Back to Journals » Clinical Epidemiology » Volume 15
Transforming the Information System for Research in Primary Care (SIDIAP) in Catalonia to the OMOP Common Data Model and Its Use for COVID-19 Research
Authors Raventós B , Fernández-Bertolín S, Aragón M , Voss EA , Blacketer C, Méndez-Boo L , Recalde M, Roel E, Pistillo A , Reyes C, van Sandijk S, Halvorsen L , Rijnbeek PR, Burn E , Duarte-Salles T
Received 23 May 2023
Accepted for publication 3 August 2023
Published 13 September 2023 Volume 2023:15 Pages 969—986
DOI https://doi.org/10.2147/CLEP.S419481
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Vera Ehrenstein
Berta Raventós,1,2,* Sergio Fernández-Bertolín,1,* María Aragón,1 Erica A Voss,3– 5 Clair Blacketer,3– 5 Leonardo Méndez-Boo,6 Martina Recalde,1 Elena Roel,1,2 Andrea Pistillo,1,7 Carlen Reyes,1 Sebastiaan van Sandijk,8 Lars Halvorsen,9 Peter R Rijnbeek,4,5 Edward Burn,1,10 Talita Duarte-Salles1,4
1Fundació Institut Universitari per a la recerca a l’Atenció Primària de Salut Jordi Gol i Gurina (IDIAPJGol), Barcelona, Spain; 2Universitat Autònoma de Barcelona, Bellaterra (Cerdanyola del Vallès), Barcelona, Spain; 3Janssen Pharmaceutical Research and Development, Titusville, NJ, USA; 4Department of Medical Informatics, Erasmus University Medical Center, Rotterdam, the Netherlands; 5OHDSI Collaborators, Observational Health Data Sciences and Informatics (OHDSI), New York, NY, USA; 6Sistemes d’Informació dels Serveis d’Atenció Primària (SISAP), Institut Català de la Salut, Barcelona, Spain; 7Universitat Pompeu Fabra, Barcelona, Spain; 8Odysseus Data Services s.r.o., Prague, Czech Republic; 9edenceHealth NV, Kontich, Belgium; 10Centre for Statistics in Medicine, University of Oxford, Oxford, UK
*These authors contributed equally to this work
Correspondence: Talita Duarte-Salles, Fundació Institut Universitari per a la recerca a l’Atenció Primària de Salut Jordi Gol i Gurina (IDIAPJGol), Gran Via Corts Catalanes, 587 àtic, Barcelona, 08007, Spain, Tel +34935824342, Email [email protected]
Purpose: The primary aim of this work was to convert the Information System for Research in Primary Care (SIDIAP) from Catalonia, Spain, to the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM). Our second aim was to provide a descriptive analysis of COVID-19-related outcomes among the general population.
Patients and Methods: We mapped patient-level data from SIDIAP to the OMOP CDM and we performed more than 3,400 data quality checks to assess its readiness for research. We established a general population cohort as of the 1st March 2020 and identified outpatient COVID-19 diagnoses or tested positive for, hospitalised with, admitted to intensive care units (ICU) with, died with, or vaccinated against COVID-19 up to 30th June 2022.
Results: After verifying the high quality of the transformed dataset, we included 5,870,274 individuals in the general population cohort. Of those, 604,472 had either an outpatient COVID-19 diagnosis or positive test result, 58,991 had a hospitalisation, 5,642 had an ICU admission, and 11,233 died with COVID-19. A total of 4,584,515 received a COVID-19 vaccine. People who were hospitalised or died were more commonly older, male, and with more comorbidities. Those admitted to ICU with COVID-19 were generally younger and more often male than those hospitalised and those who died.
Conclusion: We successfully transformed SIDIAP to the OMOP CDM. From this dataset, a general population cohort of 5.9 million individuals was identified and their COVID-19-related outcomes over time were described. The transformed SIDIAP database is a valuable resource that can enable distributed network research in COVID-19 and beyond.
Keywords: electronic health records, medical ontologies, secondary data use, common data model, OMOP
Introduction
Spain has been one of the European countries hit hardest by the ongoing Coronavirus disease 2019 (COVID-19) pandemic. The first COVID-19 cases in Spain were identified in late February 2020, and by the 1st June of that year there had been more than 32,000 COVID-19 deaths in the country.1 Further waves of infections have since followed and, although the advent of effective vaccines has dramatically improved the outlook, COVID-19 cases continue to accrue and the effects of the disease for many of the people previously infected are likely to be long-lasting.
