Back to Journals » Journal of Hepatocellular Carcinoma » Volume 10
TRIM55 Promotes Proliferation of Hepatocellular Carcinoma Through Stabilizing TRIP6 to Activate Wnt/β-Catenin Signaling
Authors Lu X, Yuan Y, Cai N, Rao D, Chen M, Chen X, Zhang B, Liang H, Zhang L
Received 20 April 2023
Accepted for publication 19 July 2023
Published 3 August 2023 Volume 2023:10 Pages 1281—1293
DOI https://doi.org/10.2147/JHC.S418049
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Imam Waked
Xun Lu,1– 3 Yue Yuan,4 Ning Cai,1– 3 Dean Rao,1– 3 Min Chen,4 Xiaoping Chen,1– 3 Bixiang Zhang,1– 3 Huifang Liang,1– 3 Lei Zhang3,5
1Hepatic Surgery Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China; 2Hubei Key Laboratory of Hepato-Pancreato-Biliary Diseases, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China; 3Clinical Medical Research Center of Hepatic Surgery, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China; 4Division of Gastroenterology, Department of Internal Medicine at Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China; 5Key Laboratory of Hepatobiliary and Pancreatic Diseases of Shanxi Province (Preparatory), Shanxi Bethune Hospital, Shanxi Academy of Medical Sciences, Shanxi Medical University; Shanxi Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Taiyuan, People’s Republic of China
Correspondence: Lei Zhang, Key Laboratory of Hepatobiliary and Pancreatic Diseases of Shanxi Province (Preparatory), Shanxi Bethune Hospital, Shanxi Academy of Medical Sciences, Shanxi Medical University; Shanxi Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Taiyuan, 030032, People’s Republic of China, Email [email protected] Huifang Liang, Hepatic Surgery Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, No. 1095 Jiefang Avenue, Wuhan, 430030, People’s Republic of China, Email [email protected]
Purpose: Tripartite motif containing 55 (TRIM55) is a member of the TRIM family and functions as an E3 ubiquitin ligase. It acts as a cancer promoter or suppressor in the malignant processes of multiple cancers. However, its proliferative function in hepatocellular carcinoma (HCC) has been poorly studied, and its underlying molecular mechanism remains unclear. In the present study, we investigated the role of TRIM55 in HCC and its mechanism of promoting HCC proliferation.
Materials and Methods: Protein expression levels of TRIM55 were measured in paired HCC and normal tissue samples using immunohistochemical (IHC) staining. The correlation between TRIM55 and clinical features was evaluated by statistical analysis. At the same time, overexpression and knockdown experiments, cycloheximide (CHX) interference experiments, ubiquitination, co-immunoprecipitation and immunofluorescence staining experiments, as well as animal experiments were used to evaluate the potential mechanism that TRIM55 promotes proliferation of hepatocellular carcinoma in vitro and in vivo.
Results: TRIM55 expression in HCC specimens was higher compared with the corresponding non-tumor tissues. The overall survival and disease-free survival time of patients with high TRIM55 expression were shorter than those with low expression of TRIM55. Functionally, TRIM55 promoted the proliferation of HCC cells and accelerated the growth of HCC xenografts. Mechanistically, TRIM55 interacted with thyroid receptor interacting protein 6 (TRIP6) and regulate its stability by influencing the ubiquitination process, thereby affecting the Wnt signaling pathway.
Conclusion: Our results indicate that TRIM55 promotes HCC proliferation by activating Wnt signaling pathways by stabilizing TRIP6. Therefore, targeting TRIM55 may be an effective therapeutic strategy to inhibit HCC growth.
Keywords: TRIM55, TRIP6, hepatocellular carcinoma, proliferation
Introduction
Worldwide, hepatocellular carcinoma (HCC) is the fourth leading cause of death from tumors and ranks sixth in the number of incident cases.1,2 Based on annual projections, the World Health Organization guesses that more than one million patients will die of liver cancer by 2030.3 As we all know, the current treatment is mainly surgery, but the prognosis is still not ideal.2,4 Hence, molecular targeted therapy has become a strategy for the prevention and treatment of HCC.5,6 The molecular mechanism of HCC needs further study.
