Back to Journals » Drug Design, Development and Therapy » Volume 17
Tuo-Min-Ding-Chuan Decoction Alleviates Airway Inflammations in the Allergic Asthmatic Mice Model by Regulating TLR4-NLRP3 Pathway-Mediated Pyroptosis: A Network Pharmacology and Experimental Verification Study
Authors Lyu M , Qin J , Huang S, Shao D, Huang G, Yang F , Gong X , Zhang S, Zhang Z, Wang J , Cui H
Received 1 March 2023
Accepted for publication 4 May 2023
Published 2 June 2023 Volume 2023:17 Pages 1613—1630
DOI https://doi.org/10.2147/DDDT.S406483
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Tin Wui Wong
Mingsheng Lyu,1,2 Jingbo Qin,3 Shuaiyang Huang,2 Dongmei Shao,4 Guirui Huang,2 Fan Yang,5,6 Xuefeng Gong,7 Shiyu Zhang,2 Zhijie Zhang,2 Ji Wang,6 Hongsheng Cui2
1Center of Respiratory Disease, Dongzhimen Hospital, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 2Department of Respiratory, The Third Affiliated Hospital, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 3Department of Geratology, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, People’s Republic of China; 4Department of Infectious Disease, Shunyi Hospital, Beijing Hospital of Traditional Chinese Medicine, Beijing, People’s Republic of China; 5College of Traditional Chinese Medicine, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 6National Institute of Traditional Chinese Medicine (TCM) Constitution and Preventive Medicine, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 7Department of Traditional Chinese Medicine, Beijing Chaoyang Hospital, Capital Medical University, Beijing, People’s Republic of China
Correspondence: Hongsheng Cui, Department of Respiratory, The Third Affiliated Hospital, Beijing University of Chinese Medicine, Beijing, People’s Republic of China, Email [email protected]; Ji Wang, National Institute of Traditional Chinese Medicine Constitution and Preventive Medicine, Beijing University of Chinese Medicine, Beijing, People’s Republic of China, Email [email protected]
Background: Tuo-Min-Ding-Chuan Decoction (TMDCD) is an effective traditional Chinese medicine (TCM) formula granule for allergic asthma (AA). Previous studies proved its effects on controlling airway inflammations, while the specific mechanism was not clear.
Methods: We conducted a network pharmacology study to explore the molecular mechanism of TMDCD against AA with the public databases of TCMSP. Then, HUB genes were screened with the STRING database. DAVID database performed GO annotation and KEGG functional enrichment analysis of HUB genes, and it was verified with molecular docking by Autodock. Then, we built a classic ovalbumin-induced allergic asthma mice model to explore the mechanism of anti-inflammation effects of TMDCD.
Results: In the network pharmacology study, we found out that the potential mechanism of TMDCD against AA might be related to NOD-like receptor (NLR) signaling pathway and Toll-like receptor (TLR) signaling pathway. In the experiment, TMDCD showed remarkable effects on alleviating airway inflammations, airway hyperresponsiveness (AHR), and airway remodeling in the asthmatic mice model. Further molecular biology and immunohistochemistry experiments suggested TMDCD could repress TLR4-NLRP3 pathway-mediated pyroptosis-related gene transcriptions to inhibit expressions of target proteins.
Conclusion: TMDCD could alleviate asthmatic mice model airway inflammations by regulating TLR4-NLRP3 pathway-mediated pyroptosis.
Keywords: Tuo-Min-Ding-Chuan Decoction, allergic asthma, TLR4, NLRP3, pyroptosis, network pharmacology, traditional Chinese medicine
Introduction
Bronchial asthma is a common respiratory disease characterized by chronic airway inflammations. Epidemiological data show that asthma has a high incidence all over the world, with nearly 300 million patients worldwide and 45.7 million adult patients in China.1,2 Recurrent asthma attacks not only seriously reduce the quality of life of asthmatic patients but also bring serious medical burdens to the world. Usually, the phenotypes of bronchial asthma are classified into early-onset AA, late-onset persistent eosinophilic asthma, exercise-induced asthma, obesity-related asthma, neutrophilic asthma, smoking asthma, etc.3 AA accounts for more than 50% of the adult asthma patients and 80% of the childhood patients.4 The pathogenesis of AA is complex; previous studies hold the view that the adaptive immune system mainly participates in the dysregulation of allergy and induces airway inflammations, which is the basis of AHR and airway remodeling. The activation of type 2 helper T cell (Th), the crucial player in humans’ adaptive immune responses, brings releases of type 2 inflammatory cytokines such as interleukin (IL)-4, IL-5, and IL-13. Nevertheless, recognition of environment antigens by innate immune cells should not be ignored for the activation of adaptive immune responses, which play a key role in priming naive T cells and generating adaptive immunities.5 Following recent studies, we know that the primary means of inducing innate immune responses were associated with ligations and activations of pattern recognition receptors (PRRs). As important representatives of PRRs family, TLR4 and NLRP3 could identify pathogens or allergens by engaging damage-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs).6 Meanwhile, activations of TLR4 and NLRP3 contribute to the development of chronic inflammation-mediated complicated diseases and lead to pyroptosis, the latter is a lytic cell death induced by pathogen infection or endogenous challenge.7–9 Typically, pyroptosis occurred in severe asthma with the characteristic of neutrophil infiltrations,10 while recent studies also reported pyroptosis was found in classical eosinophilic allergic asthma.11,12 Given pyroptosis was an important part of innate immune activation-induced inflammation, which was the key mechanism of asthma attacks, it was necessary to conduct research between TLR4-NLRP3 pathway-mediated pyroptosis and allergic asthma.
With the therapeutic properties of multi-target and few side effects, TCM has developed over 2000 years in China and has exerted medical influences to 183 countries and regions.13 TMDCD is an effective TCM formula granule in Phase 2 clinical trial for treating AA created by Professor Wang Qi from Beijing University of Chinese Medicine (BUCM), who is also known as the national Chinese medical science master and the academician of the Chinese Academy of Engineering. TMDCD is composed of 12 Chinese herbal medicine (CHM) including Ephedrae Herba, Armeniacae Semen Amarum, Gypsum, Glycyrrhizae Radix Et Rhizoma, etc. Our results suggested TMDCD showed remarkable clinical effects against AA in reducing the dosage of inhaled corticosteroid (ICS), controlling asthma acute exacerbations, and improving the quality of life (full data was not reported yet). Previous studies about TMDCD also proved it could suppress allergic inflammations and regulate the immune balance between regulatory T cells (Tregs) and Th17 in the AA mice model, besides it inhibits the degranulation of mast cells by in vitro experiments,14,15 while more specific potential mechanisms of TMDCD against AA were not figured out yet. In this study, we preliminarily explored the effecting mechanism of TMDCD on AA with the method of network pharmacology, then investigated the anti-inflammation effects of TMDCD on regulating TLR4-NLRP3 pathway-mediated pyroptosis with an animal experiment verification. The detailed workflow of the study is shown in Figure 1.