Similar to many European countries, Spain has a universal coverage healthcare system with general practitioners (GPs) acting as the gatekeepers to care.2 This role has largely been maintained during the COVID-19 pandemic. Indeed, GPs have played a pivotal role throughout the pandemic as the first point of contact for COVID-19 confirmed and probable cases and for providing short- and long-term follow-up care to the majority of COVID-19 patients. With regard to the identification of potential cases, clinical diagnoses by primary care professionals played an important role during the first of the pandemic in Spain, with the use of SARS-CoV-2 testing initially restricted to the most severe cases, such as those hospitalised and groups considered to be at particularly high-risk, such as care home residents.3 Once vaccines became available, primary care professionals were also involved in delivering the COVID-19 immunisation campaign, which achieved one of the highest vaccination rates in the European Union.4
In such a context, primary care records can provide an important foundation for COVID-19 research, particularly when linkage to testing, hospitalisation, and vaccination data is available. The Information System for Research in Primary Care (SIDIAP; www.sidiap.org) is a primary care records database covering approximately 75% of the population of Catalonia, Spain. The provenance of the data has been well documented, and the population captured has been found to be representative in terms of age, sex, and geographic distribution.5 Data from SIDIAP has previously been used in a wide range of epidemiological research studies, including COVID-19 related research.6–9 Individual-level linkage of hospital data has previously been established for SIDIAP, and further linkage to SARS-CoV-2 test results and COVID-19 vaccine records is also possible. However, the usability of SIDIAP in federated analyses is limited due to issues surrounding interoperability, as the schemas and clinical terminologies often differ across healthcare systems and datasets. In order to standardize both the language and structure of health data, the Observational Health Data Sciences and Informatics (OHDSI) developed the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM),10 which has been adapted by numerous health care databases.11–14
Our first aim was to convert data from SIDIAP to the OMOP CDM to facilitate distributed network research related to the pandemic. Our second aim was to summarise the occurrence of COVID-19-related outcomes observed and describe the characteristics of those affected and vaccinated against this disease.
Materials and Methods
Overview
Primary care data collected in SIDIAP between 1st January 2006 (when the computerization of records was complete) and 30th June 2022 (the last available date of data collection) was linked, at a patient-level, to COVID-19 testing, hospitalisation, and vaccination data. The data were mapped to the OMOP CDM following an extract, transform, and load (ETL) process which we first describe below. Using these mapped data, a cohort of the general population was followed up from 1st March 2020, with COVID-19 outcomes (outpatient COVID-19 diagnoses and positive tests, hospitalisations with COVID-19, ICU admissions with COVID-19, and COVID-19 deaths) and first dose vaccination against COVID-19 observed over follow-up until the 30th June 2022.
Mapping to the OMOP CDM: Extract, Transform, and Load
The ETL process was based on the approach put forward by the OHDSI community which involves four distinct steps: 1) designing the ETL, 2) creating the code mappings, 3) implementing the ETL, and 4) quality control to assess whether the database was fit for use. Any issues identified during quality control are addressed by updating the ETL where possible.15
Designing the ETL
The OHDSI White Rabbit tool was used to scan and characterize the source data.15 Based on this, a design was created using the Rabbit-in-a-Hat tool in which source data tables were mapped to the OMOP CDM person, observation period, visit occurrence, condition occurrence, procedure occurrence, drug exposure, measurement, observation, provider, location and death tables (see Supplementary Figure 1). The derived drug and condition era tables were also created.
Creating the Code Mappings
Mapping to the OMOP CDM requires mapping terminology to standard vocabularies in the OMOP Vocabularies.15 Examples of code mappings are given in Table 1. The Systematized Nomenclature of Medicine (SNOMED), for example, is a standard vocabulary for conditions, while RxNorm codes are a standard vocabulary for drug exposures.
 |
Table 1 Example of Mappings Implemented in the Information System for Research in Primary Care to the OMOP CDM |
COVID-19 diagnoses and patient comorbidities could first be identified from the source table Problemes (health problems, from Catalan), containing International Statistical Classification of Diseases and Related Health Problems, Tenth Revision, Clinical Modification (ICD-10-CM) codes recorded during primary care interactions. An example of a COVID-19 diagnosis code mapping for this table was from the ICD-10-CM code B34.2 (“Coronavirus infection, unspecified site”) to the OMOP concept id 439676 (“Coronavirus infection”), while an example of a COVID-19 symptom code is the mapping of the ICD-10-CM code R06.02 (“Shortness of breath”) to the concept id 312437 (“Dyspnea”). ICD-10-CM codes of health problems causing sick leaves were also identified from the source table Baixes (sick leaves), and were mapped to the condition_occurrence table of the OMOP CDM.
Prescriptions and dispensations of medications were identified from the source table Prescripcio (prescription) and Facturacio (dispensing), respectively, with this information mapped to the drug exposure table of the OMOP CDM. To map each drug national code from the Agencia Española de Medicamentos y Productos Sanitarios (AEMPS) in the source table to the best corresponding standard concept id in the OMOP CDM drug exposure table an intermediate source_to_concept_map table was used, as defined by the OMOP CDM data model. COVID-19 and non-COVID-19 vaccines were identified from two different source tables (Vacunes_covid and Vacunes_orig, respectively), and were mapped to the drug exposure table in a similar manner.
SARS-CoV-2 test results were identified from a source table including all COVID-19 tests (Covid_tests). This source table was created following the advent of the COVID-19 pandemic and was linked to SIDIAP patient data at the individual-level. These results were mapped to the measurement table in the OMOP CDM. Each polymerase chain reaction (PCR) test record in this source table was mapped to a measurement concept id of 586310 (“Measurement of Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Genetic material using Molecular method”), while antigen tests were mapped to 37310257 (“Measurement of Severe acute respiratory syndrome coronavirus 2 antigen”) and antibody tests to 37310258 (“Measurement of Severe acute respiratory syndrome coronavirus 2 antibody”). If the result was coded as Positiu (positive) in the source table, the value as concept id was then set as the concept id of 45884084 (“Positive”).
Additional information specific to the SIDIAP database was identified from the source table Variable_geo_sanitaries (geographic health data) and was mapped to the observation table. Such information included variables on socioeconomic deprivation indexes or Basic Health Areas, both based on the primary care center assigned to each patient. Information on sick leave occurrences was identified from the source table Baixes (sick leaves) and was also mapped to the observation table. This information had no equivalency in the OMOP CDM vocabulary, but was included in the CDM so it could be used in further research.