Tripartite motif containing 55 (TRIM55) is a Protein Coding gene. It was reported that TRIM55 is involved in muscle sarcomere assembly and functions as an E3 ubiquitin ligase.7,8 Most of the tripartite motif-containing (TRIM) family proteins have E3 ubiquitin ligase activity.9 More and more researches have shown that TRIM proteins negatively and positively regulate carcinogenesis.10 Changes of TRIM protein expression are closely related to the malignancy of cancers and prognosis.11 Nevertheless, there are few studies on the effect of TRIM55 in tumors. From previous studies, it is known that overexpression of TRIM55 inhibited the migration and invasion of HCC cells.12 However, the effect of TRIM55 on HCC cell proliferation is unclear.
Ubiquitination is an essential posttranslational modification that covalently links the 76-amino acid ubiquitin protein to a target protein.13 Ubiquitination largely determines the stability and activity of proteins.14 Single ubiquitin molecules have seven surface Lys residues (K6, K11, K27, K29, K33, K48, and K63), and ubiquitin molecules can be sequentially added to one of their own Lys residues, known as polyubiquitination.15 Various types of chains play different roles in different cellular processes, the most widely studied of which are K48, K63, and linear ubiquitin chains. As TRIM55 is an E3 ubiquitin ligase, the specific mechanism of ubiquitination is unknown.
In this study, we concentrated on the role and mechanism of TRIM55 in HCC proliferation. Further functional studies demonstrated that TRIM55 significantly promoted HCC growth. The tumor-promoting effect of TRIM55 was associated with the activation of Wnt signaling pathway by binding to thyroid receptor interacting protein 6 (TRIP6). Binding of TRIM55 to TRIP6 in cytoplasm suppressed ubiquitin-mediated degradation of TRIP6 in an indirect way. Furthermore, TRIM55 overexpression was correlated with poor survival of patients with HCC. Therefore, this study revealed a novel mechanism of TRIM55 to promote proliferation in HCC, which is expected to be a new therapeutic target for HCC patients.
Materials and Methods
Patients and Tissue Specimens
A cohort of 110 pairs of paraffin-embedded HCC tissues and peritumor normal liver tissues were collected from HCC patients who performed hepatic resection from 2012 to 2016 at the Hepatic Surgery Center, Tongji Hospital. All patients provided written consent for the research. Ethical approval was obtained from the Ethics Committee of Tongji Hospital. The study was conducted in accordance with the Declaration of Helsinki.
Cell Lines and Culture Conditions
Normal-type hepatocyte HL7702 cells, HCC cell lines HepG2, Alex, HLF and MHCC-97H were obtained from the Hepatic Surgery Center, Tongji Hospital, Huazhong University of Science and Technology, Wuhan, China. All cell lines were approved by the Ethics Committee of Tongji Hospital. HEK 293 cells were purchased from China Center for Type Culture Collection. All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2.
Reagents and antibodies
Puromycin was purchased from Cayman Chemical (Ann Arbor, Michigan, USA). The protein A/G agarose were purchased from Sigma (St louis, MO, USA). cycloheximide (CHX) and Mg132 were obtained from MedChemExpress (Shanghai, China). Lipofectamine 3000 Reagent were purchased from Invitrogen (Life Technologies, Carlsbad, CA, USA). All antibodies used are listed in Supplementary Table 1.
Plasmids and Lentivirus
To construct TRIM55 overexpressing cell lines, the human TRIM55 cDNA and negative control were cloned into the pLenti-CMV-Puro plasmid (Addgene #17448). Vectors containing Flag-tag were used as negative controls, respectively. To construct TRIM55-knockdown cell lines, the shRNAs targeting TRIM55 and negative control were cloned into the pLKO.1 vector (pLKO.1 puro, Addgene Plasmid #8453). Recombinant vectors encoding TRIM55 and TRIP6 were sub‐cloned into pcDNA3.1 plasmids inserted by HA-, Myc- or Flag-tagged. Lentiviruses were generated in 293T cells by co-transfection with psPAX2 (Addgene #12260), pMD2.G (Addgene#12259) and pLKO.1-shRNA or pLenti-CMV-Puro plasmids. The production of viruses, infection and establishment of stable cell clones have been described previously.16 The target sequence of the specific gene was as follows: shTRIM55-1#:5’-GCTACTTCTCAGGAGTTAGTA-3’; shTRIM55-2#:5’-GCTTTGTGAGAAGTTTGATTA-3’;
Immunoblotting and Immunoprecipitation
Immunoblotting and co-immunoprecipitation (co-IP) assays were performed as described previously.17 In a word, cells were collected and lysed in immunoprecipitation (IP) lysis buffer with protease inhibitor and phosphatase inhibitor on ice for 30 min. The lysates were incubated with protein-A/G agarose for 2 h and then immunoprecipitated with the antibody overnight at 4°C. Then, the immune complex was precipitated with protein-A/G agarose and washed five times for Western blot analysis.