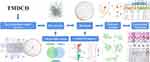 |
Figure 1 The workflow of the study. |
Materials and Methods
Components and Bioactive Ingredients in TMDCD
TMDCD was composed of Ephedrae Herba (“Mahuang” in Chinese, MH), Armeniacae Semen Amarum (“Xingren” in Chinese, XR), Gypsum (“Shigao” in Chinese, SG), Glycyrrhizae Radix Et Rhizoma (“Gancao” in Chinese, GC), Saposhnikoviae Radix (“Fangfeng” in Chinese, FF), Ganoderma (“Lingzhi” in Chinese, LZ), Mume Fructus (“Wumei” in Chinese, WM), Cicadae Perios Tracum (“Chantui” in Chinese, CT), Fagopyri Dibotryis Rhizoma (“Jinqiaomai” in Chinese, JQM), Polygoni Multiflori Caulis (“Shouwuteng” in Chinese, SWT), Gastrodiae Rhizoma (“Tianma” in Chinese, TM), and Bombyx Batryticatus (“Jiangcan” in Chinese, JC). The natural components of TMDCD were obtained from the traditional Chinese medicine systems pharmacology database (TCMSP, https://old.tcmsp-e.com/index.php, updated on May 31, 2014), traditional Chinese medicines integrated database (TCMID, http://www.megabionet.org/tcmid, updated on Nov. 12, 2012), and Shanghai institute of organic chemistry of Chinese academy of sciences chemistry database (http://www.organchem.csdb.cn, updated on Jun. 10, 2018). If the herb from TMDCD was not included in above databases, China national knowledge infrastructure (CNKI, https://www.cnki.net, updated on Apr. 10, 2022) database was searched by taking the herb name and component as keywords (such as “Gastrodiae Rhizoma” + “component”). These databases almost contained all the 499 Chinese herbs registered in the Pharmacopoeia of the People`s Republic of China (2010 edition),16 from which we could obtain all the information on these components.
Based on previous studies, the criteria of oral bioavailability (OB) ≥30% and drug likeness (DL) ≥0.18 were used to screen bioactive ingredients from the TCMSP database,13 OB and DL reflected the CHM absorption in the gastrointestinal tract. For those components obtained from other databases, Swiss ADME web service performed the screening with the method of computing the physicochemistry and estimating the pharmacokinetics, druglikeness, and medicinal chemistry friendliness of small molecules.17
Targets Related to Bioactive Ingredients
We used the TCMSP database to get the majority of bioactive ingredients-related targets, which was based on the method of experiments validated drug–target interactions and similarity-based drug–target interactions.16,18 For those components which did not exist in the TCMSP database, predicted targets were acquired from Swiss Target Prediction web service by using the 2-dimensional structure of bioactive ingredients obtained from PubChem, with parameters of the organism for “Homo sapiens” and the probability ≥0.7.19 UniProt Knowledgebase (UniProtKB) contributed to the standardization of target gene names.
AA-Related Targets and Common Targets Between TMDCD and AA
The following databases were used to collect AA-related targets with the keywords of “allergic asthma”: Online Mendelian Inheritance in Man database (OMIM),20 DisGeNET database,21 GeneCards the human gene database,22 and Therapeutic Target Database (TTD).23 After removing the repetitive targets, common targets between AA and TMDCD were screened from a Venn diagram and standardized to consistent gene names in the UniProtKB database. Then, we used Cytoscape v3.7.1 to create the “herb–ingredient–target” network,24 in which way to explore the potential effecting targets of TMDCD against AA. At last, the “degree (DC)” information was acquired from the “herb–ingredient–target” network to identify the pivotal ones from bioactive ingredients of TMDCD. Detailed steps of the “Network analysis” tool in Cytoscape were as previously reported.13
Constructions of the Protein–Protein Interaction Network
The protein–protein interactions (PPI) network was conducted by Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), a web service for implementing a functional system based on known and predicted PPI,25 with parameters of the organism for “Homo sapiens” and the minimum required interaction score ≥0.9. To optimize the complex PPI network, the topology analysis method of the PPI network was conducted to get HUB genes as previously reported.13 Specifically, HUB genes were defined as nodes with DC over twofold the median degree of all the nodes, as well as “betweenness centrality (BC)”, “closeness centrality (CC)”, and “stress” were higher than the corresponding median values of their own. Furthermore, the molecular complex detection (MCODE) plug-in performed the topology analysis based on HUB genes, the following parameters were set: DC was set to 2, cut-off score was set to 0.2, K core score was set to 2, and the mix distance was set to 100. At last, visual processing was made by Cytoscape.
Gene Ontology and KEGG Pathway Enrichment Analysis
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis of HUB genes were conducted by Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8.26 Results of biological processes (BPs), cell components (CCs), molecular functions (MFs), and KEGG enriched pathways were considered as a significant difference with the P-value ≤0.05, which helped to understand the biological processes of HUB genes. Hiplot web service (https://hiplot.com.cn) contributed to visualizations.
Molecular Docking
To prove the interaction between HUB genes and bioactive ingredients, AutoDock v4.2.6 was used to conduct molecular docking.27 We selected the top 10 genes from HUB genes as the core target receptor proteins by sorting the DC value, and pivotal ingredients, usually called ligands, were screened as mentioned before. The 3-dimensional structures of core target receptor proteins and ligands were downloaded from the RCSB Protein Data Bank database (PDB),28 TCMSP, and PubChem. Detailed steps of molecular docking were as previously reported.13 At last, we evaluated whether the binding energy was less than 0 kcal/mol to confirm molecules could spontaneously bind to each other.13 Visualization of receptor protein–ligand interactions was performed by Pymol v2.4.