Lastly, hospitalisation data, from the Conjunt Mínim Bàsic de Dades d’Alta Hospitalària (CMBD, minimum basic set of hospital discharge data) collated by the Data Analysis Program for Health Research and Innovation (PADRIS) in Catalonia, was also linked at the individual-level. This dataset included both diagnosis and procedures registered during hospital admissions for all public and private hospitals in Catalonia. ICD-10-CM codes used to register diagnoses at hospitals were mapped to the OMOP CDM, as was done with the Problemes table, with the procedure occurrence table in the CDM also populated. Lastly, hospital and ICU inpatient admission and discharge dates were mapped to the visit_occurrence table.
The linkage of data to public sources was carried out by the Institut Català de la Salut (ICS, Catalan Health Institute) and PADRIS, with PADRIS specifically handling hospitalisation data. A unique personal identifier was used for the linkage process. Both institutions were responsible for executing the linkage and delivering the new dataset already pseudo-anonymised.5
Implementing the ETL
The SIDIAP tables are stored in MariaDB database, and the OMOP CDM v5.3.1 tables are accessible through PostgreSQL. Docker containers were used to host and to deploy the full system. Python code was used for implementing the mapping in the original SIDIAP database, with Bitbucket used for version control. After completing the transformations, CDM tables were migrated to the final PostgreSQL environment. A schematic of the ETL framework used is provided in Supplementary Figure 2.
Quality Control
A range of database constraints defined by OHDSI were created in the Postgres database to prevent errors such as duplicate rows or unmatched ids across interrelated tables. Data quality was also considered systematically using the Data Quality Dashboard (DQD) R package.15,16 This tool was run on the data after conversion to the OMOP CDM to test how well the resulting CDM instance complied with OHDSI standards. DQD runs over 3,300 data quality checks paired with prespecified failure thresholds to address conformance, completeness and plausibility of the data. Conformance checks measure database’s conformance to the model specifications. Completeness checks address the quality of the mapping by looking at the frequency of values in the dataset, including checks to evaluate the proportion of source values that were not mapped to standard concepts. Lastly, plausibility checks evaluate the credibility of the values in the dataset.16 The results of DQD were used to evaluate whether the database was fit for use. Failed checks were reviewed, and the ETL was updated to address them where necessary.
Summarising the Occurrence of COVID-19-Related Outcomes and Describing the Characteristics of Those Affected
Study Population and Follow-Up
Individuals present in SIDIAP as of 1st March 2020 were identified as the study population. Any individuals who had a clinical diagnosis or positive test result for SARS-CoV-2 between the 1st January and 29th February 2020 were excluded, as were any individuals in hospital on 1st March 2020. These two exclusions were to ensure that the cohort identified from SIDIAP was representative of the general population at risk of subsequent incident COVID-19. Follow-up began on 1st March 2020 (the index date for all individuals) and ended on 30th June 2022 (the last available date of data collection).
COVID-19-Related Outcome Cohorts
Five COVID-19-related outcomes were considered: outpatient COVID-19, hospitalised with COVID-19, ICU admission with COVID-19, died with COVID-19, and vaccinated against COVID-19. Outcome cohorts only considered the first occurrence of infection and the first-dose vaccination in the patients’ history and were not mutually exclusive. Cohort entry was cohort-specific and was based on the date of occurrence of the entry event of each cohort (date of COVID-19 diagnosis, hospitalisation, ICU admission, death, or vaccination).
An outpatient diagnosis of COVID-19 was identified on the basis of a compatible clinical code or positive SARS-CoV-2 test (antigen or PCR), whichever came first, with no hospital admission with COVID-19 observed prior to or on the same day as this diagnosis. Hospitalisation with COVID-19 was identified as a hospital admission where the individual had a compatible COVID-19 clinical code or positive SARS-CoV-2 test over the 21 days prior to their admission up to three days after admission. We used the same temporal criteria to identify ICU admission with COVID-19. A COVID-19 death was defined as a death where an individual had a compatible COVID-19 clinical code or positive SARS-CoV-2 test recorded in the 28 days preceding their death, with deaths identified at the individual-level. First-dose vaccination against COVID-19 was identified on the basis of standard concept ids compatible with any of the vaccines administered in the immunisation campaign in Spain (BNT162b2, ChAdOx1, mRNA-1273, and Ad26.COV2.S).
To assess the impact of using alternative definitions for outpatient diagnosis of COVID-19, we explored three further definitions: PCR positive test, PCR or antigen positive test, and COVID-19 diagnosis (narrow definition). While the initial definition for diagnoses (referred in this paper as broad definition) allowed for broader included codes such as “Coronavirus infection” and “Suspected COVID-19”, the narrow definition only included codes specific to COVID-19, such as “Disease caused by 2019 novel coronavirus”. Supplementary Tables 1 and 2 present the full specifications of narrow and broad definitions for COVID-19 diagnosis using standard concepts within the OMOP CDM.