RNA Interference
SiRNA was transfected with Lipofectamine 3000 (Invitrogen). After transfection at 37°C for 48 h, proteins were collected and analyzed by Western blot. SiRNA targeting TRIP6 and negative controls were purchased from RiboBio (Guangzhou, China). The sequence of siRNA targeting TRIP6 was as follows: si-TRIP6: 5’-GGAGGAGACTGTGAGAATT-3’.
Cell Growth Assay
Cells were uniformly seeded into the 96-well plate and the medium was changed every 2 days. Cells were periodically detected with Cell Counting Kit-8 reagent (CCK-8, Dojindo Crop, Japan) according to the manufacturer’s instructions.18 For the colony formation assay, cells were uniformly seeded into each well of six-well plates. After 15 days, cells were stained with 1% crystal violet and photographed.
Ubiquitination Assay
In vitro TRIP6 ubiquitination assay, the plasmids HA-TRIP6, Flag-TRIM55 and Myc-Ub were transfected into 293T cells for 24 h and treated with 20 μM MG132 for 6 h. Then, cells were collected and lysed in IP lysis buffer with protease inhibitor on ice for 30 min. Immunoprecipitation was performed with anti-TRIP6 antibodies, followed by immunoblotting with corresponding antibodies.
Immunohistochemical Assay
Immunohistochemical (IHC) assay was performed as described previously.19–21 Immunohistochemical staining score was performed according to the percentage of positive staining tumor cells and staining intensity score. The score criteria of positive stained cells were as follows: less than 5% is 0, 5–25% is 1, 26–50% is 2, 51–75% is 3 and 76–100% is 4. In addition, the staining intensity was evaluated as follows: the score scale was 0 to 3, 0 is negative, 1 is light brown, 2 is brown and 3 is dark brown. The total score obtained by multiplying the above two was used to judge the level of TRIM55 and TRIP6 expression. Overall scores of >6 and ≤6 was defined as high level and low level, respectively.
Dual-Luciferase Reporter Assay
For the dual-luciferase reporter assay, cells were co-transfected with TOPFlash or FOPFlash luciferase reporter. Transfection efficiency was normalized by co-transfection with a pRL-TK reporter containing a Renilla luciferase gene. After 32 h transfection, Firefly luciferase and Renilla luciferase activities were detected using the dual-luciferase reporter assay system (Promega, Madison, WI).
Tumorigenicity Assay
All animals were cared for and used in accordance with the Guide for the Care and Use of Laboratory Animals. Male BALB/c nude mice (four-week-old) were raised under specific pathogen-free (SPF) conditions. The experimental protocol was approved by the Committee on the Ethics of Animal Experiments of the Tongji hospital. For the subcutaneous xenograft assay, 1×106 cells were injected into the axillary region of nude mice. For orthotopic implantation experiment, 2×106 cells were injected into the liver of nude mice. After four weeks, all of the nude mice were sacrificed. The tumor volume was measured and photographed. Then the tumor was fixed with 4% paraformaldehyde for immunohistochemical analysis.
Statistical Analysis
The GraphPad Prism 6.0 software was used for statistical analyses. The Chi-square test was used to analyze the relationship between the expression of TRIM55 and clinicopathological features. Survival analysis was performed using Kaplan-Meier method. Pearson’s correlation test was used to analyze the correlation between TRIM55 and TRIP6 expression in HCC tissues. The differences between the groups were analyzed by two-tailed Student’s t-test. P < 0.05 was considered statistically significant.
Results
TRIM55 Expression Was Upregulated in HCC
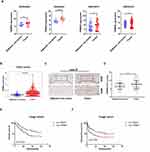
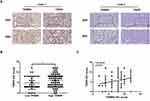
To explore the possible role of TRIM55 in HCC, we studied TRIM55 levels in tumor samples and adjacent non-tumor tissues from the GSE84005,22 GSE64041,23 GSE39791,24 GSE36376,25 and TCGA datasets. The results indicated that TRIM55 expression was upregulated in HCC tissues (Figure 1A and B). To further confirm the overexpression of TRIM55 in HCC, the expression of TRIM55 was examined by immunohistochemistry staining in a tissue microarray of 110 pairs of HCC and adjacent non-tumor tissues (Figure 1C). The results indicated that TRIM55 was upregulated in HCC specimens compared with the corresponding non-tumor tissues (Figure 1D). It was consistent with the above database results. In addition, we also explored the associations between TRIM55 and clinicopathological characteristics of HCC patients. The Chi-square test indicated that TRIM55 expression was correlated with serum AFP (P = 0.043) and tumor size (P = 0.039, Supplementary Table 2). Kaplan-Meier survival analysis indicated that patients with high expression of TRIM55 had poorer overall survival (OS: P = 0.023, Figure 1E) and disease-free survival (DFS: P = 0.039, Figure 1F) compared with low expression of TRIM55. Taken together, these data show that TRIM55 was upregulated in human HCC tissues and associated with poor prognosis of HCC patients.
