Back to Journals » Journal of Pain Research » Volume 16
The Role and Potential Mechanism of Complement Factor D in Fibromyalgia Development
Authors Lei X, Song X, Fan Y, Chen Z, Zhang L
Received 11 September 2023
Accepted for publication 4 December 2023
Published 19 December 2023 Volume 2023:16 Pages 4337—4351
DOI https://doi.org/10.2147/JPR.S439689
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Wendy Imlach
Xinhuan Lei,1,* Xiaoting Song,1,* Yongyong Fan,1,* Zhen Chen,1 Liwei Zhang1,2
1Department of Orthopedics, Taizhou Hospital of Zhejiang Province, Zhejiang University, Taizhou, Zhejiang, People’s Republic of China; 2Bone Metabolism and Development Research Center, Taizhou Hospital of Zhejiang Province, Zhejiang University, Taizhou, Zhejiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liwei Zhang; Zhen Chen, Email [email protected]; [email protected]
Purpose: Fibromyalgia (FM) is a syndrome characterized by chronic musculoskeletal pain. Its clinical symptoms include both somatic and psychiatric symptoms, making the treatment of FM extremely challenging. The cause of FM is still unknown, and some patients do not improve their symptoms even after long-term active treatment. Thus, the development of new targeted therapies is important to reduce pain and improve quality of life for FM patients.
Methods: In this study, we screened genes and secreted factors that play key roles in FM through bioinformatics and big data analysis. Furthermore, we performed CCK-8, qRT-PCR, glucose, ATP and lactate content testing, dual luciferase reporter gene assay to investigate the potential mechanism of complement factor D in fibromyalgia development.
Results: In bioinformatics and big data analysis, we identified CFD was negatively correlated with the pro-inflammatory factor IL-6 and positively correlated with the anti-inflammatory factor IL-4, which suggested that CFD may be an anti-inflammatory factor. Through cellular assays, we verified that CFD reversed the decrease in IL-4 expression and the increase in IL-6 expression in BV2 cells caused by ATP.
Conclusion: In summary, based on bioinformatic methods and big data mining we obtained a new target CFD for FM, and further experiments verified that CFD has significant inhibition of ATP-induced neuropathic pain. These findings provide a new theoretical basis for the clinical translation of CFD.
Keywords: fibromyalgia, complement factor D, bioinformatics, inflammation
Introduction
Fibromyalgia (FM) is a syndrome characterized by chronic musculoskeletal pain.1–3 The diagnostic criteria for FM are still being updated over time, and the current criteria issued by the American College of Rheumatology (ACR) include: chronic (>3 months) widespread or multisite pain, fatigue, sleep disturbances, cognitive problems, other somatic symptoms (headaches, abdominal pain or bloating, dizziness, paresthesia), diffuse and significant soft tissue tenderness on examination.4 The cause of FM is still unknown. Researches to date has proven that genetic factors, female sex, and pre-existing pain of the body are the risk factors of FM.5,6 The principles of treating FM focus on symptomatic treatment and suggest a combination of individualized and multimodal treatment approach, lacking specific therapeutic targets. Study shows some disturbances in pain processing and regulation in patients with FM, with altered levels of pain-related neuropeptides and neurotransmitters.1,7 Changes in secretory factor levels in FM patients make the exploration of targeted therapeutics meaningful. The current treatment of FM is mostly symptomatic, so the development of new targeted therapies is important to reduce pain and improve the quality of life of patients with FM.
Inflammation of the nervous system is thought to be a potential cause of FM, and pro-inflammatory cytokines such as IL-6, IL-8 are increased in FM patients relative to healthy controls.8 Immune cells can act as mediators in the development of inflammation,9 for example macrophages in muscle have been shown to contribute to the development of chronic muscle pain.10 Pro-inflammatory cytokines can activate and sensitize injury receptors, induce pain and trigger nociceptive sensitization.11 Anti-nuclear antibody (ANA) positive FM patients have a more pronounced inflammatory response than ANA negative controls, suggesting that autoimmunity may be involved in the inflammatory pain of FM.12 Therefore, it is believed that there is an association between FM and neuroinflammation. Besides, some FM patients respond to treatment with N-methyl-D-aspartate (NMDA) glutamate receptor antagonists,13 and a low glutamate diet has been shown to reduce FM symptoms.14 Elevated glutamate levels may be involved in the development of FM. However, FM patients do not always show good tolerance to treatment with glutamate receptor antagonist drugs.15 Currently, therapeutic drugs for FM include tricyclic antidepressants, serotonin–norepinephrine reuptake inhibitors, gabapentinoids, and analgesics.1,2 The current clinical treatment for FM is numerous and heterogeneous, with very limited efficacy for some patients, and lack of specific drugs to relieve symptoms. Since neuroinflammation is inextricably linked to FM, it is clinically relevant to explore specific therapeutic factors that inhibit the inflammatory response in FM.