Variables
The age, as of 1st March 2020 (the index date for all individuals), and sex of study participants were extracted. Using their most recent observation, individuals´ body mass index (BMI) and smoking status (classified as never smoker, former smoker, or current smoker) were also obtained. Individuals´ comorbidities and medication use were also summarised relative to the 1st March 2020. The comorbidities included were: autoimmune diseases, asthma, malignant neoplastic disease, diabetes mellitus, heart disease, hypertensive disorder, renal impairment, chronic obstructive lung disease (COPD), and dementia. These health conditions were based on an individual’s entire observed history prior to the index date. In addition, for those individuals in the outpatient diagnosis of COVID-19 cohort, symptoms recorded within two days prior to two days after index date were also identified. The following symptoms were considered: cough, dyspnea, diarrhea, headache, fever, one of anosmia, hyposmia, or dysgeusia, either malaise or fatigue, and pain.
Descriptive Analysis
The characteristics of the study population as a whole and each of the COVID-19-related outcome cohorts were summarised, with counts and percentages for categorical variables and median and interquartile ranges (IQR) for continuous variables. Cohort entry over time is plotted for the study cohorts. The proportion of persons in the outpatient COVID-19 cohort with a symptom of interest is summarised and stratified by calendar month.
All analytical code and detailed definitions of algorithms for identifying events (including COVID-19 and comorbidities) have been made publicly available at: https://github.com/SIDIAP/CovidCdmSummary
Results
Mapping to the OMOP CDM
The SIDIAP CDM contained information on 8,265,343 unique individuals. These people had 252,201,881 records in the condition occurrence table, 1,623,418,192 records in the drug exposure table, and 1,575,796,906 records in the measurement table. We mapped a total of 244,070,592 (96.8%) records on the condition domain, 1,537,021,869 (94.7%) records in the drug domain, and 1,575,431,674 (100%) records in the measurements domain. Details on the source codes and records mapped can be found in Table 2. Of the 3,484 data quality checks run against the database, 3,440 passed (98.7%). The remaining 44 checks that failed were considered not to require immediate action. Most of those stemmed from the source data due to potential data entry errors (eg, male-specific conditions recorded for females) or to the nature of our data (eg, prescription end date after the last day of data availability). Minor errors on unit measurements were detected and require further exploration for future use. Each of these checks is summarised in Supplementary Table 3.
 |
Table 2 Mapping Coverage for Terms and Registries in the Main OMOP CDM Domains |
COVID-19 Outcomes
We identified 5,932,342 individuals present in the database as of 1st March 2020. We excluded 8,580 individuals who were hospitalised on the study start date. None of the individuals had a history of COVID-19 between the 1st January and 29th February 2020. A total of 5,923,762 individuals were included in the general population cohort of people alive and not hospitalised in the database as of 1st March 2020. Over follow-up, 2,270,939 had either an outpatient COVID-19 diagnosis or positive test result, 95,018 were hospitalised with COVID-19, 12,340 had an ICU admission with COVID-19,17,678 had a COVID-19 death, and 4,585,515 received at least one dose of a COVID-19 vaccine.
The distribution of age for each study cohort, stratified by sex, is shown in Figure 1. The average age of the general population study cohort was 43 years (IQR: 25 to 59), and 50.7% were female. Median age was higher among outcome cohorts, most notably among those with a COVID-19 death who had an average age of 85 (78 to 90). Patients admitted to ICU, however, were younger than those admitted to hospital (63 [53 to 71] compared to 65 [51 to 78]). While the outpatient COVID-19 cohort was majority female (53.7%), those hospitalised and admitted to ICU were more typically male (54.5% and 67.2%, respectively). Patients with a COVID-19 death were close to equally distributed by sex (49.1% were female). Vaccine recipients were typically younger than those admitted to hospital or ICU and with a COVID-19 death. Comorbidities were generally more common among those with a COVID-19 outcome compared to the general population, except for the outpatient COVID-19 cohort, see Table 3. For example, the prevalence of diabetes and hypertension were 23.1% and 44.5%, respectively, among those hospitalised with COVID-19 compared to 7.3% and 16.7% in the general population.
 |
Table 3 Characteristics of the COVID-19 Study Cohorts |
 |
Figure 1 Histogram of age, stratified by sex, for the general population and each COVID-19 outcome cohort. |
Cohort entry over calendar time, stratified by age, is shown in Figure 2. The figure illustrates the various waves of COVID-19, along with the much greater number of cases of COVID-19 hospitalisations, ICU admission, and deaths among the older age groups. The highest number of outpatient COVID-19 cases did though occur among the younger age group.
 |
Figure 2 COVID-19 outcome cohort entry over calendar time, stratified by age group. |
Capture of COVID-19 symptoms over calendar time is shown in Figure 3 and stratified by age group in Supplementary Figures 3 and 4, respectively. Cough and fever were the most common symptoms identified, but all symptoms had a prevalence of less than 6% with substantial changes over time.
 |
Figure 3 Symptoms recorded at time of outpatient COVID-19 diagnosis or positive test. |
The impact of different definitions for outpatient COVID-19 is shown in Figure 4, where cohort entry over calendar time is depicted. While from September 2020 definitions were generally in accordance, the first wave of COVID-19 in Catalonia was only identified when including the broad COVID-19 diagnosis definition.
 |
Figure 4 Outpatient COVID-19 cohort entry over calendar time. |
First-dose vaccinations over time overall and by vaccine product are shown in Figure 5, and stratified by age group in Supplementary Figure 5. Plots over time mirror the order in which age groups were prioritised for vaccination (starting with the elderly), and are in accordance with the different nationwide guidelines for the provision of the different COVID-19 vaccine products in Spain. For instance, the majority of ChAdOx1 recipients were aged 60 to 69 years.