Animals
An in vivo experiment was conducted to verify the predicted pathways obtained from the network pharmacology study. Thirty-six female BALB/c mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China), weighted 18–20 g at 7–8 weeks of age. The mice were bred in the scientific research and experiment center of BUCM [certification SCXK (Beijing) 2016-0006]. This study was approved by the Research Ethics Committee of BUCM [No. BUCM-4-2020092001-3146] and followed the laboratory animal guideline for ethical review of animal welfare [GB/T 35892–2018].
Drugs and Reagents
The following drugs and reagents were used in this study: ovalbumin (OVA) [Sigma Aldrich, A5503], acetyl-β-methylcholine chloride [Sigma Aldrich, A2251], Imject alum adjuvant [Thermo Fisher Scientific, 77161], Trizol Reagent [Thermo Fisher Scientific, 15596026], modified Masson trichrome stain kit [Beijing Solarbio, G1340], glycogen periodic acid Schiff (PAS/hematoxylin) stain kit [Beijing Solarbio, G1281], tris buffered saline buffer with Tween 20 (TBST) [Beijing Solarbio, T1082], 5% non-fat powdered milk [Beijing Solarbio, D8340], ECL Western blotting substrate kit [Beijing Solarbio, PE0010], peroxidase-conjugated goat anti-rabbit IgG [ZSGB-BIO, PV-9001], 3,3`-diaminobenzidine (DAB) [ZSGB-BIO, ZLI-9018], normal goat serum [Beijing Biotopped, SYX1100], Omni-Easy one-step PAGE gel fast preparation kit [Shanghai Epizyme, R4011-03], Reverse Transcription System [Promega, A3500], qPCR Master Mix [Promega, A6002], radio-immunoprecipitation assay (RIPA) lysis buffer [Beijing Applygen, C1053], anti-TLR4 antibody [Abcam, ab13867], anti-TRAF6 antibody [Abcam, ab33915], anti-NLRP3 antibody [Abcam, ab263899], anti-Caspase-1 antibody [Abcam, ab138483], anti-GSDMD antibody [Abcam, ab219800], ASC/TMS1 rabbit mAb [CST, 67824], β-Actin rabbit mAb [CST, 4970S], mouse OVA-specific immunoglobulin E (OVA-sIgE) enzyme linked immunosorbent assay (ELISA) kit [Jiangsu Meimian, MM0816], mouse IL-1β ELISA kit [Beijing Dongge, DG30057M], mouse IL-4 ELISA kit [Beijing Dongge, DG30030M], mouse IL-5 ELISA kit [Beijing Dongge, DG30031M], mouse IL-6 ELISA kit [Beijing Dongge, DG30062M], mouse IL-13 ELISA kit [Beijing Dongge, DG30613M], mouse IL-18 ELISA kit [Beijing Dongge, DG30082M], mouse tumor necrosis factor (TNF)-α ELISA kit [Beijing Dongge, DG30048M], mouse interferon (IFN)-γ ELISA kit [Beijing Dongge, DG30056M]. Dexamethasone (DEX) acetate tablets were brought from Zhejiang Xianju Pharmaceutical Co., Ltd (Zhejiang, China)[200327]. The ingredients of TMDCD, containing 12 kinds of CHM including MH 10 g, XR 10 g, SG 30 g, GC 6 g, FF 10 g, LZ 10 g, WM 20 g, CT 10 g, JQM 15 g, SWT 15 g, TM 10 g, and JC 10 g, were purchased from Beijing Tong Ren Tang Technology Development Co., Ltd. (Beijing, China). TMDCD solution was prepared as previously described, the active constituents of TMDCD were measured by LC-MS/MS.15 Detailed batch information of ingredients is listed in Supplementary Table 1.
OVA-Induced Asthmatic Model and Therapeutic Interventions
The model was established according to our previous studies.14,15 Briefly, mice were allowed to acclimatize themselves for 1 week to adapt to the experimental environment, then they were equally and randomly divided into six groups: control group, OVA-induced asthma (OVA) group, DEX group, high-dose TMDCD (TMDCD-H) group, middle-dose TMDCD (TMDCD-M) group, and low-dose TMDCD (TMDCD-L) group. Each group contained six mice. Mice in the OVA group, DEX group, and TMDCD-H/M/L group were intraperitoneally injected with a 0.2 mL PBS mixture containing 2 μg OVA and 20 μg aluminum hydroxide on day 0 and day 14 for sensitization. From day 21 to day 25, all mice except the control group were challenged with the 1% OVA nebulization inhalant solution using a compression atomizer [Omron Health Care China Co., Ltd, NE-C900] every day at the rate of 2 mL/min for 30 minutes. The control group mice were intraperitoneally injected and inhaled the same amount of PBS. Based on the method of preventive treatment of TCM, mice in the TMDCD-H/M/L group were respectively administered three-dose TMDCD (40.56, 20.28, and 10.14 g/kg/d) through gastric administration at 1 hour before the OVA challenge once a day from day 14 to day 25. Meanwhile, with the dose of 1 mg/kg/d, DEX was given to the DEX group mice from day 21 to day 25, and the control group mice were orally given the same amount of PBS. All mice were sacrificed on day 26. The sensitization, challenge, and therapeutic interventions are shown in Figure 2.
Measurement of AHR
Mice were anesthetized and intubated 24 hours after the last OVA challenge. Then, Buxco FinePointe RC system (St. Paul, MN, USA) was used to measure the AHR of all mice. Parameters of the RC system and ventilator were set as our previous study reported, and four gradient concentrations of methacholine (0, 3.125, 6.25, and 12.5 mg/mL) were used to perform the pulmonary resistance and compliance by recording pulmonary resistance (RL) and dynamic pulmonary compliance (Cdyn) values.15
Detection of Serum and Bronchoalveolar Lavage Fluid (BALF) Cytokines
BALF was collected by syringe three times with 1 mL PBS after the pulmonary resistance and compliance test. Serum was obtained by centrifugation after blood collection. Cytokine levels of IL-4, IL-5, IL-13, IL-1β, IL-18, IL-6, TNF-α, and IFN-γ in BALF, and IL-1β, IL-18, OVA-sIgE in serum were detected by above ELISA kits strictly following the instructions.