In this study, we screened differentially expressed genes (DEGs) by analyzing and examining RNA-seq data in the fibromyalgia and healthy groups from an open-source data, establishing functional enrichment analysis and protein interaction networks, and further screening for 5 signature genes of FM, which were then validated by cellular experiments. This study explores the specific changes of FM at the molecular level and provides a reliable theoretical basis for the targeted treatment of FM.
Materials and Methods
Data Sources
The GSE67311 dataset was obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and contained 142 samples, including 67 FM group and 75 control group whole blood gene expression data. All datasets were subjected to background elimination to remove missing, low-expressed, and non-corresponding gene probes,16 resulting in a total of 23,306 genes annotated.
Cell Culture
Mouse microglial cell line BV2 and human renal epithelial cell line 293T were purchased from the China Typical Culture Collection (Wuhan, China). The cells were inoculated in DMEM medium containing 10% fetal bovine serum and 1% penicillin – streptomycin and cultured in a constant temperature incubator at 37 °C with 5% CO2. When the cell fusion reached 70–80%, the cells were passaged, and cells in logarithmic growth phase were taken for cell experiments.
Main Instruments and Reagents
DMEM culture medium, PBS solution, 0.25% Trypsin-EDTA trypsin were purchased from HyClone Laboratories (Logan, USA). Adenosine triphosphate (ATP) was obtained from Sigma-Aldrich (St. Louis, USA). Human recombinant CFD protein was obtained from Solarbio Science & Technology (Beijing, China). miRNA-mimic, primer sequences for IL4 and IL6 were brought from Sangon Biotech (Shanghai, China). Extraction reagents, reverse transcription kits and fluorescent quantitative PCR reagents were purchased from Takara Bio (Kyoto, Japan).
Differentially Expressed Genes
In this study, the RNA-seq data of GSE67311 (fibromyalgia group vs healthy group) were subjected to t-test and Bayesian test using the limma package in R language. Differentially expressed genes (DEGs) were screened for |Log2(FC)| >0.25 and P < 0.05, with negative numbers representing down-regulation and positive numbers representing up-regulation. Pearson correlation coefficients were calculated to determine the gene-to-gene correlations, with |R|>0.2 and P<0.05 as filtering correlation conditions.
Functional Enrichment Analysis of Differentially Expressed Genes and Protein Interaction Network (PPI)
The software packages “org.Hs.eg.db”, “clusterProfiler”17 and “enrichplot”18 of R were applied Gene ontology functional analysis (GO) and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis (KEGG) analysis were performed and the results were visualized. Prediction of differentially expressed protein interactions was performed using STRING online database, setting the effective binding fraction >0.7.
Molecular Typing and Weighted Co-Expression Network Analysis of Fibromyalgia Samples
The NMF molecular typing model was constructed using the “Consensus ClusterPlus” analysis package in R language. The NMF hierarchical clustering was performed using the adjusted and unified dataset with the clusters k-value ranging from 2 to 8, and the k-value with better clustering stability were selected for the molecular typing according to the clustering effect.19 Similarly, the “WGCNA” toolkit in R language was used to construct a scale-free weighted gene co-expression network to identify co-expressed genes and modules associated with clinical traits.20
Characteristic Gene Screening
Characterization pre-selection of FM differentially expressed genes by LASSO regression using R package glmnet.17 LASSO combines feature selection and model construction by adding penalty constraints to the algorithm. In this study, all FM differential gene expression data were applied as feature selection inputs to the LASSO logistic regression model. We used gradient feature selection to evaluate the expression values of each gene. Features were selected when a t-test result of P < 0.05. The regularization parameter λ is selected by 10 times cross-validation so that the error of the selected model is within one standard deviation from the minimum error.21 The obtained differentially expressed characteristic genes were crossed with the secreted protein library (https://www.proteinatlas.org/humanproteome/proteinclasses) and the resulting ones were the characteristic genes of secreted protein.
