Back to Journals » Infection and Drug Resistance » Volume 15
The Value of Metagenomic Next-Generation Sequencing in Hematological Malignancy Patients with Febrile Neutropenia After Empiric Antibiotic Treatment Failure
Authors Zhang M, Wang Z, Wang J, Lv H, Xiao X, Lu W, Jin X, Meng J, Pu Y, Zhao M
Received 1 March 2022
Accepted for publication 30 June 2022
Published 7 July 2022 Volume 2022:15 Pages 3549—3559
DOI https://doi.org/10.2147/IDR.S364525
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Meng Zhang,1,2,* Zhao Wang,2,* Jiaxi Wang,1 Hairong Lv,2 Xia Xiao,2 Wenyi Lu,2 Xin Jin,2 Juanxia Meng,2 Yedi Pu,2 MingFeng Zhao2
1First Central Clinical College, Tianjin Medical University, Tianjin, 300192, People’s Republic of China; 2Department of Hematology, Tianjin First Central Hospital, Tianjin, 300192, People’s Republic of China
*These authors contributed equally to this work
Correspondence: MingFeng Zhao, Department of Hematology, Tianjin First Central Hospital, No. 24, Fukang Road, Nankai District, Tianjin, 300192, People’s Republic of China, Tel +16622082788, Email [email protected]
Background: It was crucial to use empirical antibiotics in febrile neutropenia (FN) patients. However, most patients still died from infection due to poor efficacy. Metagenomic next-generation sequencing (mNGS) is a rapid microbiological diagnostic method. The value of mNGS in patients with FN remains to be studied, especially after empiric antibiotic treatment.
Methods: We retrospectively analyzed the differences between mNGS and the traditional methods in 192 patients with hematological malignancies who have received empiric antibiotic treatment. Samples were collected when patient had chills or half an hour before peak body temperature. And we compared the differences between FN and non-FN patients, mainly including types of pathogens and the diagnostic value of different pathogens.
Results: Despite receiving empirical treatment, the pathogen detection rate of mNGS was significantly higher than the traditional method (80.21% vs 25.00%, P< 0.001). And it has obvious advantages in detecting mixed pathogens infection (80.21% vs 4.17%, P< 0.001). Then, we found that mNGS saw more pathogens in the FN than in the non-FN group, especially fungus. 21/33 (63.63%) of FN patients was diagnosed with fungal infections. The fungal detection rate in FN was significantly higher than non-FN group (32.35% vs 12.22%, P=0.001). Besides, the sensitivity of mNGS was higher than the traditional methods in both FN and non-FN group (P< 0.001), but no significant difference in specificity (P> 0.05). In the FN group, empiric antibiotic treatment of 46/102 (45.10%) patients did not treat all the pathogens detected by mNGS. After adjusting the antimicrobial regimen according to the results of mNGS, the effective rate at 72 hours and 7 days was 22/46 (47.83%) and 24/102 (52.17%), respectively.
Conclusion: mNGS had a significant impact on the diagnosis of infection and the second-line antimicrobial therapy in FN. mNGS plays a more important role in FN patients, especially in the diagnosis of fungal infections.
Purpose: Firstly, we compared the difference between mNGS and the traditional methods in the diagnosis of infection. Secondly, we assessed the value of mNGS in FN patients by comparing it with non-FN patients, including types of pathogens and the diagnostic value of different pathogens. In order to show that mNGS plays a more important role in FN.
Keywords: metagenomic next-generation sequencing, febrile neutropenia, infection, fever, empiric antibiotic treatment
Background
FN, which is a common complication for more than 80% of patients with hematological malignancies, is caused by the primary disease, high-dose radiotherapy and chemotherapy, immunotherapy, and transplantation.1 It is considered a medical emergency and prompts immediate medical attention for evaluating and administrating empiric broad spectrum antibiotics.2 The clinical symptoms and syndromes of FN patients are often not obvious, and most patients only have a fever. The pathogens and the site of infection are also not clear.2 Besides, the infection-related mortality rate of FN is relatively high, among which bloodstream infection is the most common reason (42%).3 Therefore, the empiric antibiotic treatment is needed as soon as possible, to reduce the infection-related mortality rate.2 However, some patients still die from infection due to the poor efficacy of empirical treatment. The use of unnecessary antibiotics not only leads the drug resistance, but also may affect the therapeutic effect.4 At present, clinicians still lack effective methods to help them adjust antibiotics in time.
