, while CR-KP01 only expressed blaNDM-1 and blaSHV-2. Ampc gene and the loss of membrane porin were not detected in CR-KP (Supplementary Table 1). The virulence-associated genes detected by PCR for each isolate are listed in (Supplementary Table 2). Most CRKP isolates expressed fimH, wab G and ent B (Supplementary Table 1). None of strains detected serotypes K1, K2, KS, K54, K57, K20.
String Test, Biofilm Formation, Serum Bactericidal Assay and Virulence Behavior in the Galleria mellonella Infection Model
None of strains exhibited the hypermucoviscous phenotype during the string test and all strains were weak-biofilm producing (Figure 2A). However, the survival of CR-KP01 after 3 h of incubation with serum was 98%, which suggested that they were resistant to the kill of serum independence of biofilm. The strains isolated from blood showed intermediate to the kill of serum (35–50%), while others were susceptible (Figure 2B). We infected G mellonellalarvae with three typical K. pneumoniae from Cluster A, Cluster B and Cluster C with an inoculum of 1×108 CFU, 1×106 CFU and 1×104 CFU. The mortality rate of G mellonella infected with CR-KP01 was 100.00% after 24h with an inoculum of 1×108 CFU and 1×106 CFU (Figure 3A). CR-KP 14 isolated from blood caused 70% mortality of the G mellonella after 48h with an inoculum of 1×108 CFU (Figure 3B). CR-KP-02 from Cluster B was less virulent than the other strains, killing only 15% of the G mellonella after 72h with an inoculum of 1×108 CFU (Figure 3C).
Two Phenotypes of Klebsiella pneumoniae ST147 from Blood
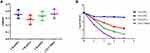
The 14-day and 21-day survival rates for CR-KP were 78.57% and 71.42%, respectively. Sepsis caused by CR-KP eventually caused 40.00% of newborn deaths and have the highest mortality rate during 7 to 14 days of hospitalization. To our surprise, the strains (35.71%, 5/14) isolated from the blood had two different phenotypes, Type I was more similar to CR - KP01, which possessed higher virulence with white and sticky colony and Type II was gray colony. PFGE and MLST confirmed that the two phenotypes were derived from the same clone ST147 (Figure 4A). Both two phenotypes were negative for the string test and were weak-biofilm producing (Figure 4B). While the survival of Type I after 3 h of incubation with serum was 65%, which was much higher than Type II (0.00%) (Figure 4C). In vivo experiments confirmed that Type I was more likely to cause death in G mellonella (Figure 4D).
Transcriptome Profiling of Two Phenotypes of Klebsiella pneumoniae ST147
In order to explore the mechanism of virulence difference between the two phenotypic, we have carried out a comparison of these two phenotypic transcriptome data. The results had shown 115 genes with differential expression (false discovery rate, ≤0.001; > 2-fold change), of which 44 were up-regulated and 71 were down-regulated. Based on COG annotations, the functional categories “carbohydrate transport and metabolism category”, “amino acid transport and metabolism” and “Transcription” were significantly enriched in Type I (Figure 5A). However, 20 gene functional categories were enriched based on GO annotations, including “integral component of membrane”, “amino acid metabolism”, and “catalytic activity” (Figure 5B).
Next, a total of 115 DEGs were assigned to 41 KEGG pathways. KEGG pathway analysis showed that 10 pathways were obviously changed (P-value <0.05) in Type I, compared with Type II (Supplementary Table 2). The majority of the up-regulation genes were mainly involved in arginine and proline metabolism (blaastA, blaastB, blaastC, blaastD and blaptuA). Additionally, the genes encoding fatty acid degradation, geraniol degradation and phosphotransferase system were also overexpressed (Figure 6).
Discussion
To the best of our knowledge, this is the first outbreak in neonates of a K. pneumoniae ST147 clone that produced NDM-1 and showed resistance against to tigecycline due to overexpression of the efflux pump gene acrR and its regulatory gene ramA. It was first detected at Chongqing Health Center for Women and Children in September 2017 from a intravenous integrated catheter sample in NICU and quickly spread to other neonates. Various studies have been published on the spread of carbapenem-resistant Enterobacteriaceae, but data on neonates are scarce. In the past few years, CR-KP carrying blaKPC-2 had been found in neonates in many countries, especially in China and India. NDM-1-producing K. pneumoniae causing neonatal infection was first reported in a teaching hospital in mainland China, 201518 and then outbreaks of blaNDM-1- possessing K. pneumoniae in neonatal units have been reported intermittently in other parts of China.13,19–21
The study of Datta et al reported that sepsis caused by K. pneumoniae did not result in a higher mortality rate.22 A study from China reported five CR-KP carrying blaNDM-1 were susceptible to gentamicin, amikacin, aztreonam, ciprofloxacin, and levofloxacin and the prognosis of the newborn was milder. While in our study, with very limited therapeutic options, these pathogens eventually leaded to nearly 30% of newborn deaths and mostly from sepsis. Two newborns who survived sepsis, one was treated with meropenem combined with fosfomycin and the other with a high dose of meropenem, which suggested these two regimens may be effective treatment for NDM - producing K. pneumoniae sepsis.