Determination of BV2 Cell Viability by CCK-8
Logarithmically grown BV2 cells were inoculated in 6-well plates and divided into blank group, ATP model control group and treatment group (ATP+human recombinant protein CFD), with three parallel sub-wells of 1.5×103 cells per well in each group, and the cells were placed in a 37°C, 5% CO2 incubator overnight. On the second day, 1 µL/mL DMSO solution was added to the control group, 300 µmol/L ATP to the ATP model control group and 300 µmol/L ATP and 1µL/mL rCFD to the treatment group, respectively. After incubation for 48 h, the supernatant was aspirated and 100 µL of culture medium was added to each well, followed by 10 µL of CCK-8 solution. The cells were then incubated in a cell incubator protected from light. After incubation for 2 h in the incubator, the absorbance was read at 450 nm.
Detection of IL4 and IL6 Gene Expression by Quantitative Real-Time Fluorescence Polymerase Chain Reaction (qRT-PCR)
BV2 cells were inoculated in 6-well plates with 1×105 cells per well, and the cells were incubated at 37 °C in a 5% CO2 incubator. After cell fusion reached 60–70%, the medium was changed and DMSO solution (1 µL/mL), ATP solution (300 µmol/L) and ATP solution (300 µmol/L) + rCFD (1 µL/mL) were added to each group, respectively. Incubation was continued for 24–48 hours. PCR operations were then performed.
Total cellular RNA was extracted by TRIzol method, reverse transcribed into cDNA and then subjected to fluorescent quantitative PCR. Reaction system: SYBR 5 µL, PCR upstream and downstream primers 0.8 µL each, DNA template 1 µL, ddH2O 3.2 µL, total volume 10 µL. Reaction conditions: preheated denaturation at 95 °C for 30s; cyclic system 95 °C for 15s, 58 °C for 1 min, 40 cycles in total. The relative expression levels of mRNA were normalized by GAPDH and the relative expression was calculated by the 2−ΔΔCt method.22 The primer sequences are shown in Table 1.
 |
Table 1 List of Primers for qRT-PCR |
Glucose, ATP and Lactate Content Testing
Measure key products of glycolysis, including glucose, ATP and lactate, to understand changes in glycolysis. Cell precipitates (1×107 cells or more) were collected for subsequent sonication and used for glucose, ATP and lactate micro-titration assays according to the kit procedure (Solarbio Science & Technology, Beijing, China).
Dual Luciferase Reporter Gene Assay
Firstly, the 3′UTR sequence of CFD gene was obtained from the gene library to construct the wild-type fragment. The wild-type fragment included the binding site of miRNA-3193. The purified PCR product was cloned into the psiCHECK-2 vector named psiCHECK-2-CFD-3′UTR. Subsequently, the firefly luciferin signal and sea kidney luciferin signal were measured in the cells using a fluorescence luminescence detector according to the instructions of the dual luciferase reporter gene assay kit (Hanheng Bio, Shanghai, China).
Statistical Analysis
GraphPad Prism 8 (GraphPad Software, San Diego, USA) software was used for data analysis. The mean data conforming to normal distribution were expressed as mean ± standard deviation (x ± s). The comparison of the means between 2 groups was performed by independent sample t-test, and the comparison between multiple groups was performed by ANOVA analysis. The difference was considered statistically significant at P<0.05.
Results
Screening for FM-Related Differentially Expressed Genes
By preprocessing the GEO-GSE67311 transcriptional profile and normalizing the data, a total of 23,306 mRNAs were identified. 70 differentially expressed genes were screened in the FM group compared with the healthy population (|Log2(FC)| >0.25 and P < 0.05), of which 38 genes were significantly upregulated and 32 genes were downregulated (Figure 1A). Next, GO function annotation and KEGG pathway enrichment analysis of 70 differentially expressed genes revealed that the GO functions of differentially expressed genes were mainly enriched in antioxidant response, tyrosine phosphorylation, myofibrillogenesis and contraction, actin assembly, endopeptidase activity, growth factor activity regulation and receptor ligand activation (Figure 1B–D). KEGG was mainly enriched in IL-17 signaling pathway, tumor necrosis factor signaling pathway, cytokine receptor interaction, PI3K-Akt signaling pathway and complement system (Figure 1E). Then, to further clarify the role of these genes in fibromyalgia, this study used the STRING website to construct a protein-protein interaction network for these 62 differentially expressed genes. The results showed that IFIT1, RSAD2, EPB42, IFI44, IFI44L and SLC4A1 were the key node genes (Figure 2A and B).