Conventional microbiological testing mainly includes microbial culture technology, polymerase chain reaction (PCR), and immunology technology. But there are many problems, such as taking a long time, a lower positive rate, and limited types of tests.5 mNGS is a rapid microbial diagnosis method, which determines pathogenic by analyzing the DNA/RNA content and abundance of microorganisms in clinical specimens. It can increase the sensitivity of pathogen detection, shorten detection time, and detect multiple pathogenic at the same time.6 In addition, mNGS also has advantages in diagnosing rare pathogen infections.7
In recent years, numerous researches have shown that mNGS owned essential value in hematological patients with FN. It could effectively improve the efficacy of pathogen detection and efficiently develop the precise treatment plan after clarifying that the pathogens can reduce mortality and avoid antibiotic abuse.8
However, research on the use of mNGS in FN remains scarce, especially after empirical antibiotic treatment. This article, which is uses a retrospective study, compares the difference between mNGS and the traditional methods in the diagnostic value after empiric antibiotic treatment failure. Besides we also assess the value of mNGS in FN patients by comparing some of their characteristics with non-FN patients, including types of pathogens and the diagnostic value of different pathogens. In order to show that mNGS plays a more important role in FN.
Materials and Methods
Study Patients
We retrospectively analyzed 192 patients who have received empiric antibiotic treatment. They were diagnosed with hematological malignancies in Tianjin First Central Hospital, Tianjin, China, between January 2019 and July 2021. A total of 102 patients were FN, and the remaining 90 patients were non-FN who did not develop neutropenia but had a fever.
The included patients need to meet the following criteria (1) Diagnosed with hematological malignancy; (2) Single oral temperature ≥38.3°C (axillary temperature ≥38.0°C), or oral temperature ≥38.0°C (axillary temperature ≥37.7°C for more than 1 h2); (3) Poor efficacy of empirical antibiotics treatment (the peak fever drop less than 0.5°C or has no significant change after receiving empirical treatment 72–96 hours); (4) Completed the traditional methods, such as culture and PCR tests (including EBV, CMV, BK, HBV, etc), and mNGS at the same time; (5) FN patients needed to meet: absolute neutrophil count (ANC) in peripheral blood <0.5 × 109/L.2 (6) non-FN patients needed to meet: ANC>0.5 × 109/L.
Sample Sequencing and Data Analysis
Using TIAN amp Micro DNA kit (DP316, Tiangen Biochemical Technology) kit and QIA amp Viral RNA Mini kit (52906 Qiagen) kit to extract DNA and RNA. Using SuperScript II reverse Transcription Kit (18064–014, Invitrogen) kit to reverse transcription of RNA into double-stranded complementary DNA (ds cDNA), which was ultrasonically broken into 200–300bp fragments. The sequence of the linker was circularized into a single-stranded circular structure. The circularized library was copied by a rolling circle to generate DNB nanospheres and loaded onto the sequencing chip. Using the BGISEQ-100 gene sequencer for sequencing. In order to obtain high-quality sequencing data, we removed the low-quality and short-read (<35bp) sequences. In addition, it was compared with the bacterial library, fungal library, and virus library after removing the human-derived sequence. The comparative classification reference database could be downloaded from the National Center for Biotechnology (ftp://ftp.ncbi.nlm.nih.gov/genomes/).
Diagnostic Assessment for a Positive mNGS Result
Samples were collected when the patients had chills or half an hour before peak body temperature. And they were sent to the GBI laboratory within 12h. If the result meet any of the following criteria, it was considered positive.9 (1) bacterium or virus whose coverage rate scored 10-fold greater than that of any other microbes, (2) fungi whose coverage rate scored 5-fold higher than that of any other fungus, (3) >30% relative abundance at the genus level in bacterium, virus, or fungi. If the detected pathogens were commonly reported infectious pathogens, they were considered causative agents. If the detected pathogens were uncommonly reported pathogens, the mNGS results were interpreted according to the patients’ clinical features; otherwise, the detected reads were classified as nonpathogenic microbe sequences. In addition, we had negative controls. The detection results of negative control products should not detect pathogens. If relevant pathogens were detected, it indicate that there may be DNA pollution sources in the environment. Positive contained specific microbic DNA.