The virulence of the isolated strains was detected in vitro and in vivo. Although the results showed that the isolated strain was not typical hypervirulent K. pneumoniae (hvKp) with lower biofilm formation and serum resistance, CR-KP from blood still caused higher mortality rate compared standard strains in vivo. Although we did not unequivocally verify the hypervirulent character of this clone, most CR-KP isolates expressed fimH, wab G and ent B. However, ent B, one of four genes related to siderophores, is widespread among the classic pathotype Klebsiella (cKp). Esra Deniz found 72% of K. pneumoniae was positive for entB.23 FimH, encoded type 1 fimbriae and mediated bacterial adhesion, was also prevalent in cKp.24 Luis Izquierdo had demonstrated that wabG gene is involved in attachment to alpha-L-glycero-D-manno-heptopyranose II (L,D-HeppII) at the O-3 position of an alpha-D-galactopyranosyluronic acid (alpha-D-GalAp) residue related to the synthesis of LPS and its mutations can lead to changes in the morphology of colonies.25
Current studies have defined the whole genome characterization of CR-KP. The transcriptional behavior of CR-KP is quite few. In this study, we found two different phenotypes of K. pneumoniae ST147 outbreak from neonatal sepsis and compared the pathogenicity and transcriptome differences between these two phenotypes. This transcriptional study provides additional clues to understand the key molecular mechanisms involved in the pathogenesis of CR-KP. We found that compared with Type II, 10 pathways were obviously changed in Type I especially amino acid metabolism. Bacterial pathogens are able to infect a broad spectrum of host tissues, in part due to the flexibility of metabolic procedures such as amino acid metabolism. Potter had demonstrated that synthesis of aspartate is absolutely essential for the invasive infection of Staphylococcus aureus.26 Another study had investigated the role of pyruvate carboxylase, which was important across multiple infection models and confirmed a connection between growth and resistance to host cell killing.27 Nuxoll had found that CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism and a mutation in argH (arginine metabolism related genes) attenuated virulence in a mouse kidney abscess model in comparison to wild type JE2.
The transcriptional study showed that arginine and proline metabolism was significantly changed between Type I and Type II. Arginine is an energy-rich amino acid that provides nitrogen, carbon, and energy to a variety of bacteria in a variety of environments. Arginine can be broken down through a variety of pathways such as arginine pathway, arginine deaminase (ADI) pathway, arginine succinyltransferase (AST) pathway, etc.28 We have demonstrated that the transcription level of AST pathway in type I was significantly higher than that in type II. AST pathway could provide nitrogen during nitrogen restriction,29 promote polyamine homeostasis by controlling levels of intracellular arginine and ornithine,30 which is vital to the physiological function of bacteria. Of greater significant, Hiroki Shirai thought that AST intermediates could be potential antimicrobial agents against pathogen.31
In addition to affecting physiological functions, arginine metabolism is also related to host immune responses. Chelsie E hadfond that Twin arginine translocation, ammonia incorporation, and polyamine biosynthesis are crucial for Proteus mirabilis fitness during bloodstream infection.32 Helicobacter pylori has been proved to inhibit the production of nitric oxide through producing arginase and reduce the lethality of macrophages.33 Furthermore, the researchers found L-Arginine Enhances Intracellular killing of carbapenem-resistant Klebsiella pneumoniae ST258 by murine neutrophils.34 Therefore, we speculate that arginine and proline metabolism play a critical role in the pathogenesis of K. pneumoniae. However, there is an obvious a shortcoming of the work that is the lack of full genome sequencing data for the two lineages. Next, we will conduct whole genome sequencing for the two types to further verify our conjecture.
Conclusion
Our study showed that CR-KP was a serious emerging challenge among hospitalized neonates. Most important of all, we found two phenotypes of K. pneumoniae with different virulent. Compared to Type II, Type I was significant up-regulated in arginine metabolic pathway which is essential for the survival and pathopoiesis of bacteria. Our study is of great value in solving the pathogenesis of K. pneumoniae and providing a new therapeutic target.