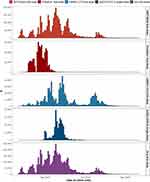 |
Figure 5 COVID-19 vaccination cohort entry over calendar time. |
Discussion
We have extracted and transformed the SIDIAP, a database of population-wide primary care electronic health records (EHR) with more than eight million individuals in Catalonia, to the OMOP CDM. With more than 3,400 data quality checks performed to assess data quality, the resulting database can be considered fit for use to inform appropriate research questions. Demonstrating the breadth of data captured, a descriptive analysis of various COVID-19-related outcomes among the general population has been performed, providing a broad overview of COVID-19 in Catalonia and the characteristics of the individuals affected and vaccinated with a first vaccine dose against this disease.
While the initial implementation of OMOP CDM posed challenges, the resources available through the OHDSI community and the European Health Data & Evidence Network’s (EHDEN) Innovative Medicines Initiative (IMI) 2 consortium facilitated the mapping of the SIDIAP data to the OMOP CDM. The SIDIAP mapping workflow is now automated, simplifying the procedure and enabling the transformation of each data update (every six months). The adoption of the OMOP CDM will enable the use of the growing body of open-source tools available for OMOP formatted-data, and will facilitate its use for both single database studies and distributed network research. Indeed, the mapping of SIDIAP data to the OMOP CDM has already facilitated its use in several international network studies on COVID-19. Examples include characterization studies of individuals tested for SARS-CoV-2,17 and patients with COVID-19, including hospitalised adults,18 children and adolescents,19 and patients with comorbidities.20–23 Other network studies have explored the susceptibility to COVID-19 among patients using renin-angiotensin system and alpha-1 blockers,24,25 and have validated models for predicting COVID-19 outcomes.26,27 In addition, several studies have been developed to characterize the background incidence rates of adverse events of special interest for COVID-19 vaccines,28,29 and to estimate the risk of some of these events following COVID-19 vaccination and infection.30–33 Other studies based solely on SIDIAP data mapped to the OMOP CDM have focused on different aspects of the COVID-19 pandemic in Catalonia, including COVID-19 patient trajectories in Catalonia,34 and the impact of cancer and obesity on these trajectories.35,36 Additionally, other studies have described the impact of the pandemic on trends of mental health diagnoses,37,38 and have assessed inequalities in COVID-19 vaccination and infection.39
In our descriptive analysis, we found that individuals with a COVID-19 outcome were typically older and had more comorbidities than the general population. This was particularly pronounced for the most severe outcomes studied. This is in concordance with research to date, with numerous studies finding older age to be associated with worse outcomes in COVID-19.40–43 While those with an outpatient COVID-19 diagnosis or positive test were more often female in our data, those hospitalised were majority male, as were 67.2% of those admitted to ICU. People who died with COVID-19 were almost equally distributed by sex. Previous research studies have reported mixed results for diagnoses and positive tests, for example two studies from the UK which reported a higher risk of testing positive for SARS-CoV-2 among men,44,45 while a study from China found there to have been a higher attack rate among women.46 A range of studies have though previously found males to be at an increased risk of severest outcomes.41,42,47 Regarding COVID-19 vaccines, first dose vaccine recipients were almost equally distributed by sex and were slightly older than those in the general population, which was in concordance with official statistics.48
The importance of appropriate phenotyping when using routinely collected data is also demonstrated when comparing alternative definitions of an outpatient COVID-19 case in our data. A definition that relied solely on testing for SARS-CoV-2 or using a narrow set of diagnosis codes would have missed many of the COVID-19 cases from the first wave, a time when testing was not widely available and medical vocabularies had not yet introduced COVID-19 specific codes.
Strengths and Limitations
Much of the COVID-19 literature is based on studies where study populations have been drawn from people hospitalised with COVID-19, tested for infection, or who volunteered to participate in a study. Such studies can be subject to a number of biases, in particular collider bias which can lead to the reporting associations that do not exist in the general population or by attenuating, inflating or reversing the sign of true associations.49 This underscores the importance of developing comprehensive datasets to generate the reliable evidence required to inform decision-making related to the pandemic. With more than four million first dose vaccine recipients, two million outpatient cases of COVID-19 captured, and a breadth of data capture that allows for comparisons with the general population and subsequent hospital care to be described, the mapped SIDIAP database described here is one such resource.
While EHR data bring numerous opportunities, with the data collected for non-research purposes careful curation is required. Using a well-established common data model meant that existing open-source tools could be used to evaluate data quality and that research studies can be run in a distributed manner. This has allowed the database to already have been used in a number of international network research studies, with standardised analytic packages and only aggregated results sets shared.
One limitation of the dataset has been seen with the likely underreporting of COVID-19 cases and symptoms, which reflects the nature of routinely collected health care data not designed for specific research questions. The underreporting of COVID-19 cases also reflects how COVID-19 testing strategies and the subsequent recording of cases in EHRs have evolved throughout the pandemic due to varying public health strategies, and it will unavoidably miss undetected asymptomatic cases or detected cases not reported to the Catalan public health system. Since 28 March 2022, free testing in primary care has been restricted to specific groups, including the elderly, people who are pregnant or immunocompromised, and those working with vulnerable populations. In addition, people who test positive with a self-test are no longer advised to notify these results to their GP or to seek medical advice unless symptoms worsen or if medical leave is required. Therefore, future COVID-19 studies, and particularly those using data beyond this date, will require careful consideration to minimize and adjust for potential misclassification bias.50 The estimates drawn from this database in terms of COVID-19 symptoms are much lower than reported in studies informed by self-reported patient data.51,52 The underreporting of COVID-19 symptoms has already been described in another network study based on routinely collected real-world data,53 and would likely be reduced if free text data recorded during primary care visits was also mapped to the OMOP CDM. Other limitations include lack of cause of death, hospital prescribing of medicines and lab data, while SARS-CoV-2 variants and contact tracing are also not captured. Lastly, it is important to note that our analysis of COVID-19-related outcomes was limited to individuals present in our database as of March 2020. Consequently, data on individuals who moved into the catchment area of SIDIAP or were born after the start of the study were not included.