Lung Histopathology and Immunohistochemistry (IHC)
Lung tissues paraffin sections (4 μm) were made and dyed with hematoxylin/eosin (H&E), Masson, and PAS staining following the standard protocol. Firstly, two independent observers scored H&E staining sections’ inflammatory degrees from 0 to 4, which reflected the severity of the pathological tissue.13 IHC was conducted to identify the target proteins’ positive expressions. Detailed steps of IHC staining followed the guidance of antibody manufacturers, mainly including antigen retrieval (pH 6.0 or pH 9.0) for 45 min, blocking proteins with normal goat serum for 1 hour to prevent the non-specific antibody binding, primary antibodies incubations overnight at 4 °C, secondary antibodies incubations at room temperature for 1 hour, finally detected and amplified with horseradish peroxidase (HRP)-DAB. The following primary antibodies were used: TLR4 1:500, TRAF6 1:1000, NLRP3 1:500, ASC 1:100, Caspase 1 1:1000, GSDMD 1:1000. Image acquisitions of H&E, Masson, PAS, and IHC staining were performed by Olympus BX-60 upright microscope (Tokyo, Japan), then ImageJ software (version 1.53, National Institutes of Health) conducted the image quantitative analysis and the calculation of the positive area of goblet cell (AREA%), collagen volume fractions, the percentage of total airway wall area (Wat) to the area enclosed by outer airway perimeter (TAt%), and the percentage of Wat to internal airway perimeter (Wat/Pi). Detailed methods followed our previous study.13
Protein Preparations and Western Blot Analysis
Mice lung tissues were homogenized and lysed in RIPA lysis buffer containing protease inhibitors, then the lysates were centrifuged to get the supernatants and mixed with 5× loading buffer. Boil them for denaturation. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Tris-Glycine running buffer were used to separate the protein samples with the parameters at 120 V for 90 min, then samples were transferred to polyvinylidene fluoride (PVDF) [Sigma Aldrich, IPVH00010] membranes using a transfer buffer at 200 mA for 2 hours. Then the PVDF membranes were blocked with TBST containing 5% non-fat milk for 2 hours. The membranes were washed with TBST three times, incubated with primary antibody overnight at 4 °C, and followed by HRP-labeled secondary antibody incubations at room temperature for 1 hour. The following primary antibodies were used: TLR4 1:500, TRAF6 1:2000, NLRP3 1:1000, ASC 1:1000, Caspase 1 1:1000, GSDMD 1:1000, β-actin 1:8000. ECL substrate kit performed the visualization of target proteins.
Real-Time Quantitative Polymerase Chain Reaction (Real-Time PCR)
Total RNA was extracted and purified from mice lung tissues with the Trizol method. Then, it was reverse-transcribed to cDNA using Promega Reverse Transcription System with Gene Amp PCR 9700 (Thermo Fisher Scientific, Waltham, MA, USA). Stratagene Mx3000P (Agilent, La Jolla, CA, USA) performed real-time PCR with qPCR Master Mix according to the manufacturer’s instructions, detailed fluorescence quantitation steps were as follows: hot start polymerase activation at 95°C for 2 min, followed by 45 cycles of PCRs: denaturation at 95°C for 15s, annealing and extension at 60°C for 60s, successively, then extension at 72°C for 60s by one cycle. The relative expressions of target genes were calculated using the 2−ΔΔCt method. Detailed primers are listed in Table 1.
 |
Table 1 Primers of Target Genes |
Data Analysis and Statistics
Statistical analysis was performed by SPSS 20.0 statistical software (SPSS Inc., USA). The data conforming to normal distribution would be expressed as mean ± standard error of the mean (SEM). Median (quartile 25%, 75%) [M (Q1, Q3)] was used for data not conforming to normal distribution. One-way analysis of variance (ANOVA) was used to compare the unpaired parametric data between groups, the least-significant difference (LSD) performed post hoc tests, and the nonparametric test was used for two groups’ nonparametric data analysis. The P-value of less than 0.05 was considered statistically significant.
Results
Screening of Common Targets Between TMDCD and AA
A total of 179 bioactive ingredients were identified in TMDCD after removing repetitive ingredients, of which 23 were from MH, 19 were from XR, 92 were from GC, 18 were from FF, 61 were from LZ, 8 were from WM, 15 were from JQM, 3 were from CT, 9 were from SWT, 5 were from TM, and 12 were from JC. The bioactive ingredient of Gypsum was magnesium sulfate, and it was rejected because it has no corresponding target information. Detailed information was listed in Supplementary Table 2.
TMDCD-related targets were predicted by TCMSP database and Swiss Target Prediction web service. Respectively, 228, 73, 234, 84, 40, 191, 207, 29, 83, 16, and 30 potential target genes were predicted for MH, XR, GC, FF, LZ, WM, JQM, CT, SWT, TM, and JC. After removing repetitive targets, 360 genes were left.
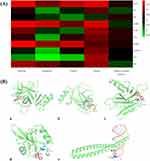
In total, 3267 AA-related targets were obtained by screening the above databases and removing repetitive targets, of which 2524 were from Genecards, 904 were from OMIM, 371 were from Disgenet, 9 were from Drugbank, and 4 were from TTD. In total, 200 common targets were get by comparing the targets between TMDCD and AA (Supplementary Table 3). With the method mentioned above, the “herb–ingredient–target” network was built by Cytoscape and shown in Figure 3A, which comprised 421 nodes and 2851 edges. The relationship between TMDCD bioactive ingredients and target genes was described by edges in the “herb–ingredient–target” network, the larger the area of the node represented the closer connected nodes. Besides, the top five pivotal ingredients were obtained as follows: quercetin (MOL000098), kaempferol (MOL000422), luteolin (MOL000006), wogonin (MOL000173), and 7-Methoxy-2-methyl isoflavone (MOL003896). The common targets network was performed by the Jvenn plug-in (Figure 3B).29
PPI Network Construction, GO Function and KEGG Pathway Analysis
STRING database conducted common targets PPI network analysis (Figure 3C). With 200 nodes, 885 edges, and an average node degree of 8.89, the PPI network performed the interaction of common targets and depicted the biological processes of TMDCD against AA. Then, we used the “Network analysis” tool to perform the topology analysis and obtain 35 HUB genes of TMDCD based on the parameters results of “DC”, “BC”, “CC”, and “stress” (Table 2). The median degree of DC, BC, CC, and stress was 8, 0.3872549, 0.00304641, and 672. Visual processing of HUB genes was performed by Cytoscape, larger sizes and brighter colors represented high degree values, and smaller sizes and darker colors represented low degree values (Figure 3D). Results of MCODE indicated each HUB gene had good correlations and similar functions, it was shown in Figure 3E.