Author Contributions
HZ and YS designed the study, YS and CL performed the experiments. QH analyzed data. HZ wrote this manuscript. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported in part by the Health Bureau in Chongqing (Grant 2013-12-133).
Disclosure
The authors declare that they have no competing interests.
References
1. Hibberd PL, Hansen NI, Wang ME, et al. Trends in the incidence of possible severe bacterial infection and case fatality rates in rural communities in Sub-Saharan Africa, South Asia and Latin America, 2010–2013: a multicenter prospective cohort study. Reprod Health. 2016;13(1):65. doi:10.1186/s12978-016-0177-1
2. Ahmed I, Ali SM, Amenga-Etego S; Alliance for M, Newborn Health Improvement mortality study g. Population-based rates, timing, and causes of maternal deaths, stillbirths, and neonatal deaths in south Asia and sub-Saharan Africa: a multi-country prospective cohort study. Lancet Glob Health. 2018;6(12):e1297–e308. doi:10.1016/S2214-109X(18)30385-1
3. Li JY, Chen SQ, Yan YY, et al. Identification and antimicrobial resistance of pathogens in neonatal septicemia in China-a meta-analysis. Int J Infect Dis. 2018;71:89–93. doi:10.1016/j.ijid.2018.04.794
4. Smith A, Anandan S, Veeraraghavan B, Thomas N. Colonization of the preterm neonatal gut with carbapenem-resistant Enterobacteriaceae and its association with neonatal sepsis and maternal gut flora. J Glob Infect Dis. 2020;12(2):101–104. doi:10.4103/jgid.jgid_104_19
5. Singh NP, Choudhury DD, Gupta K, et al. Predictors for gut colonization of carbapenem-resistant Enterobacteriaceae in neonates in a neonatal intensive care unit. Am J Infect Control. 2018;46(6):e31–e35. doi:10.1016/j.ajic.2018.01.007
6. Mukherjee S, Mitra S, Dutta S, Basu S. Neonatal sepsis: the impact of carbapenem-resistant and hypervirulent Klebsiella pneumoniae. Front Med. 2021;8:634349. doi:10.3389/fmed.2021.634349
7. Ahmad N, Ali SM, Khan AU. Molecular characterization of novel sequence type of carbapenem-resistant New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in the neonatal intensive care unit of an Indian hospital. Int J Antimicrob Agents. 2019;53(4):525–529. doi:10.1016/j.ijantimicag.2018.12.005
8. Bor M, Ilhan O. Carbapenem-resistant Klebsiella pneumoniae outbreak in a neonatal intensive care unit: risk factors for mortality. J Trop Pediatr. 2021;67(3). doi:10.1093/tropej/fmaa057
9. Mohammad ali Tabrizi A, Badmasti F, Shahcheraghi F, Azizi O. Outbreak of hypervirulent Klebsiella pneumoniae harbouring blaVIM-2 among mechanically-ventilated drug-poisoning patients with high mortality rate in Iran. J Glob Antimicrob Resist. 2018;15:93–98. doi:10.1016/j.jgar.2018.06.020
10. Huang YH, Chou SH, Liang SW, et al. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018;73(8):2039–2046. doi:10.1093/jac/dky164
11. Lu Y, Feng Y, McNally A, Zong Z. Occurrence of colistin-resistant hypervirulent Klebsiella variicola. J Antimicrob Chemother. 2018;73(11):3001–3004. doi:10.1093/jac/dky301