Molecular Typing and Weighted Co-Expression Network Analysis of FM-Related Differentially Expressed Genes
Traditionally, the diagnosis of disease relies on pathology,23 but this hardly reflects the biological nature of the disease. New approaches to disease typing based on molecular typing can provide global characterization of disease at the genetic level, greatly deepen the understanding of molecular pathological information, thus this typing approach has an important clinical role. To verify whether the above 70 FM-associated differentially expressed genes have clinical translational potential, we used the GEO-Merge dataset for molecular typing studies of FM.
Based on the enrichment scores of 70 genes, we performed a clustering analysis of 68 FM specimens using K-Means consistency clustering. However, stable clustering results could not be obtained when k=2-9 (Figure 2C–E), ie, the 70 differentially expressed genes could not be used for typing of dilated cardiomyopathy samples, and their mechanisms of action need further investigation.
Next, we performed WGCNA construction of the GEO-GSE67311 dataset. The weighted co-expression network was constructed using the WGCNA package in the R toolkit, however, our further study revealed that the gene co-expression modules (Figure 2F–H) were not obtained by WGCNA analysis, which also suggests that there may be more complex mechanisms of action for these genes that need further investigation.
FM-Related Trait Gene Screening
The 70 FM-related differentially expressed genes were characterized by the LASSO algorithm,17 and 20 factors were screened as candidate feature genes (Figure 3A). Subsequently, we intersected these genes with the secreted proteins in the secreted protein library and finally obtained five FM-related differentially secreted genes, namely CPA3, IL3RA, CFD, OCLN and IGKV6-21 (Figure 3B). We found that secreted factors CPA3, IL3RA, CFD, OCLN and IGKV6-21 were all down expressed in FM (Figure 3D). Further ROC curves also suggested AUC values of 0.745, 0.670, 0.640, 0.578, and 0.648 for CPA3, IL3RA, CFD, OCLN, and IGKV6-21, respectively (Figure 3C). Based on this, we screened 5 genes of secreted proteins by bioinformatics analysis, which suggested that these genes may have a role in inhibiting the process of FM.
Construction of ceRNA Network Upstream of 5 Secretory Factors
In this study, the miRNAs and lncRNAs upstream of CPA3, IL3RA, CFD, OCLN and IGKV6-21 were analyzed by TargetScan, miRanda and miRDB databases. A total of 36 CPA3-associated miRNAs, 2 IL3RA-associated miRNAs, 1 CFD-associated miRNA, 31 OCLN-associated miRNAs and 0 IGKV6-21-associated miRNAs were intersected by the three databases (Figure 4). Next, lncRNA prediction of miRNAs upstream of CPA3, IL3RA, CFD and OCLN was performed using the spongeScan database, and a total of 36 lncRNAs were obtained. We performed ceRNAs network construction of these predicted miRNAs and lncRNAs using Cytoscape software. The results showed that hsa-miR-590-3p and hsa-miR-377-3p were the core node genes of OCLN (Figure 4).
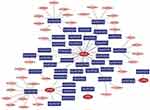 |
Figure 4 Construction of ceRNA network upstream of 5 secretory factors. Construction of ceRNA networks for CPA3, IL3RA, CFD and OCLN genes. |
Correlation Analysis of 5 Characteristic Secretory Genes with IL-4, IL-6, IL-10 and TNF Genes
The correlations of CPA3, IL3RA, CFD, OCLN and IGKV6-21 with IL4, IL6, IL10 and TNF genes were analyzed using the GEO-GSE67311 dataset (Figure 5A–D). We found that complement factor D (CFD) was negatively correlated with the pro-inflammatory factor IL-6 (Figure 5A) and positively correlated with the anti-inflammatory factor IL-4 (Figure 5C). Therefore, we suggested that CFD may play an important role in the pain-causing of FM.