Conclusion
We successfully harmonised SIDIAP to the OMOP CDM, and we illustrated its potential to perform COVID-19 distributed network research, as it captures COVID-19 diagnoses, SARS-CoV-2 test results, hospitalisations, deaths, and vaccinations in Catalonia, Spain. In this study, we have summarised the mapping of this dataset and described observed COVID-19-related outcomes and the characteristics of those individuals affected and vaccinated against this disease. In addition, we have provided insights regarding important considerations for future research in our setting, including the impact of different outpatient COVID-19 definitions and significant testing-related information. The transformed SIDIAP database is a valuable resource that can enable distributed network research in COVID-19 and beyond.
Data Sharing Statement
In accordance with current European and national law, the data used in this study are only available for the researchers participating in this study. Thus, we are not allowed to distribute or make publicly available the data to other parties. However, researchers from public institutions can request data from SIDIAP if they comply with certain requirements. Further information is available online (https://www.sidiap.org/index.php/menu-solicitudesen/application-proccedure) or by contacting SIDIAP ([email protected]). Details on the code used for the ETL can be obtained through direct contact with the corresponding author.
Ethics Approval and Informed Consent
This study was approved by the Clinical Research Ethics Committee of the IDIAPJGol (project code: 21/052-PCV).
Acknowledgments
BR and SF-B are joint first authors. EB and TD-S are joint senior authors. We would like to acknowledge the patients who suffered or died from this devastating disease, and these patient’s families and carers. We would also like to thank the healthcare professionals in the Catalan healthcare system involved in the management of COVID-19 during these challenging times, from primary care to intensive care units; the Institut de Català de la Salut and the la Recerca i la Innovació en Salut for providing access to the different data sources accessible through SIDIAP.
Funding
This study was carried out as part of the doctoral program in methodology of biomedical research and public health at the Autonomous University of Barcelona. This project is funded by the Health Department from the Generalitat de Catalunya with a grant for research projects on SARS-CoV-2 and COVID-19 disease organised by the Direcció General de Recerca i Innovació en Salut. This project has received support from the European Health Data and Evidence Network (EHDEN) project. EHDEN received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 806968. The JU receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. MR was funded by Wereld Kanker Onderzoek Fonds (WKOF, grant number: 2017/1630), as part of the international grants from the World Cancer Research Fund. ER was supported by Instituto de Salud Carlos III, Spain (grant No CM20/00174). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Disclosure
EAV and CB are employees of Janssen Research and Development LLC and shareholders of Johnson & Johnson (J&J) stock. LH reports edenceHealth, as a certified EHDEN SME, received payment for certifying the earlier OMOP CDM transformation work done by SIDIAP in their function as an EHDEN Data Partner, during the conduct of the study. TD-S reports grants from Innovative Medicines Initiative 2, during the conduct of the study. The authors report no other conflicts of interest in this work.
References
1. Roser M, Ortiz-Ospina E. Spain: what is the daily number of confirmed deaths? Available from: OurWorldInData.org;
2. Gérvas J, Pérez Fernández M, Starfield BH. Primary care, financing and gatekeeping in Western Europe. Fam Pract. 1994;11(3):307–317. doi:10.1093/fampra/11.3.307
3. Borras-Bermejo B, Martínez-Gómez X, Gutierrez-San Miguel M, et al. Asymptomatic SARS-CoV-2 infection in nursing homes, Barcelona, Spain, April 2020. Emerg Infect Dis. 2020;26(9):2281–2283. doi:10.3201/eid2609.202603
4. Mathieu E, Ritchie H, Ortiz-Ospina E, et al. What share of the population has received at least one dose of vaccine?. Available from: OurWorldInData.org;
5. Recalde M, Rodríguez C, Burn E, et al. Data resource profile: the information system for research in primary care (SIDIAP). Int J Epidemiol. 2022;51(6):e324–e336. doi:10.1093/ije/dyac068
6. Ramos R, Comas-Cufí M, Martí-Lluch R, et al. Statins for primary prevention of cardiovascular events and mortality in old and very old adults with and without type 2 diabetes: retrospective cohort study. BMJ. 2018;362:k3359. doi:10.1136/bmj.k3359
7. Alexander M, Loomis AK, van der Lei J, et al. Non-alcoholic fatty liver disease and risk of incident acute myocardial infarction and stroke: findings from matched cohort study of 18 million European adults. BMJ. 2019;367. doi:10.1136/bmj.l5367
8. Burn E, Murray DW, Hawker GA, Pinedo-Villanueva R, Prieto-Alhambra D. Lifetime risk of knee and Hip replacement following a GP diagnosis of osteoarthritis: a real-world cohort study. Osteoarthritis and Cartilage. 2019;27:1627–1635. doi:10.1016/j.joca.2019.06.004
9. Prieto-Alhambra D, Balló E, Coma E, et al. Filling the gaps in the characterization of the clinical management of COVID-19: 30-day hospital admission and fatality rates in a cohort of 118 150 cases diagnosed in outpatient settings in Spain. Int J Epidemiol. 2021;49(6):1930–1939. doi:10.1093/ije/dyaa190