 |
Table 2 DC, BC, CC, and Stress Information of HUB Genes |
DAVID database conducted the analysis of the GO function and KEGG pathway on 35 HUB genes. After deleting data with the P-value ≥0.05 and unrelated to AA, 283 BPs, 26 CCs, 52 MFs, and 119 KEGG pathways related to AA were obtained. The top 10 significantly enriched clusters in BPs, CCs, and MFs were selected for analysis (Figure 4A), as well as the top 20 significant KEGG pathways were enriched (Figure 4B). BPs enrichment analysis demonstrated that the HUB genes were related to positive regulation of transcription from RNA polymerase II promoter, positive regulation of transcription, DNA-templated, negative regulation of apoptotic process, and lipopolysaccharide-mediated signaling pathway. The main MFs of HUB genes were enzyme binding, transcription factor binding, identical protein binding, RNA polymerase II core promoter proximal region sequence-specific DNA binding, and chromatin binding. The CCs of HUB genes were enriched in cytosol, nucleoplasm, protein complex, nuclear chromatin, and mitochondrion. Results of KEGG pathways analysis indicated that the potential mechanism of TMDCD against AA might be associated with NOD-like receptor (NLR) signaling pathway, prolactin signaling pathway, VEGF signaling pathway, T cell receptor signaling pathway, and Toll-like receptor (TLR) signaling pathway, etc. As listed in the KEGG map, NLR and TLR signaling pathways were closely related to inflammations and would induce pyroptosis, which suggests that TLR4-NLRP3 pathway-mediated pyroptosis might be the potential mechanism of TMDCD against AA.
Molecular Docking Prediction
Molecular docking was finished by the top 10 core targets and five pivotal ingredients, the former was identified by sorting DC values from 35 HUB genes by size, and the latter was obtained from the “herb–ingredient–target”. Detailed information on docking results is listed in Table 3 and Figure 5A. The binding energy of less than −5 kcal/mol indicates a stronger connection between ligands and receptor proteins.13,30 Hence, we screened out the receptor protein–ligand interactions with the lowest energy, and Pymol contributed to the visualization (Figure 5B). These results indicated that potential pivotal ingredients of TMDCD had excellent abilities to spontaneously bind the receptor proteins of AA.
 |
Table 3 Binding Energy Between Receptor Proteins and Ligands from Molecular Docking Results (kcal/mol) |
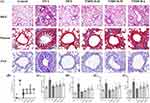
TMDCD Alleviated OVA-Induced Pulmonary Damages and AHR
In this study, H&E, PAS, and Masson staining were used to evaluate OVA-induced pulmonary damage. It was observed that OVA group mice showed typical pathological changes around the peribronchial and perivascular regions including airway epithelial defect, inflammatory cell especially eosinophil infiltration, mucus hypersecretion, airway smooth muscle cells (ASMs) proliferation, and collagen deposition, which suggested gradual progression of airway inflammation, goblet cell hyperplasia, and airway remodeling (Figure 6A). On the contrary, TMDCD attenuated these pathological changes and helped in decreasing inflammatory degrees, AREA%, collagen volume fractions, TAt%, and Wat/Pi (P<0.05) (Figure 6B–F). Some quantitative analysis results did not show statistical differences between the DEX group and the TMDCD-H/M/L group.
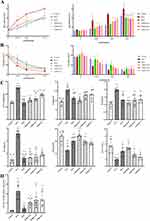
Besides, we used Buxco FinePointe RC system to measure the AHR of mice. It was indicated that OVA group mice showed a significant deterioration in different doses of methacholine induced RL and Cdyn (P<0.05), and they were attenuated by treating with TMDCD (P<0.05) (Figure 7A and B). These data suggested that TMDCD could decrease airway inflammation, airway remodeling, and AHR in asthmatic mice.
TMDCD Mitigated OVA-Induced Type 2 Inflammations
Usually, AA was induced by Th2 and characterized by type 2 inflammation. Hence, we measured the level of Th2-related cytokines by ELISA. As illustrated in Figure 7C, TMDCD decreased the abnormal release of OVA-sIgE in serum and IL-4, IL-5, and IL-13 in BALF compared with the OVA group (P<0.001). Meanwhile, it increased the level of IFN-γ in BALF and the ratio of IFN-γ/IL-4 (P<0.05) (Figure 7C), which suggested that TMDCD might regulate the imbalance of Th1 and Th2. Furthermore, we evaluated the mRNA level of GATA-binding protein 3 (GATA3) to validate the inhibitory effects on the transcription factor of Th2-related cytokines. It was shown that oral administration of TMDCD significantly inhibited GATA3 mRNA relative expressions in OVA-induced asthmatic mice (Figure 7D), which were consistent with the above findings in cytokines. Collectively, TMDCD mitigated OVA-induced type 2 inflammation in a dose-dependent manner, especially TMDCD-H could be regarded as the best dose.
TMDCD Inhibited Airway Inflammations by Regulating TLR4-NLRP3 Pathway-Mediated Pyroptosis
Given airway inflammation was the main pathological manifestation of asthmatic mice, the inhibition of airway inflammation is conducive to decreasing airway remodeling and AHR. As mentioned earlier, we suggested the predicted mechanism of TMDCD against AA might be related to NLR and TLR signaling pathways and confirmed that TMDCD-H was the best dose. In this part, we evaluated levels of target genes and proteins of the TLR4-NLRP3 pathway-mediated pyroptosis in asthmatic mice and the effects of the TMDCD-H on regulating them.