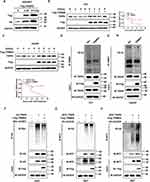
TRIM55 Promotes HCC Cell Proliferation in vitro and in vivo
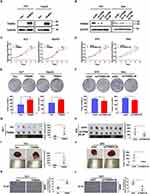
The overexpression of TRIM55 in HCC prompted us to explore its potential role in HCC cell proliferation. Hence, based on the expression of TRIM55 in HCC cell lines (Supplementary Figure 1), we constructed stable TRIM55 overexpression and knockdown HCC cell lines to investigate the cell proliferation ability. HLF cells and HepG2 cells were transfected with TRIM55 lentivirus expression vector. 97H and ALEX cells were transfected with a small hairpin RNA (shTRIM55) specifically targeting TRIM55. The efficiency of TRIM55 overexpression and knockdown is shown in Figure 2A and B. ShTRIM55-2# with higher knockout efficiency will be used for further studies. Then, we performed CCK8 and colony formation assays. Results showed that overexpression of TRIM55 in HLF and HepG2 cells significantly increased cell proliferation and clonogenicity (Figure 2C and E). On the contrary, knockdown of TRIM55 in 97H and ALEX cells significantly decreased cell proliferation and clonogenicity (Figure 2D and F). To further validate the effect of TRIM55 on tumor growth in vivo, a subcutaneous xenograft tumor model and an intrahepatic tumor implantation model in nude mice were employed. As expected, the TRIM55 overexpression group displayed significantly bigger tumors in nude mice compared with the control (Figure 2G and I), while downregulation of TRIM55 led to smaller tumors in nude mice (Figure 2H and J). Moreover, the IHC analysis revealed that Ki-67 expression in xenograft tumor tissue was enhanced in the overexpression group of TRIM55 compared with the control group (Figure 2K), while downregulation of TRIM55 led to decrease the expression of Ki-67 (Figure 2L). Taken together, these results indicate that TRIM55 acts as a tumor promoter in the development of HCC.
TRIM55 Interacts and Colocalizes with TRIP6
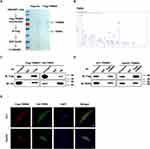
To identify novel TRIM55-mediated protein–protein interactions in HCC, we performed mass spectrometry. HEK293T cells transfected with FLAG-vector or FLAG-TRIM55 were immunoprecipitated with an anti-Flag antibody to identify TRIM55 candidate binding (Figure 3A and B), and then differentially expressed proteins were found (Supplementary Table 3). These differentially expressed proteins were detected by exogenous Co-IP in HEK293T cells. These results revealed that TRIM55 had a strong interaction with TRIP6 (Figure 3C). Their interaction was further confirmed by endogenous Co-IP experiments of HLF and HepG2 cells (Figure 3D). In addition, we also detected the intracellular distribution of TRIM55 and TRIP6 by confocal immunofluorescence microscopy. The result revealed that TRIM55 and TRIP6 were co-localized in the cytoplasm of HLF and HepG2 cells after transfection of Flag-TRIM55 and HA-TRIP6 (Figure 3E).
Tumor Promoting Effect of TRIM55 is Dependent on TRIP6
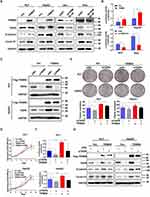
After confirming the interaction between TRIM55 and TRIP6, we investigated the effects of TRIM55 on TRIP6. We found that TRIM55 overexpression increased the protein expression of TRIP6 and downregulation of TRIM55 reduced the protein expression of TRIP6 (Figure 4A). Since TRIP6 can promoted cell proliferation and activated Wnt pathway,26 the effect of TRIM55 on the Wnt/β-Catenin pathway was evaluated by TOP/FOP luciferase reporter assay and Western blot assay. We found that TRIM55 overexpression significantly promoted the activity of Wnt pathway as demonstrated by TOP/FOP luciferase reporter assay, while TRIM55 downregulation reduced the activity of Wnt pathway (Figure 4B). Consistently, TRIM55 overexpression efficiently enhanced protein expression of β-Catenin and Cyclin D1 in HLF and HepG2 cells, while downregulation of TRIM55 decreased the protein level of β-Catenin and Cyclin D1 in 97H and Alex cells (Figure 4A). The results showed that TRIM55 enhanced the protein expression of TRIP6 and promoted the activation of the Wnt pathway.