Effect of Recombinant Protein CFD (rCFD) on the Viability of ATP-Induced BV2 Cells
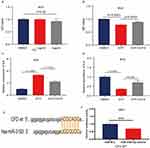
Based on the findings, we considered that the secreted factor CDF may have an important role in neuropathic pain of FM. First, the expression of CFD in healthy individuals and FM groups was compared using the GEO-GSE67311 dataset. The results showed that the CFD was significantly decreased in the FM group (Figure 3C), suggesting that CFD may be a pain suppressor in FM. Then, we applied 1ug/mL and 2ug/mL rCFD to BV2 cells and calculated the cell viability by CCK8 assay. The results showed that the concentration of rCFD less than 2ug/mL was not toxic to BV2 cells (Figure 6A). Third, ATP-induced BV2 cells were applied to establish a neuropathic pain model,24 and the cell survival rate was examined after the addition of rCFD. We found that 1ug/mL rCFD had an anti-ATP-induced change in neuronal cell viability compared to the control (Figure 6B).
Next, we studied the effect of secreted factor rCFD on the expression of IL-4 and IL-6 genes in BV2 cells induced by ATP. The results showed that ATP could significantly increase the expression of IL-6 in BV2 cells; the addition of 1ug/mL rCFD significantly inhibited the expression of IL-6 compared with the ATP group (Figure 6C). Similarly, ATP significantly decreased the expression of IL-4 in BV2 cells; while the addition of 1ug/mL rCFD significantly promoted the expression of IL-4 compared with the ATP group (Figure 6D). These findings suggested that secreted factor CFD can affect neuropathic pain and inflammatory responses of FM.
Subsequently, to investigate the upstream regulatory mechanism of CFD, combining the results of TargetScan, miRanda and miRDB database analysis, we verified the targeting relationship between CFD and miRNA-3193. The results showed that CFD was a direct target gene of miRNA-3193 (Figure 6E and F).
Effect of rCFD on Glucose Metabolism in BV2 Cells
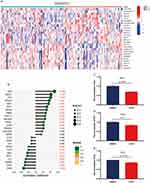
There are few studies on glucose metabolism and FM, and it is unknown whether fibromyalgia can regulate glucose metabolism in glial cells through secreted factors and thus affect inflammatory response and pain. To clarify the pathogenic mechanism of FM, we extracted genes related to glucose metabolism from the MSigDB website (Systematic name: M1870) combined with the GEO-GSE67311 dataset for differential analysis. The results showed that 26 genes showed differential expression in FM compared to the controls (Figure 7A). Next, we analyzed the correlation of CFD with these 26 genes. Our study revealed a negative correlation of CFD with MIOX, CHPF and ESRRB (Figure 7B), and a positive correlation with GOT1, RAE1, ZNF292, ZNF292, NSDHL, ECD, EIF6, VCAN, PDHA1, FAM162A, CHST12, PMM2 and CXCR4 (Figure 7B). In conclusion, our findings suggested that CFD may have a function in regulating glucose metabolism in FM.
To verify this result, the levels of glucose, ATP and lactate in BV2 cells were explored in this study. The levels of glucose (Figure 7C), ATP (Figure 7D) and lactate (Figure 7E) were significantly decreased in BV2 cells after the addition of rCFD compared to the control. These results suggested that CFD may affect neuropathic pain and inflammatory responses through glucose metabolism.
Discussion
Fibromyalgia can have a significant impact on patients both physically and psychologically.25 At present, FM lacks specific treatment and is often treated symptomatically, requiring consideration of both somatic and psychological factors, which may be related to the fact that the etiology and pathogenesis of fibromyalgia are still unclear.26 The search for molecular markers specific to FM has facilitated the development of its therapeutic modalities.27
Bioinformatics combined with big biological data analysis has allowed researchers to gain a deeper understanding of disease at the genetic level, especially in cancer, where a range of biomarkers have been identified to predict cancer recurrence, metastasis, chemosensitivity, and prognosis,28 and these technological advances have provided unprecedented opportunities to explore the pathogenesis of FM. We performed differential expression gene analysis using the GSE67311 dataset derived from the GEO database (67 FM cases versus 75 control cases) and screened 70 differentially expressed genes, which provides a basis for further focusing and screening of new molecular targets.