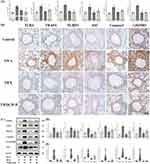
First of all, we measured the level of IL-1β and IL-18 in serum and BALF by ELISA, which represented the activation of TLR4-NLRP3 pathway-mediated pyroptosis. It was observed that the level of IL-1β and IL-18 in serum and BALF in the OVA group were all statistically higher than that in the control group (P<0.001), while they were attenuated by treating with the TMDCD-H group (P<0.01) (Figure 8A). Besides, TMDCD-H also reduced the production of inflammatory cytokines such as IL-6 and TNF-ɑ, they were related to IL-1β and IL-18 (P<0.001) (Figure 8A). Secondly, we hypothesized that TMDCD-H might prevent IL-1β derived inflammations through the inhibition of TLR4-NLRP3 pathway-mediated pyroptosis, which played a key role in enlarging inflammatory signalings and tissue damage. As evidenced by IHC of the lung tissues in OVA-induced asthmatic mice, we proved that the main target proteins of TLR4-NLRP3 pathway-mediated pyroptosis, including TLR4, TRAF6, NLRP3, ASC, Caspase 1, and GSDMD, were obviously expressed and marked in brown around airway epithelial cells, airway basement membranes, alveolar cells, and vascular around regions. After applying TMDCD, these positive expressions were inhibited (Figure 8B). Similar findings were also observed by Western blot, these results all proved that TMDCD could downregulate the target protein level of TLR4-NLRP3 pathway-mediated pyroptosis (P<0.05) (Figure 8C and D). Further real-time PCR experiments proved that the respective relative gene expressions of the above target protein were reduced in the TMDCD group (P<0.05), so it was suggested that TMDCD could repress target gene transcriptions to inhibit expressions of target proteins (Figure 8E). Overall, these observations suggested that TMDCD inhibited airway inflammation by regulating TLR4-NLRP3 pathway-mediated pyroptosis.
Discussion
As a common phenotype of bronchial asthma, AA is supposed by conventional wisdom to belong to Th2-driven type 2-high asthma, adaptive immune responses exert prime functions in the initiation of chronic airway inflammations of AA.31 When asthmatic patients were challenged with allergens from the environment, the cleaved airway epithelial barrier promoted antigen-presenting cells (APCs) to present antigens and generate adaptive immunities, the crosstalk between the airway epithelial barrier and APCs is crucial to inhaled allergens in Th2 immunity.32 With increasing levels of cytokines like IL-4, IL-5, and IL-13 released by Th2 and type 2 innate lymphoid cells (ILC2s), a cascade of downstream events was driven including IgE-triggered hypersensitivity, developments and activations of inflammatory cells like eosinophils, goblet cell metaplasia, airway narrowing mediated by ASMs contractions, and remodeling of the epithelium and subepithelial matrix.33 Finally, airway inflammations, AHR, and airway remodeling were induced. GATA3 was recognized as the Th2-related specific transcription factor, knocking down or deletions of GATA3 resulted in diminished Th2 differentiation.34 As previous studies reported that GATA3 expressions were critical for maintaining Th2 identities,35 we explored the effects of TMDCD on regulations of GATA3 in asthmatic models and proved that TMDCD alleviated OVA-induced Th2-related inflammations, AHR, and airway remodeling by inhibiting GATA3 expressions to reduce Th2-related cytokines releases, which met with our previous findings.14
It is quite difficult to elucidate the potential effecting mechanisms of TCM formulas, but the development of the network pharmacology study in the recent two decades brings the dawn. By establishing the “compound–protein/gene–disease” network based on high-throughput data, network pharmacology reveals the regulation principles of small molecules, which helps to predict herb targets, understands the biological foundation of diseases, and identifies new biomarkers to distinguish different TCM syndromes.36,37 In this study, we found that the beneficial effects of TMDCD against AA might be related to NLR and TLR signaling pathways. TLR and NLR were located in the cell membrane and cytoplasm, respectively, which participated in the recognition of bacteria, viruses, and allergens. TLR4 and NLRP3 were the main components in TLR/NLR family and were widely distributed in many cells, including macrophages, airway epithelial cells (AECs), neutrophils, etc. When PAMP and DAMP were recognized by TLR4, the myeloid differentiation factor 88 dependent pathway was activated, which involved the IL-1R-associated kinases, TRAF-6, and mitogen-activated kinases (MAPK) and culminated with the transcription of nuclear factor kappa B (NFκB).38,39 Meanwhile, the NLRP3 inflammasome triggered Caspase 1-mediated ligand bound pro-IL-1β and pro-IL-18 to produce bioactive IL-1β and IL-18. The up-regulation of the TLR4-NLRP3 pathway led to innate immunities activations characterized by releases of cytokines and chemokines, such as IL-1β, IL-18, IL-25, IL-33, and thymic stromal lymphopoietin (TSLP), not only which directly promoted local innate inflammatory cells recruitments and infiltrations around the airway, such as mast cells, eosinophils, and neutrophils, but also enhanced APCs motilities and Th2/Th17 differentiation.8,40–43 In addition, TLR4-NLRP3 pathway activations also induced pyroptosis, a form of programmed pro-inflammatory cell death, which was first found to be a self-protection and defense behavior of the host cell when clearing infected cells.44 Different from apoptosis, excessive pyroptosis promoted inflammatory responses.44 GSDMD pores played as the effectors of pyroptosis. Cleaved by Caspase 1, N-GSDMD was integrated into the cell membrane to form a pore, which was large enough to release small proteins like IL-1β and to bring in sodium with water, finally resulting in the cell volume to increase and membrane rupture.45 Our findings indicated TMDCD could repress TLR4-NLRP3 pathway-related gene transcriptions to inhibit expressions of target proteins and then decreased pyroptosis, which echoes hypotheses from the previous network pharmacology study. However, limited by funds, morphological changes of pyroptosis in different groups were not observed by the transmission electron microscope. We hope to conduct more experiments in the future.
Previous studies around the TLR4-NLRP3 pathway mainly focus on severe asthma and steroid-resistant asthma,10,46 and rarely on classical AA. Some studies claimed that pyroptosis was found in toluene diisocyanate-induced asthma.47 In this study, we proved that TLR4-NLRP3 pathway-related proteins and gene expressions were statistically increased in the OVA-induced AA mice model, which supported related studies.7,48 Further experiments proved the effects of TMDCD on alleviating these pathological changes. Results from our previous studies indicated that TMDCD had effects on down-regulating differential expression proteins such as IL-25 and TSLP, which were representative innate immune cytokines.15 Given TMDCD might act as the negative regulator of innate immune cytokines, we carried out this study for further verification. Besides, IL-1β and IL-18 could promote Th17 differentiation, it was found in this study TMDCD could reduce the level of IL-1β and IL-18 in serum and BALF by inhibiting TLR4-NLRP3 pathway activations, which meets with our previous findings about Th17 percentage in the spleen and RORγt mRNA level in the lung were decreased after applying TMDCD on the BALB/c eosinophilic asthma model,14 supporting our findings in another way.