To explore the effect of TRIP6 on the cell proliferation mediated by TRIM55, HLF and HepG2 cells with TRIM55-overexpression or control cells were transfected with siRNA against TRIP6 (Figure 4C). Knockdown of TRIP6 significantly decreased the promoting effect of TRIM55 on cell proliferation (Figure 4D) and clonogenicity (Figure 4E) ability of both HLF and HepG2 cells. In addition, we explored whether TRIP6 affected the promotion effect of TRIM55 on the Wnt pathway. Luciferase reporter assay confirmed that knockdown of TRIP6 also decreased the TRIM55-activated Wnt signaling in HLF and HepG2 cells (Figure 4F). Western blot analysis revealed that TRIP6 knockdown could inhibit the TRIM55-mediated upregulation of the Wnt pathway (Figure 4G, Supplementary Figure 2). Taken together, these results showed that tumor promoting effect of TRIM55 and its effect on Wnt pathway are dependent on TRIP6.
TRIM55 Stabilizes TRIP6 Protein by Inhibiting K48-Linked Ubiquitination
To further investigate how TRIP6 mediates the effect of TRIM55, we investigated the effect of TRIM55 on TRIP6 expression. We found that the expression of HA-TRIP6 in 293T cells with constant HA-TRIP6 transfection level increased with the increase of Flag-TRIM55 transfection level (Figure 5A). It indicated that TRIM55 increase the stability of TRIP6. To further confirm whether TRIM55 impacted the stability of TRIP6. We treated HLF and HepG2 cells with or without TRIM55 overexpression with the protein synthesis inhibitor CHX. TRIP6 was more stable in HLF and HepG2 cells with TRIM55 overexpression (Figure 5B and C). As TRIM55 is an E3 ubiquitin ligase, we then investigated the effect of TRIM55 on TRIP6 ubiquitination. Therefore, we implemented a ubiquitination experiment and found that TRIM55 overexpression was accompanied by decreased ubiquitination in HLF and HepG2 cells (Figure 5D and E). The in vitro ubiquitination assay also verified that TRIM55 decreased the ubiquitination of TRIP6 (Figure 5F). The major types of ubiquitin chains are K48- and K63-linked polyubiquitin chains: the polyubiquitination of K48-linked mainly induces the degradation of target proteins, while the polyubiquitination of K63-linked mainly regulates the activation of target proteins.27 Therefore, we further investigated the type of TRIP6 ubiquitination regulated by TRIM55. 293T cells were co-transfected with Flag-TRIM55, Myc-TRIP6 and K48 or K63 ubiquitin plasmids. As shown in Figure 5G and H, TRIM55 inhibited the K48-linked ubiquitination of TRIP6 and promoted the K63-linked ubiquitination of TRIP6. In short, these results indicated that TRIM55 may bind to TRIP6 and increase the stability of TRIP6 by inhibiting ubiquitin-mediated degradation of TRIP6 protein in an indirect way.
TRIP6 Was Positively Correlated with TRIM55 Expression in HCC Tissues
To further explore the relationship between TRIM55 and TRIP6 in human HCC, we detected the expression of TRIM55 and TRIP6 in a tissue microarray of 110 pairs of HCC tumor samples by IHC staining (Figure 6A). We found that the TRIP6 mean score of high TRIM55 expression group was significantly higher than that of low expression group (Figure 6B). Additionally, we analyzed the correlation between TRIM55 and TRIP6 expression. These results showed that TRIP6 expression was positively correlated with TRIM55 expression in HCC tissues (Figure 6C).
In summary, these data suggest that TRIM55 promotes proliferation of hepatocellular carcinoma through stabilizing TRIP6 to activate Wnt/β-catenin signaling (Supplementary Figure 3).