Clinical Evaluation
We further evaluated the diagnostic performance of mNGS compared to clinical diagnosis, including positive predictive value (PPV), negative predictive value (NPV), specificity and sensitivity. (Sensitivity=mNGS-positive/clinical diagnosed infections. Specificity=mNGS-negative/clinical diagnosed non-infections. PPV= clinical diagnosed infections with mNGS-positive/mNGS-positive. NPV= clinical diagnosed non-infections with mNGS-negative/ mNGS-negative.) The consistency of mNGS and the traditional methods were also analyzed. If two methods detected the same pathogens, we considered it was “completely match”. “Mismatch” referred to completely different pathogens and “partly match” meant that the pathogens detected by two methods were only partially consistent. A mixed infection was defined as a non-single pathogenic infection, such as bacteria + fungus/bacteria + viruses/fungus + viruses/bacteria + fungus + viruses. The efficiency of an antibiotic was defined as patients’ peak temperature decreasing by 0.5°C, or there being no fever after being exposed antibiotic 72 hours or 7 days. Finally, according to clinical manifestations, traditional laboratory results, imaging examinations and microbiological evidences, patients were divided into the infected group, uninfected group and uncertain group.
Statistical Analysis
Comparative analysis was conducted by Pearson χ2 test or the McNemar test for discrete variables where appropriate. Normally distributed measurement data were represented by (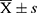 ) and analyzed using t-test. Data analyses were performed using SPSS 22.0 software. P values < 0.05 were considered significant.
) and analyzed using t-test. Data analyses were performed using SPSS 22.0 software. P values < 0.05 were considered significant.
Result
Patient Characteristics
The group included 192 enrolled subjects. 68.75% were male, and the median age was 42.88 years old (range, 10–75). The most common diagnosis was acute lymphoblastic leukemia (ALL) in 53 (27.60%) and acute myeloid leukemia (AML) in 53 (27.60%). Sixty-eight (35.42%) patients had received transplantation or Chimeric Antigen Receptor T-Cell immunotherapy (CAR-T). Furthermore, according to clinical diagnosis, all patients were divided into the infected group (145 cases, 75.52%), the non-infected group (22 cases, 11.46%), and the uncertain (24 cases,12.50%). Before mNGS, all patients received empiric antibiotic treatment. Sixty-eight (35.42%) patients received anti-bacteria, anti-fungus and anti-viruses simultaneously. Anti-fungal and anti-viral antibiotic were routinely used as prophylaxis to transplant patients. According to the number of neutrophils, 102 cases were FN, and others were non-FN. There were no significant differences in age, gender, previous treatment regimens, and empirical antibiotics between the two groups of patients (P>0.05) (Table 1).
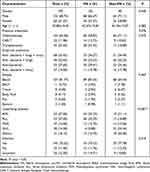 |
Table 1 Characteristics of Patients |
Pathogens Detected by mNGS
The mNGS detected 141 kinds of pathogens in 192 patients, by contrast, only 15 pathogens were detected by the traditional methods. The most common pathogens detected by mNGS and the traditional methods were both bacteria (Staphylococcus species was the most common). The most common virus was Human betaherpesvirus 5. Besides, there was no statistical difference in bacterial, fungal and viral distribution between the two groups (P=0.887) (Figures 1 and 2).
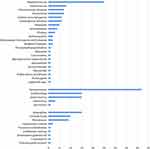 |
Figure 1 Distribution of pathogenic detected by mNGS in FN patients. |
 |
Figure 2 Distribution of pathogenic detected by mNGS in non-FN patients. |
In addition, the proportion of mixed infection detected by mNGS was also significantly higher than the traditional methods (39.61% vs 4.17%, P<0.001).