10. Hripcsak G, Duke JD, Shah NH, et al. Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–578.
11. Voss EA, Makadia R, Matcho A, et al. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J Am Med Inform Assoc. 2015;22:553–564. doi:10.1093/jamia/ocu023
12. Datta S, Posada J, Olson G, et al. A new paradigm for accelerating clinical data science at Stanford medicine. arXiv. 2020. doi:10.48550/arXiv.2003.10534
13. Junior EPP, Normando P, Flores-Ortiz R, et al. Integrating real-world data from Brazil and Pakistan into the OMOP common data model and standardized health analytics framework to characterize COVID-19 in the Global South. J Am Med Inform Assoc. 2022:ocac180. doi:10.1093/jamia/ocac180
14. Papez V, Moinat M, Voss EA, et al. Transforming and evaluating the UK Biobank to the OMOP common data model for COVID-19 research and beyond. J Am Med Inform Assoc. 2022;30(1):103–111. doi:10.1093/jamia/ocac203
15. Observational Health Data Sciences and Informatics. The Book of OHDSI. Independently published; 2019.
16. Blacketer C, Defalco FJ, Ryan PB, Rijnbeek PR. Increasing trust in real-world evidence through evaluation of observational data quality. J Am Med Inform Assoc. 2021;28(10):2251–2257. doi:10.1093/jamia/ocab132
17. Golozar A, Lai LY, Sena AG, et al. Baseline phenotype and 30-day outcomes of people tested for COVID-19: an international network cohort including >3.32 million people tested with real-time PCR and >219,000 tested positive for SARS-CoV-2 in South Korea, Spain and the United States. medRxiv. 2020;2020:1.
18. Burn E, You SC, Sena AG, et al. Deep phenotyping of 34,128 adult patients hospitalised with COVID-19 in an international network study. Nat Commun. 2020;11(1):5009. doi:10.1038/s41467-020-18849-z
19. Duarte-Salles T, Vizcaya D, Pistillo A, et al. Thirty-day outcomes of children and adolescents with COVID-19: an international experience. Pediatrics. 2021;148(3):e2020042929. doi:10.1542/peds.2020-042929
20. Tan EH, Sena AG, Prats-Uribe A, et al. COVID-19 in patients with autoimmune diseases: characteristics and outcomes in a multinational network of cohorts across three countries. Rheumatology. 2021;60(SI):SI37–SI50. doi:10.1093/rheumatology/keab250
21. Recalde M, Roel E, Pistillo A, et al. Characteristics and outcomes of 627 044 COVID-19 patients living with and without obesity in the United States, Spain, and the United Kingdom. Int J Obes. 2021;45(11):2347–2357. doi:10.1038/s41366-021-00893-4
22. Reyes C, Pistillo A, Fernández-Bertolín S, et al. Characteristics and outcomes of patients with COVID-19 with and without prevalent hypertension: a multinational cohort study. BMJ Open. 2021;11(12):e057632. doi:10.1136/bmjopen-2021-057632
23. Roel E, Pistillo A, Recalde M, et al. Characteristics and outcomes of over 300,000 patients with COVID-19 and history of cancer in the United States and Spain. Cancer Epidemiol Biomarkers Prev. 2021;30(10):1884–1894. doi:10.1158/1055-9965.EPI-21-0266
24. Nishimura A, Xie J, Kostka K, et al. International cohort study indicates no association between alpha-1 blockers and susceptibility to COVID-19 in benign prostatic hyperplasia patients. Front Pharmacol. 2022;13:945592. doi:10.3389/fphar.2022.945592
25. Morales DR, Conover MM, You SC, et al. Renin-angiotensin system blockers and susceptibility to COVID-19: an international, open science, cohort analysis. Lancet Digit Health. 2021;3(2):e98–e114. doi:10.1016/S2589-7500(20)30289-2
26. Williams RD, Markus AF, Yang C, et al. Seek COVER: using a disease proxy to rapidly develop and validate a personalized risk calculator for COVID-19 outcomes in an international network. BMC Med Res Methodol. 2022;22(1):35. doi:10.1186/s12874-022-01505-z
27. Reps JM, Kim C, Williams RD, et al. Implementation of the COVID-19 vulnerability index across an international network of health care data sets: collaborative external validation study. JMIR Med Inform. 2021;9(4):e21547. doi:10.2196/21547
28. Li X, Ostropolets A, Makadia R, et al. Characterising the background incidence rates of adverse events of special interest for covid-19 vaccines in eight countries: multinational network cohort study. BMJ. 2021;373:1435. doi:10.1136/bmj.n1435
29. Burn E, Li X, Kostka K, et al. Background rates of five thrombosis with thrombocytopenia syndromes of special interest for COVID-19 vaccine safety surveillance: incidence between 2017 and 2019 and patient profiles from 38.6 million people in six European countries. Pharmacoepidemiol Drug Saf. 2022;31(5):495–510. doi:10.1002/pds.5419
30. Li X, Burn E, Duarte-Salles T, et al. Comparative risk of thrombosis with thrombocytopenia syndrome or thromboembolic events associated with different covid-19 vaccines: international network cohort study from five European countries and the US. BMJ. 2022;379:e071594. doi:10.1136/bmj-2022-071594