Discussion
TRIM proteins have been shown to play a wide range of functions in various tumor processes. Whether TRIM family proteins act as oncogenes or tumor suppressors depends on the specific signaling pathway and tumor type.10 Affecting different signaling pathways can have different effects. For instance, TRIM7 and TRIM29 exhibit opposite effects in HCC according to different studies.28–31 They negatively or positively regulate tumorigenesis by influencing signaling pathways. A previous study revealed that overexpression of TRIM55 inhibited the migration and invasion of HCC cell via EMT and MMP2 pathway. However, this study found that TRIM55 promoted HCC growth by activating the Wnt/β-catenin signaling pathway. It has been well documented that activation of the Wnt/β-catenin signaling pathway promotes HCC cell proliferation, migration and invasion.32 In addition, tumor type affects whether TRIM family proteins act as oncogenes or tumor suppressors.10 The expression level and function of TRIM55 are different in the development of different human malignancies. This evidence may support the dual role of TRIM55 as an oncogene or tumor suppressor gene, depending on the tumor type. The TCGA dataset and our study show that TRIM55 expression was upregulated in HCC tissues. However, a previous study revealed TRIM55 was downregulated in lung adenocarcinoma and colorectal cancer. Thus, TRIM55 may have different roles in different types of tumors. To sum up, TRIM55 acts as oncogenes or tumor suppressors depending on the specific signaling pathway and tumor type.
To explore the molecular mechanism of TRIM55, TRIP6 was identified as an interacting partner of TRIM55 by mass spectrometry analysis. The interaction between TRIM55 and TRIP6 was verified by Western blot analysis of co-IP products. The co-localization of TRIM55 and TRIP6 in cytoplasm was confirmed by confocal immunofluorescence microscopy. TRIP6 has been reported to promote tumor growth in several cancers and is involved in Wnt signaling pathway.33–35 In keeping with this, we found that knockdown of TRIP6 in HCC cells could decrease the effects of TRIM55 on cell proliferation, colony formation and Wnt signaling. It is inferred that tumor promoting effect of TRIM55 in HCC is, at least in part, dependent on TRIP6.
Our study demonstrated that TRIM55 stabilized TRIP6 protein by inhibiting the K48-linked polyubiquitin state of TRIP6. However, TRIM55, as an E3 ubiquitin ligase, degrades proteins mainly through ubiquitination.36,37 This result seems to make it difficult to explain its E3 ligase activity. This suggest that TRIM55 indirectly interact with TRIP6. So, we speculate that TRIM55-TRIP6 complex may recruit lipid or proteins that inhibit ubiquitin-ligase activity, thereby inhibiting ubiquitin-mediated degradation of TRIP6.38 The specific mechanism by which this process is inhibited needs to be further studied.
TRIM55 is a member of the TRIM family, and the proteins of this family have been reported to play important roles in cancer progression. Overexpression of many proteins was significantly correlated with clinicopathological features, such as tumor size and AFP levels. These factors are important reasons leading to poor clinical prognosis.39 In this study, we found that TRIM55 was upregulated in human HCC tissues and significantly correlated with tumor size, serum AFP and poor prognosis in HCC patients. Moreover, further results indicated that overexpression of TRIM55 significantly increased cell proliferation and clonogenicity. Even though therapeutic targeting of TRIM55 is an underdeveloped area of research, all these data suggest that overexpression of TRIM55 may promote the development of tumor, suggesting TRIM55 as a potential target in cancer therapy.
In general, our study reveals a new mechanism whereby TRIM55 plays a role in the pathogenesis and proliferation of HCC by binding to and stabilizing TRIP6 as a tumor-promoting factor. Our findings provide new insights into the development of HCC and provide potential therapeutic targets for the treatment and management of HCC.
Data Sharing Statement
All data generated or analyzed during this study are included in the manuscript and its additional files.
Ethics Approval and Consent to Participate
Animal experiments in our study were approved by the Medical Ethics Committee of Wuhan Tongji hospital (TJH-202212013).
Funding
This study was funded by the National Natural Science Foundation of China (No. 81874189, 81202300 and 82073090), the National Key Research and Development Program of China (2018YFA0208904) and Shanxi Province“136” Revitalization Medical Project Construction Funds.
Disclosure
The authors declare no competing interest in this work.
References
1. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. doi:10.1038/s41572-020-00240-3
2. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi:10.1038/s41575-019-0186-y
3. World Health Organization. Projections of mortality and causes of death, 2016 to 2060. Available from: http://www.who.int/healthinfo/global_burden_disease/projections/en/.