This study has some limitations. We just focused on the classical TLR4-NLRP3 pathway-mediated pyroptosis, while non-classical pyroptosis pathway players were omitted such as Caspase 11. Besides, this was just an exploring study, we planned to conduct more in vitro experiments around AECs or macrophages to prove the effects of TMDCD on regulating innate immune cells.
Conclusion
With the method of network pharmacology, we supposed the effecting mechanisms of TMDCD against AA were related to NLR and TLR pathways. Further experiments verified that TMDCD alleviated airway inflammations, AHR, and airway remodeling in the OVA-induced asthmatic mice model by regulating TLR4-NLRP3 pathway-mediated pyroptosis. In this paper, we claimed that TMDCD could be a complementary and alternative therapy for AA.
Abbreviations
AA, allergic asthma; AECs, airway epithelial cells; AHR, airway hyperresponsiveness; ANOVA, analysis of variance; APCs, antigen-presenting cells; AREA%, positive area of goblet cell; ASC, apoptosis-associated speck-like protein containing CARD; ASMs, airway smooth muscle cells; BALF, bronchoalveolar lavage fluid; BC, betweenness centrality; BPs, biological processes; Caspase, cysteinyl aspartate-specific proteinase; CC, closeness centrality; CCs, cell components; Cdyn, dynamic pulmonary compliance; CHM, Chinese herbal medicine; CT, Chantui; DAMPs, damage-associated molecular patterns; DC, degree; DEX, dexamethasone; DL, drug likeness; ELISA, enzyme-linked immunosorbent assay; FF, Fangfeng; GATA3, GATA-binding protein 3; GC, Gancao; GO, Gene Ontology; H&E, hematoxylin/eosin; HRP, horseradish peroxidase; ICS, inhaled corticosteroid; IFN, interferon; IgE, immunoglobulin E; IHC, immunohistochemistry; IL, interleukin; ILC2s, type 2 innate lymphoid cells; JC, Jiangcan; JQM, Jinqiaomai; KEGG, Kyoto Encyclopedia of Genes and Genomes; LSD, least-significant difference; LZ, Lingzhi; MFs, molecular functions; MH, Mahuang; NLR, NOD-like receptor; NLRP, NLR family pyrin domain-containing; OB, oral bioavailability; OVA, ovalbumin; PAMPs, pathogen-associated molecular patterns; PAS, periodic acid Schiff; PPI, protein–protein interactions; PRRs, pattern recognition receptors; PVDF, polyvinylidene fluoride; Real-time PCR, real-time quantitative polymerase chain reaction; RIPA, radio-immunoprecipitation assay; RL, pulmonary resistance; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; SEM, standard error of the mean; SG, Shigao; SWT, Shouwuteng; TAt%, the percentage of Wat to the area enclosed by outer airway perimeter; TBST, tris-buffered saline buffer with Tween 20; TCM, traditional Chinese medicine; Th, helper T cell; TLR, Toll-like receptor; TM, Tianma; TMDCD, Tuo-Min-Ding-Chuan Decoction; TNF, tumor necrosis factor; TRAF6, tumor necrosis factor receptor associated factor 6; Tregs, regulatory T cells; TSLP, thymic stromal lymphopoietin; Wat/Pi, the percentage of Wat to internal airway perimeter; Wat, total airway wall area; WM, Wumei; XR, Xingren; DAB, 3, 3`-diaminobenzidine.
Data Sharing Statement
All the relevant data is provided within the paper and its supporting information files. The datasets analysed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
All authors would like to thank Dr Wenchao Dan for his kindness help in the work of network pharmacology.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine [No: ZYYCXTD-C-202001].
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Huang K, Yang T, Xu J, et al. Prevalence, risk factors, and management of asthma in China: a national cross-sectional study. Lancet. 2019;394(10196):407–418. doi:10.1016/S0140-6736(19)31147-X
2. Stern J, Pier J, Litonjua AA. Asthma epidemiology and risk factors. Semin Immunopathol. 2020;42(1):5–15. doi:10.1007/s00281-020-00785-1
3. Xie M, Wenzel SE. A global perspective in asthma: from phenotype to endotype. Chin Med J. 2013;126(1):166–174.
4. Respiratory Allergy Group of Chinese Society of Allergy, Asthma Group of Chinese Thoracic Society, Chinese Medical Association. 中国过敏性哮喘诊治指南(第一版,2019年 [Chinese guidelines for the diagnosis and treatment of allergic asthma (2019, the first edition)]. Zhonghua Nei Ke Za Zhi. 2019;58(9):636–655. Chinese. doi:10.3760/cma.j.issn.0578-1426.2019.09.004
5. Eisenbarth SC. The innate and adaptive immune systems in allergy: a two-way street. Clin Exp Allergy. 2006;36(2):135–137. doi:10.1111/j.1365-2222.2006.02425.x
6. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi:10.1016/j.cell.2010.01.022
7. Chen X, Xiao Z, Jiang Z, Jiang Y, Li W, Wang M. Schisandrin B attenuates airway inflammation and airway remodeling in asthma by inhibiting NLRP3 inflammasome activation and reducing pyroptosis. Inflammation. 2021;44(6):2217–2231. doi:10.1007/s10753-021-01494-z
8. Perros F, Lambrecht BN, Hammad H. TLR4 signalling in pulmonary stromal cells is critical for inflammation and immunity in the airways. Respir Res. 2011;12:125. doi:10.1186/1465-9921-12-125
9. Wang K, Sun Q, Zhong X, et al. Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell. 2020;180(5):941–955.e20. doi:10.1016/j.cell.2020.02.002
10. Kim RY, Pinkerton JW, Essilfie AT, et al. Role for NLRP3 inflammasome-mediated, IL-1β-dependent responses in severe, steroid-resistant asthma. Am J Respir Crit Care Med. 2017;196(3):283–297. doi:10.1164/rccm.201609-1830OC
11. Zasłona Z, Flis E, Wilk MM, et al. Caspase-11 promotes allergic airway inflammation. Nat Commun. 2020;11(1):1055. doi:10.1038/s41467-020-14945-2
12. Ge X, Cai F, Shang Y, et al. PARK2 attenuates house dust mite-induced inflammatory reaction, pyroptosis and barrier dysfunction in BEAS-2B cells by ubiquitinating NLRP3. Am J Transl Res. 2021;13(1):326–335.