12. Institute CaLS. Performance Standards for Antimicrobial Susceptibility Testing; 2020.
13. Yu J, Wang Y, Chen Z, et al. Outbreak of nosocomial NDM-1-producing Klebsiella pneumoniae ST1419 in a neonatal unit. J Glob Antimicrob Resist. 2017;8:135–139. doi:10.1016/j.jgar.2016.10.014
14. Jin X, Chen Q, Shen F, et al. Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 during treatment with tigecycline and polymyxin. Emerg Microbes Infect. 2021;10(1):1129–1136. doi:10.1080/22221751.2021.1937327
15. Zhao J, Zhang Y, Fan Y, et al. Characterization of an NDM-5-producing hypervirulent Klebsiella pneumoniae sequence type 65 clone from a lung transplant recipient. Emerg Microbes Infect. 2021;10(1):396–399. doi:10.1080/22221751.2021.1889932
16. Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi:10.1128/jcm.33.9.2233-2239.1995
17. Contreras-Alvarado LM, Zavala-Vega S, Cruz-Cordova A, et al. Molecular epidemiology of multidrug-resistant uropathogenic Escherichia coli O25b strains associated with complicated urinary tract infection in children. Microorganisms. 2021;9(11):2299. doi:10.3390/microorganisms9112299
18. Zhang X, Li X, Wang M, et al. Outbreak of NDM-1-producing Klebsiella pneumoniae causing neonatal infection in a teaching hospital in mainland China. Antimicrob Agents Chemother. 2015;59(7):4349–4351. doi:10.1128/AAC.03868-14
19. Yu J, Tan K, Rong Z, et al. Nosocomial outbreak of KPC-2- and NDM-1-producing Klebsiella pneumoniae in a neonatal ward: a retrospective study. BMC Infect Dis. 2016;16(1):563. doi:10.1186/s12879-016-1870-y
20. Jin Y, Song X, Liu Y, et al. Characteristics of carbapenemase-producing Klebsiella pneumoniae as a cause of neonatal infection in Shandong, China. Exp Ther Med. 2017;13(3):1117–1126. doi:10.3892/etm.2017.4070
21. Huang X, Cheng X, Sun P, Tang C, Ni F, Liu G. Characteristics of NDM-1-producing Klebsiella pneumoniae ST234 and ST1412 isolates spread in a neonatal unit. BMC Microbiol. 2018;18(1):186. doi:10.1186/s12866-018-1334-1
22. Datta S, Roy S, Chatterjee S, et al. A five-year experience of carbapenem resistance in Enterobacteriaceae causing neonatal septicaemia: predominance of NDM-1. PLoS One. 2014;9(11):e112101. doi:10.1371/journal.pone.0112101
23. Candan ED, Aksoz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62(4):867–874. doi:10.18388/abp.2015_1148
24. Zhang S, Zhang X, Wu Q, et al. Clinical, microbiological, and molecular epidemiological characteristics of Klebsiella pneumoniae-induced pyogenic liver abscess in southeastern China. Antimicrob Resist Infect Control. 2019;8:166. doi:10.1186/s13756-019-0615-2
25. Izquierdo L, Coderch N, Pique N, et al. The Klebsiella pneumoniae wabG gene: role in biosynthesis of the core lipopolysaccharide and virulence. J Bacteriol. 2003;185(24):7213–7221. doi:10.1128/JB.185.24.7213-7221.2003
26. Potter AD, Butrico CE, Ford CA, et al. Host nutrient milieu drives an essential role for aspartate biosynthesis during invasive Staphylococcus aureus infection. Proc Natl Acad Sci U S A. 2020;117(22):12394–12401. doi:10.1073/pnas.1922211117
27. Kim GL, Hooven TA, Norambuena J, et al. Growth and stress tolerance comprise independent metabolic strategies critical for Staphylococcus aureus infection. mBio. 2021;12(3):e0081421. doi:10.1128/mBio.00814-21
28. Tocilj A, Schrag JD, Li Y, et al. Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli. J Biol Chem. 2005;280(16):15800–15808. doi:10.1074/jbc.M413833200
29. Kiupakis AK, Reitzer L. ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J Bacteriol. 2002;184(11):2940–2950. doi:10.1128/JB.184.11.2940-2950.2002
30. Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–176. doi:10.1146/annurev.micro.57.030502.090820
31. Shirai H, Mizuguchi K. Prediction of the structure and function of AstA and AstB, the first two enzymes of the arginine succinyltransferase pathway of arginine catabolism. FEBS Lett. 2003;555(3):505–510. doi:10.1016/S0014-5793(03)01314-0
32. Armbruster CE, Forsyth VS, Johnson AO, et al. Twin arginine translocation, ammonia incorporation, and polyamine biosynthesis are crucial for Proteus mirabilis fitness during bloodstream infection. PLoS Pathog. 2019;15(4):e1007653. doi:10.1371/journal.ppat.1007653
33. Gobert AP, McGee DJ, Akhtar M, et al. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc Natl Acad Sci U S A. 2001;98(24):13844–13849. doi:10.1073/pnas.241443798
34. Penaloza HF, Ahn D, Schultz BM, Pina-Iturbe A, Gonzalez LA, Bueno SM. L-arginine enhances intracellular killing of carbapenem-resistant Klebsiella pneumoniae ST258 by murine neutrophils. Front Cell Infect Microbiol. 2020;10:571771. doi:10.3389/fcimb.2020.571771
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.