4. Zhou H, Song T. Conversion therapy and maintenance therapy for primary hepatocellular carcinoma. Biosci Trends. 2021;15(3):155–160. doi:10.5582/bst.2021.01091
5. Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi:10.1056/NEJMra1713263
6. Wu J, Yang S, Xu K, et al. Patterns and trends of liver cancer incidence rates in Eastern and SouthEastern Asian Countries (1983–2007) and Predictions to 2030. Gastroenterology. 2018;154(6):1719–1728 e1715. doi:10.1053/j.gastro.2018.01.033
7. Perera S, Mankoo B, Gautel M. Developmental regulation of MURF E3 ubiquitin ligases in skeletal muscle. J Muscle Res Cell Motil. 2012;33(2):107–122. doi:10.1007/s10974-012-9288-7
8. Perera S, Holt MR, Mankoo BS, Gautel M. Developmental regulation of MURF ubiquitin ligases and autophagy proteins nbr1, p62/SQSTM1 and LC3 during cardiac myofibril assembly and turnover. Dev Biol. 2011;351(1):46–61. doi:10.1016/j.ydbio.2010.12.024
9. Hu MM, Shu HB. Multifaceted roles of TRIM38 in innate immune and inflammatory responses. Cell Mol Immunol. 2017;14(4):331–338. doi:10.1038/cmi.2016.66
10. Huang N, Sun X, Li P, et al. TRIM family contribute to tumorigenesis, cancer development, and drug resistance. Exp Hematol Oncol. 2022;11(1):75. doi:10.1186/s40164-022-00322-w
11. Hatakeyama S. trim family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. doi:10.1016/j.tibs.2017.01.002
12. Li X, Huang L, Gao W. Overexpression of Tripartite Motif Containing 55 (TRIM55) inhibits migration and invasion of Hepatocellular Carcinoma (HCC) cells via epithelial-mesenchymal transition and Matrix Metalloproteinase-2 (MMP2). Med Sci Monit. 2019;25:771–777. doi:10.12659/MSM.910984
13. Cockram PE, Kist M, Prakash S, Chen SH, Wertz IE, Vucic D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021;28(2):591–605. doi:10.1038/s41418-020-00708-5
14. Li Q, Zhang W. Progress in anticancer drug development targeting ubiquitination-related factors. Int J Mol Sci. 2022;23(23):15104.
15. Yang K, Xiao W. Functions and mechanisms of the Ubc13-UEV complex and lysine 63-linked polyubiquitination in plants. J Exp Bot. 2022;73(16):5372–5387. doi:10.1093/jxb/erac239
16. Liu F, Liao Z, Qin L, et al. Targeting VPS72 inhibits ACTL6A/MYC axis activity in hepatocellular carcinoma progression. Hepatology. 2023. doi:10.1097/HEP.0000000000000268
17. Liao Z, Chen L, Zhang X, et al. PTPRepsilon acts as a metastatic promoter in hepatocellular carcinoma by facilitating recruitment of SMAD3 to TGF-beta receptor 1. Hepatology. 2020;72(3):997–1012. doi:10.1002/hep.31104
18. Zhu H, Zhang H, Pei Y, et al. Long non-coding RNA CCDC183-AS1 acts AS a miR-589-5p sponge to promote the progression of hepatocellular carcinoma through regulating SKP1 expression. J Exp Clin Cancer Res. 2021;40(1):57. doi:10.1186/s13046-021-01861-6
19. Song J, Liu Y, Liu F, et al. The 14-3-3 sigma protein promotes HCC anoikis resistance by inhibiting EGFR degradation and thereby activating the EGFR-dependent ERK1/2 signaling pathway. Theranostics. 2021;11(3):996–1015. doi:10.7150/thno.51646
20. Liu X, Su K, Sun X, et al. Sec62 promotes stemness and chemoresistance of human colorectal cancer through activating Wnt/beta-catenin pathway. J Exp Clin Cancer Res. 2021;40(1):132. doi:10.1186/s13046-021-01934-6
21. Hu C, Xin Z, Sun X, et al. Activation of ACLY by SEC63 deploys metabolic reprogramming to facilitate hepatocellular carcinoma metastasis upon endoplasmic reticulum stress. J Exp Clin Cancer Res. 2023;42(1):108. doi:10.1186/s13046-023-02656-7