13. Lyu M, Wang Y, Chen Q, et al. Molecular mechanism underlying effects of wumeiwan on steroid-dependent asthma: a network pharmacology, molecular docking, and experimental verification study. Drug Des Devel Ther. 2022;16:909–929. doi:10.2147/DDDT.S349950
14. Zhou Y, Zhao H, Wang T, Zhao X, Wang J, Wang Q. Anti-inflammatory and anti-asthmatic effects of TMDCT decoction in eosinophilic asthma through Treg/Th17 balance. Front Pharmacol. 2022;13:819728. doi:10.3389/fphar.2022.819728
15. Qin J, Lv M, Jiang Z, et al. Tuo-Min-Ding-Chuan decoction alleviate ovalbumin-induced allergic asthma by inhibiting mast cell degranulation and down-regulating the differential expression proteins. Front Pharmacol. 2021;12:725953. doi:10.3389/fphar.2021.725953
16. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6(1):13. doi:10.1186/1758-2946-6-13
17. Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1):42717. doi:10.1038/srep42717
18. Ye H, Ye L, Kang H, et al. HIT: linking herbal active ingredients to targets. Nucleic Acids Res. 2011;39(Database):D1055–D1059. doi:10.1093/nar/gkq1165
19. Li X, Tang H, Tang Q, Chen W. Decoding the mechanism of huanglian jiedu decoction in treating pneumonia based on network pharmacology and molecular docking. Front Cell Dev Biol. 2021;9:638366. doi:10.3389/fcell.2021.638366
20. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(D1):D789–D798. doi:10.1093/nar/gku1205
21. Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48(D1):D845–D855. doi:10.1093/nar/gkz1021
22. Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. doi:10.1002/cpbi.5
23. Wang Y, Zhang S, Li F, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031–D1041. doi:10.1093/nar/gkz981
24. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
25. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi:10.1093/nar/gky1131
26. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
27. Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi:10.1002/jcc.21256
28. Burley SK, Bhikadiya C, Bi C, et al. RCSB protein data bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021;49(D1):D437–D451. doi:10.1093/nar/gkaa1038
29. Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. Jvenn: an interactive venn diagram viewer. BMC Bioinform. 2014;15(1):293. doi:10.1186/1471-2105-15-293
30. Li C, Du X, Liu Y, et al. A systems pharmacology approach for identifying the multiple mechanisms of action for the Rougui-Fuzi Herb pair in the treatment of cardiocerebral vascular diseases. Evid Based Complement Alternat Med. 2020;2020:5196302. doi:10.1155/2020/5196302
31. Fahy JV. Type 2 inflammation in asthma--present in most, absent in many. Nat Rev Immunol. 2015;15(1):57–65. doi:10.1038/nri3786
32. Deckers J, Branco madeira F, Hammad H. Innate immune cells in asthma. Trends Immunol. 2013;34(11):540–547. doi:10.1016/j.it.2013.08.004
33. Hammad H, Lambrecht BN. The basic immunology of asthma. Cell. 2021;184(6):1469–1485. doi:10.1016/j.cell.2021.02.016
34. Zhu J. T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine. 2015;75(1):14–24. doi:10.1016/j.cyto.2015.05.010
35. Nakayama T, Hirahara K, Onodera A, et al. Th2 cells in health and disease. Annu Rev Immunol. 2017;35(1):53–84. doi:10.1146/annurev-immunol-051116-052350
36. Wang X, Wang Z-Y, Zheng J-H, Li S. TCM network pharmacology: a new trend towards combining computational, experimental and clinical approaches. Chin J Nat Med. 2021;19(1):1–11. doi:10.1016/S1875-5364(21)60001-8
37. Zhang R, Zhu X, Bai H, Ning K. Network pharmacology databases for traditional Chinese medicine: review and assessment. Front Pharmacol. 2019;10:123. doi:10.3389/fphar.2019.00123
38. Aluri J, Cooper MA, Schuettpelz LG. Toll-like receptor signaling in the establishment and function of the immune system. Cells. 2021;10(6):6. doi:10.3390/cells10061374
39. Zakeri A, Russo M. Dual role of toll-like receptors in human and experimental asthma models. Front Immunol. 2018;9:1027. doi:10.3389/fimmu.2018.01027
40. Holgate ST. Innate and adaptive immune responses in asthma. Nat Med. 2012;18(5):673–683. doi:10.1038/nm.2731
41. Sanders NL, Mishra A. Role of interleukin-18 in the pathophysiology of allergic diseases. Cytokine Growth Factor Rev. 2016;32:31–39. doi:10.1016/j.cytogfr.2016.07.001
42. McAlees JW, Whitehead GS, Harley IT, et al. Distinct Tlr4-expressing cell compartments control neutrophilic and eosinophilic airway inflammation. Mucosal Immunol. 2015;8(4):863–873. doi:10.1038/mi.2014.117
43. Bruchard M, Rebé C, Derangère V, et al. The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat Immunol. 2015;16(8):859–870. doi:10.1038/ni.3202
44. Yu P, Zhang X, Liu N, Tang L, Peng C, Chen X. Pyroptosis: mechanisms and diseases. Signal Transduct Target Ther. 2021;6(1):128. doi:10.1038/s41392-021-00507-5
45. Kovacs SB, Miao EA. Gasdermins: effectors of pyroptosis. Trends Cell Biol. 2017;27(9):673–684. doi:10.1016/j.tcb.2017.05.005
46. Brusselle GG, Provoost S, Bracke KR, Kuchmiy A, Lamkanfi M. Inflammasomes in respiratory disease: from bench to bedside. Chest. 2014;145(5):1121–1133. doi:10.1378/chest.13-1885
47. Zhuang J, Cui H, Zhuang L, et al. Bronchial epithelial pyroptosis promotes airway inflammation in a murine model of toluene diisocyanate-induced asthma. Biomed Pharmacother. 2020;125:109925. doi:10.1016/j.biopha.2020.109925
48. Hui C, Liu X. Regulatory effect of NLRP3 on airway inflammatory response and pyroptosis in mice with asthma. Zhongguo Dang Dai Er Ke Za Zhi. 2021;23(9):959–964. doi:10.7499/j.issn.1008-8830.2106107
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.