31. Burn E, Duarte-Salles T, Fernandez-Bertolin S, et al. Venous or arterial thrombosis and deaths among COVID-19 cases: a European network cohort study. Lancet Infect Dis. 2022;22(8):1142–1152. doi:10.1016/S1473-3099(22)00223-7
32. Burn E, Roel E, Pistillo A, et al. Thrombosis and thrombocytopenia after vaccination against and infection with SARS-CoV-2 in Catalonia, Spain. Nat Commun. 2022;13(1):7169. doi:10.1038/s41467-022-34669-9
33. Li X, Raventós B, Roel E, et al. Association between covid-19 vaccination, SARS-CoV-2 infection, and risk of immune mediated neurological events: population based cohort and self-controlled case series analysis. BMJ. 2022;376:e068373. doi:10.1136/bmj-2021-068373
34. Burn E, Tebé C, Fernandez-Bertolin S, et al. The natural history of symptomatic COVID-19 during the first wave in Catalonia. Nat Commun. 2021;12(1):777. doi:10.1038/s41467-021-21100-y
35. Roel E, Pistillo A, Recalde M, et al. Cancer and the risk of coronavirus disease 2019 diagnosis, hospitalisation and death: a population-based multistate cohort study including 4 618 377 adults in Catalonia, Spain. Int J Cancer. 2022;150(5):782–794. doi:10.1002/ijc.33846
36. Recalde M, Pistillo A, Fernandez-Bertolin S, et al. Body mass index and risk of COVID-19 diagnosis, hospitalization, and death: a cohort study of 2 524 926 catalans. J Clin Endocrinol Metab. 2021;106(12):e5030–e5042. doi:10.1210/clinem/dgab546
37. Raventós B, Abellan A, Pistillo A, Reyes C, Burn E, Duarte-Salles T. Impact of the COVID-19 pandemic on eating disorders diagnoses among adolescents and young adults in Catalonia: a population-based cohort study. Int J Eat Disord. 2023;56(1):225–234. doi:10.1002/eat.23848
38. Raventós B, Pistillo A, Reyes C, et al. Impact of the COVID-19 pandemic on diagnoses of common mental health disorders in adults in Catalonia, Spain: a population-based cohort study. BMJ Open. 2022;12(4):e057866. doi:10.1136/bmjopen-2021-057866
39. Roel E, Raventós B, Burn E, Pistillo A, Prieto-Alhambra D, Duarte-Salles T. Socioeconomic Inequalities in COVID-19 Vaccination and Infection in Adults, Catalonia, Spain. Emerg Infect Dis. 2022;28(11):2243–2252. doi:10.3201/eid2811.220614
40. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi:10.1016/S0140-6736(20)30566-3
41. Docherty AB, Harrison EM, Green CA, et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ. 2020;369. doi:10.1136/bmj.m1985
42. Gupta S, Hayek SS, Wang W, et al. Factors associated with death in critically ill patients with coronavirus disease 2019 in the US. JAMA Intern Med. 2020;180(11):1436–1447. doi:10.1001/jamainternmed.2020.3596
43. Williamson EJ, Walker AJ, Bhaskaran K, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584(7821):430–436. doi:10.1038/s41586-020-2521-4
44. de Lusignan S, Dorward J, Correa A, et al. Risk factors for SARS-CoV-2 among patients in the oxford royal college of general practitioners research and surveillance centre primary care network: a cross-sectional study. Lancet Infect Dis. 2020;20(9):1034–1042. doi:10.1016/S1473-3099(20)30371-6
45. Ho FK, Celis-Morales CA, Gray SR, et al. Modifiable and non-modifiable risk factors for COVID-19, and comparison to risk factors for influenza and pneumonia: results from a UK Biobank prospective cohort study. BMJ Open. 2020;10(11):e040402. doi:10.1136/bmjopen-2020-040402
46. Qian J, Zhao L, Ye R-Z, Li X-J, Liu Y-L. Age-dependent gender differences of COVID-19 in mainland China: comparative study. Clin Infect Dis. 2020;71(9):2488–2494. doi:10.1093/cid/ciaa683
47. Petrilli CM, Jones SA, Yang J, et al. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;369. doi:10.1136/bmj.m1966
48. Generalitat de Catalunya. Salut/Dades COVID. Available from: dadescovid.cat.
49. Griffith GJ, Morris TT, Tudball MJ, et al. Collider bias undermines our understanding of COVID-19 disease risk and severity. Nat Commun. 2020;11(1):5749. doi:10.1038/s41467-020-19478-2
50. Pham A, Cummings M, Lindeman C, Drummond N, Williamson T. Recognizing misclassification bias in research and medical practice. Fam Pract. 2019;36(6):804–807. doi:10.1093/fampra/cmy130
51. Grant MC, Geoghegan L, Arbyn M, et al. The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): a systematic review and meta-analysis of 148 studies from 9 countries. PLoS One. 2020;15(6):e0234765. doi:10.1371/journal.pone.0234765
52. Menni C, Valdes AM, Freidin MB, et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med. 2020;26(7):1037–1040. doi:10.1038/s41591-020-0916-2
53. Kostka K, Duarte-Salles T, Prats-Uribe A, et al. Unraveling COVID-19: a large-scale characterization of 4.5 Million COVID-19 cases using CHARYBDIS. Clin Epidemiol. 2022;14:369–384. doi:10.2147/CLEP.S323292
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