22. Wang J, Peng R, Zhang Z, Zhang Y, Dai Y, Sun Y. Identification and validation of key genes in hepatocellular carcinoma by bioinformatics analysis. Biomed Res Int. 2021;2021:6662114. doi:10.1155/2021/6662114
23. Zhou H, Jiang L, Wang G, Su L, Hou L, Xue X. Identification of MCM4 as a prognostic marker of hepatocellular carcinoma. Biomed Res Int. 2021;2021:7479326. doi:10.1155/2021/7479326
24. Rao B, Li J, Ren T, et al. RPL19 is a prognostic biomarker and promotes tumor progression in hepatocellular carcinoma. Front Cell Dev Biol. 2021;9:686547. doi:10.3389/fcell.2021.686547
25. Hou Y, Wang Z, Huang S, et al. SKA3 Promotes tumor growth by regulating CDK2/P53 phosphorylation in hepatocellular carcinoma. Cell Death Dis. 2019;10(12):929. doi:10.1038/s41419-019-2163-3
26. Gou H, Liang JQ, Zhang L, et al. TTPAL promotes colorectal tumorigenesis by stabilizing trip6 to activate wnt/beta-catenin signaling. Cancer Res. 2019;79(13):3332–3346. doi:10.1158/0008-5472.CAN-18-2986
27. van WijkSJ, Fulda S, Dikic I, Heilemann M, van Wijk SJ. Visualizing ubiquitination in mammalian cells. EMBO Rep. 2019;20(2). doi:10.15252/embr.201846520
28. Zhu L, Qin C, Li T, et al. The E3 ubiquitin ligase TRIM7 suppressed hepatocellular carcinoma progression by directly targeting Src protein. Cell Death Differ. 2020;27(6):1819–1831. doi:10.1038/s41418-019-0464-9
29. Hu X, Tang Z, Ma S, Yu Y, Chen X, Zang G. Tripartite motif-containing protein 7 regulates hepatocellular carcinoma cell proliferation via the DUSP6/p38 pathway. Biochem Biophys Res Commun. 2019;511(4):889–895. doi:10.1016/j.bbrc.2019.02.001
30. Xu M, Hu J, Zhou B, Zhong Y, Lin N, Xu R. TRIM29 prevents hepatocellular carcinoma progression by inhibiting Wnt/beta-catenin signaling pathway. Acta Biochim Biophys Sin. 2019;51(1):68–77. doi:10.1093/abbs/gmy151
31. Du H, Xu Q, Xiao S, et al. MicroRNA-424-5p acts as a potential biomarker and inhibits proliferation and invasion in hepatocellular carcinoma by targeting TRIM29. Life Sci. 2019;224:1–11. doi:10.1016/j.lfs.2019.03.028
32. Huang G, Liang M, Liu H, et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/beta-catenin pathway. Cell Death Dis. 2020;11(12):1065. doi:10.1038/s41419-020-03276-1
33. Gur’ianova OA, Sablina AA, Chumakov PM, Frolova EI. Down-regulation of TRIP6 expression induces actin cytoskeleton rearrangements in human carcinoma cell lines. Mol Biol. 2005;39(5):905–909.
34. Zhao W, Dai Y, Dai T, et al. TRIP6 promotes cell proliferation in hepatocellular carcinoma via suppression of FOXO3a. Biochem Biophys Res Commun. 2017;494(3–4):594–601. doi:10.1016/j.bbrc.2017.10.117
35. Lin VT, Lin FT. TRIP6: an adaptor protein that regulates cell motility, antiapoptotic signaling and transcriptional activity. Cell Signal. 2011;23(11):1691–1697. doi:10.1016/j.cellsig.2011.06.004
36. Lu K, Pan Y, Huang Z, Liang H, Ding ZY, Zhang B. TRIM proteins in hepatocellular carcinoma. J Biomed Sci. 2022;29(1):69. doi:10.1186/s12929-022-00854-7
37. Zhan W, Zhang S. TRIM proteins in lung cancer: mechanisms, biomarkers and therapeutic targets. Life Sci. 2021;268:118985. doi:10.1016/j.lfs.2020.118985
38. Chen SY, Zhang HP, Li J, et al. Tripartite motif-containing 27 attenuates liver ischemia/reperfusion injury by suppressing Transforming Growth Factor beta-Activated Kinase 1 (TAK1) by TAK1 binding protein 2/3 degradation. Hepatology. 2021;73(2):738–758. doi:10.1002/hep.31295
39. Dai CY, Lin CY, Tsai PC, et al. Impact of tumor size on the prognosis of hepatocellular carcinoma in patients who underwent liver resection. J Chin Med Assoc. 2018;81(2):155–163. doi:10.1016/j.jcma.2017.06.018
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.