Detection Efficiency of mNGS
The positive rate of mNGS was significantly higher than that of the traditional methods, despite all patients receiving empiric antibiotic therapy (80.21% vs 25%, P<0.001). And the detection rate of bacteria, fungus and viruses were both significantly higher than traditional methods (P<0.001), too. According to our data, the mNGS positive rate seems less affected by prior antibiotic usage (Table 2).
 |
Table 2 Detection Efficiency of mNGS and Comparison in FN and Non-FN Groups |
The consistency between mNGS and traditional methods was also analyzed. The result of mNGS and traditional methods were both positive in 48 (25%) cases, of which 31/48 (64.58%) were partly matched, 10/48 (20.83%) were fully matched, and 7/48 (14.58%) were mismatched (Figure 3).
Comparison of Pathogens Detected by mNGS in FN and Non-FN Groups
More pathogen species were detected in the FN group than the non-FN group. And the bacterial, fungal and viral distribution were statistically different between the two groups (P=0.021). More fungus were detected in the FN group (Table 2).
The most common bacterium and virus were similar (Staphylococcus species and Human betaherpesvirus 5), but the most common fungus was different. Aspergillus was the most common fungi in the FN group, but the most common fungi in the non-FN group was Pneumocystis jirovecii.
Differences in Detection Efficiency by mNGS
The mNGS positive rate was no statistically different between FN and non-FN groups (83.33% vs 76.67%, P < 0.001). When analyzing the detection rate of various pathogens, we found that only the detection rate of fungus was significantly higher than the non-FN group (32.35% vs 12.22%, P=0.001) (Table 2).
Furthermore, regardless of the FN or non-FN group, the sensitivity of mNGS was significantly higher than traditional methods (P< 0.001). However, there was no statistical difference in specificity. The PPV and NPV of mNGS were 92.86% and 43.75%, respectively (Table 3).
 |
Table 3 Diagnostic Efficiency of Culture and mNGS Compared to Clinical Diagnosis |
The Effect on the Adjustment of Antibiotics According to mNGS in FN Patients
All enrolled patients were previously treated with empiric antibiotics, but the symptoms were not relieved after 72–96 hours. In the FN group, empiric antibiotic treatment of 46/102 (45.10%) patients did not treat all the pathogens detected by mNGS. After adjusting the antimicrobial regimen according to the results of mNGS, the effective rate at 72 hours and 7 days was 22/46 (47.83%) and 24/102 (52.17%), respectively.
As we all know, unnecessary antibiotics could lead to drug resistance.4 Futhermore, mNGS may allow doctors to reduce unnecessary antibiotics applications. As we observed, 5 patients discontinued inappropriate antibiotics and did not cause a bad clinical prognosis (Table 4). Our data partly shows that mNGS will help clinicians adjust the use of antibiotics if empiric antibiotics have poor effectiveness after 72–96 hours. In addition, those samples were collected and the results typically came back within 24 hours, which could help clinicians adjust antibiotics in time.
 |
Table 4 The mNGS Results That Led Discontinued Inappropriate Antibiotics |
Diagnostic Value of mNGS in FN Patients with Fungal Infection
mNGS could detect fungus in 33 patients with FN. Aspergillus was the most common, followed by Candida. Four cases were positive by culture or pathology, among which 3 cases fully matched with mNGS test results and 1 case partly matched. Only 2 cases were G positive and 4 cases in GM. 21/33 (63.63%) cases was diagnosed with fungal infections, of which pulmonary fungal infections were the most common. 10/21 patients adjusted the use of anti-fungal drugs according to mNGS and infection was effectively controlled (Table 5).
 |
Table 5 Diagnostic Value of mNGS in Fungal Infection in Patients with FN |
Discussion
FN is a common lethal complication for patients with hematological malignancies. The empiric antibiotic treatment is crucial to reducing the patient’s fatality rate. However, there were still some patients who received failed therapy. These patients often lack reliable methods to identify the type of pathogens quickly and accurately. Conventional detection methods (PCR, culture, etc.) have many deficiencies, such as taking a long time, a lower positive rate, and limited detection of pathogens. They may be affected by empiric antibiotics. Therefore, we try to find some new ways to provide reliable methods for using second-line antibiotics.
In recent years, mNGS has moved from scientific application to clinical practice and is changing how diseases are diagnosed and treated. Therefore, we compared the differences between mNGS and the traditional methods in 192 patients with hematological malignancies. Previous studies have shown that early mNGS can effectively improve the efficacy of pathogen detection, for FN children with hematological diseases.8 We further evaluated the value of mNGS in FN patients by comparing the characteristics of them with non-FN patients, including types of pathogens and the diagnostic value of different pathogens. In order to show that mNGS plays a more important role in FN.
First of all, the results showed that the positive rate and sensitivity of mNGS were significantly higher than the traditional methods after empirical treatment, and there was no statistical difference in specificity. Hongxia Duan’s team also found that the sensitivity of mNGS was significantly higher than that of the culture method (67.4% vs 23.6%, P < 0.001).9 In comparison, the specificity of mNGS was not significantly different from the culture methods (68.8% vs 81.3%, P = 0.41). It is worth noting that the mNGS positive rate seems less likely to be affected by previous antimicrobial use,10 and the results seems similar to the patients who did not use empiric therapy in earlier reports.12 But some studies have shown that if patients received effective antimicrobial treatment for more than 4 days, the detection rate of mNGS would then decrease to a great extent.13 However, mNGS also has some limitations, such as human background, background bacterial contamination, and no uniform standards for detailed experimental procedures. Therefore, further research is still required.
Secondly, the team of Parize P confirmed that mNGS has obvious advantages in detecting mixed pathogen infections in 2017.11 And we also found that mNGS could detect bacteria, fungus and viruses simultaneously. mNGS can see about 90% of the pathogens detected by traditional methods. And 25% of the patients’ mNGS results were entirely consistent with traditional methods.
FN patients are prone to fungal infections because of their weakened immune function. Due to a lack of reliable evidence, the fungus is rarely identified as the cause of early fever in FN. A research showed that the incidence of fungal infection would be was about 60% if neutropenia persisted for 3 weeks.14 However, methods for identifying fungal infections are limited and often lead to missed diagnoses. Our results confirmed that mNGS could detect more pathogens in the FN group, and the fungal detection rate was significantly higher than in the non-FN group (32.35% vs 12.22%, P=0.021). As study has demonstrated that mNGS of lung tissue and bronchoalveolar lavage fluid could help diagnose fungal infection in lungs.15 Thus, mNGS plays a more important role in FN patients, especially in the diagnosis of fungal infections.
Finally, early and rapid results of mNGS can provide clinical clues to the next step in diagnosis and treatment, especially in avoiding the overuse of antibiotics. In our result, 45.1% of all patients adjusted antibiotics according to mNGS. 52.17% of patients’ symptoms were effectively controlled. mNGS may provide evidence for adjustment of adjusting second-line antibiotics therapies for FN patients.
In our research, 5 patients discontinued inappropriate antibiotics and did not adversely affect the clinical outcomes, which could reduce the incidence of drug-resistant bacterial infections. Besides, the results of mNGS are reported very quickly, allowing clinicians to adjust antibiotics rapidly and effectively. Currently, the optimal duration of empirical treatment is unclear. Previous studies have shown that empirical antibiotic can be discontinued after 72h of apyrexia.16 If the antibiotic was stopped under the guidance of mNGS, it may be safer.
In short, mNGS can detect multiple pathogens simultaneously and had a significant impact on therapy in which the second-line antibiotic therapy of FN. mNGS examination for FN patients at early stage or empirical antibiotic failure can help doctors to identify pathogens. mNGS plays a more important role in FN patients, especially in the diagnosis of fungal infections.
Abbreviations
FN, febrile neutropenia; mNGS, Metagenomic next generation sequencing; CMV, Human betaherpesvirus 5; BK, Human polyomavirus 1; HBV, Hepatitis B; EBV, Human gammaherpesvirus 4; PCR, polymerase chain reaction; PPV, positive predictive value; NPV, negative predictive value; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia.
Ethics Statement
The study was approved by the Ethics Committee of Tianjin First Central Hospital. The need for informed consent was waived due to the retrospective nature of the study and because the data were anonymously analyzed.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by grants from the General Project of National Natural Science Foundation of China (81970180 to MZ), and the Key Science and Technology Support Project of Tianjin Science and Technology Bureau (20YFZCSY00800 to MZ), as well as Tianjin Key Medical Discipline (Specialty) Construction Project.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Keng MK, Sekeres MA. Febrile neutropenia in hematologic malignancies. Curr Hematol Malig Rep. 2013;8(4):370–378. doi:10.1007/s11899-013-0171-4
2. Freifeld A, Bow E, Sepkowitz K, et al. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the infectious diseases society of America. Clin Infect Dis. 2011;52(4):e56–e93. doi:10.1093/cid/cir073
3. Yan C, Wang Y, Mo X, et al. Incidence, risk factors, microbiology and outcomes of pre-engraftment bloodstream infection after haploidentical hematopoietic stem cell transplantation and comparison with HLA-identical sibling transplantation. Clin Infect Dis. 2018;67:S162–S173. doi:10.1093/cid/ciy658
4. Mikulska M, Viscoli C, Orasch C, et al. Aetiology and resistance in bacteraemias among adult and paediatric haematology and cancer patients. J Infect. 2014;68(4):321–331. doi:10.1016/j.jinf.2013.12.006
5. Fenollar F, Raoult D. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int J Antimicrob Agents. 2007;S7–15. doi:10.1016/j.ijantimicag.2007.06.024
6. Grumaz S, Stevens P, Grumaz C, et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8(1):73. doi:10.1186/s13073-016-0326-8
7. Doan T, Wilson M, Crawford E, et al. Illuminating uveitis: metagenomic deep sequencing identifies common and rare pathogens. Genome Med. 2016;8(1):90. doi:10.1186/s13073-016-0344-6
8. Guo F, Kang L, Zhang L. mNGS for identifying pathogens in febrile neutropenic children with hematological diseases. Int J Infect Dis. 2022;116:85–90. PMID: 34929357. doi:10.1016/j.ijid.2021.12.335
9. Duan H, Li X, Mei A, et al. The diagnostic value of metagenomic next⁃generation sequencing in infectious diseases. BMC Infect Dis. 2021;21(1):62. PMID: 33435894; PMCID: PMC7805029. doi:10.1186/s12879-020-05746-5
10. Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67:S231–S240. doi:10.1093/cid/ciy693
11. Parize P, Muth E, Richaud C, et al. Untargeted next-generation sequencing-based first-line diagnosis of infection in immunocompromised adults: a multicentre, blinded, prospective study. Clin Microbiol Infect. 2017;23(8):
12. Benamu E, Gajurel K, Anderson J, et al. Plasma microbial cell-free DNA next generation sequencing in the diagnosis and management of febrile neutropenia. Clin Infect Dis. 2021;74(9):1659–1668.
13. Zhang Y, Cui P, Zhang H, et al. Clinical application and evaluation of metagenomic next-generation sequencing in suspected adult central nervous system infection. J Transl Med. 2020;18(1):199. doi:10.1186/s12967-020-02360-6
14. Pagano L, Valentini C, Fianchi L, Caira M. The role of neutrophils in the development and outcome of zygomycosis in haematological patients. Clin Microbiol Infect. 2009;15:33–36. doi:10.1111/j.1469-0691.2009.02977.x
15. Yang L, Song J, Wang Y, Feng J. Metagenomic next-generation sequencing for pulmonary fungal infection diagnosis: lung biopsy versus bronchoalveolar lavage fluid. Infect Drug Resist. 2021;14:4333–4359. doi:10.2147/IDR.S333818
16. Aguila’ Guisado M, Espigado I, Martín-Peña A, et al. Optimisation of empirical antimicrobial therapy in patients with haematological malignancies and febrile neutropenia (How Long study): an open-label, randomised, controlled Phase 4 trial. Lancet Haematol. 2017;4(12):e573–e583. doi:10.1016/S2352-3026(17)30211-9
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.