Secreted proteins are mainly a group of proteins with important physiological functions such as cytokines, growth factors, complement, degrading enzymes, antibodies, peptide hormones and immunoglobulins. More attention is currently being paid to secreted proteins in the field of oncology, as they have become a major source of potential tumor markers that can be used as targets to guide tumor diagnosis, treatment and prognosis.29,30 Studies on the correlation between FM and secreted proteins have been mainly limited to cytokines,31 IL-4, IL-6, IL-10 and TNF factors are involved in the polarization of microglia, where IL-6 and TNF factors have the ability to promote the inflammatory response, while IL-4 and IL-10 have the ability to inhibit the inflammatory response.2 However, there are relatively few studies to further investigate whether secreted proteins can be used as potential markers or therapeutic targets for FM. In this study, we screened FM differentially characterized genes by LASSO and obtained a total of 5 FM differentially expressed secreted proteins. We analyzed the correlation between 5 secreted factors and IL-4, IL-6, IL-10 and TNF factors. Since CFD was negatively correlated with the pro-inflammatory factor IL-6 and positively correlated with the anti-inflammatory factor IL-4, this suggested that CFD may be an anti-inflammatory factor and may have a suppressive effect on the development of FM.
CFD, also known as adipsin, is mainly synthesized by adipose tissue and sciatic nerve tissue. Current studies report that CFD plays an important regulatory role in the physiological and pathological processes of systemic sclerosis, diabetes (diabetic nephropathy), obesity, dyslipidemia, and hypertension.32,33 Recent studies have also suggested the possible involvement of CFD in respiratory disorders,34 neurological disorders35 and even preterm abortion.36 However, the role of CFD in FM has not been studied. As mentioned above, we observed that CFD is involved in the regulation of the levels of inflammation-related factors IL-4 and IL-6. Besides, pro-inflammatory cytokines, such as IL-1β, IL-6, and TNF-α, can activate and sensitize injury receptors, and another potential source of them is adipose tissue. There is also evidence that diffuse or multifocal pain is more common in obese individuals.37 Weight loss can improve symptoms of FM,38 and weight loss of at least 10% is associated with greater improvement in FM symptoms and an increase in IL-10.39 Besides, researches have indicated that FM patients showed reduced responses to adrenocorticotropic hormone, epinephrine, cortisol and growth hormone compared to healthy controls, suggesting insulin resistance,40 and that FM can be prevented by improving the control of blood glucose levels.41 Based on the above findings, we suggest that CFD is likely to be involved in regulating the occurrence of FM through the regulation of energy metabolism. Through cellular assays, we verified that CFD reversed the decrease in IL-4 expression in BV2 cells caused by ATP; similarly, CFD reversed the increase in IL-6 expression in BV2 cells caused by ATP. Therefore, it is reasonable to believe that CFD can be used as a therapeutic target for FM or as an inhibitor of inflammatory factors in combination with conventional FM drugs for the treatment of FM. At the last, though we have specified that CFD was negatively correlated with the pro-inflammatory factor IL-6 and positively correlated with the anti-inflammatory factor IL-4, further experiment needs to be performed to elucidate the detailed mechanism and signal pathway underlying the effect of CFD on anti-inflammation.
Conclusion
In summary, based on bioinformatic methods and big data mining we obtained a new target CFD for FM, and further experiments verified that CFD has significant inhibition of ATP-induced neuropathic pain. These findings provide a new theoretical basis for the clinical translation of CFD.
Ethical Approval
The patients involved in the GEO have obtained ethical approval. The study was approved by the ethics review board of Taizhou Hospital of Zhejiang Province (Approval Number: 2022-12-15-01). And all procedures and analyses performed in this study involving human data were in accordance with the ethical standards and with the Helsinki Declaration.
Acknowledgments
This study was supported in part by grants from the National Natural Science Foundation of China (82202738), the Basic and Public Research Project of Zhejiang Province (LGF20H250004).
Disclosure
The authors declare that they have no competing interest.
References
1. Bair MJ, Krebs EE. Fibromyalgia. Ann Intern Med. 2020;172(5):Itc33–itc48. doi:10.7326/AITC202003030
2. Siracusa R, Paola RD, Cuzzocrea S, et al. Fibromyalgia: pathogenesis, mechanisms, diagnosis and treatment options update. Int J Mol Sci. 2021;22(8):3891. doi:10.3390/ijms22083891
3. Winslow BT, Vandal C, Dang L. Fibromyalgia: diagnosis and management. Am Fam Physician. 2023;107(2):137–144.
4. Wolfe F, Clauw DJ, Fitzcharles M-A, et al. 2016 Revisions to the 2010/2011 fibromyalgia diagnostic criteria. Semin Arthritis Rheum. 2016;46(3):319–329. doi:10.1016/j.semarthrit.2016.08.012
5. Kerr JI, Burri A. Genetic and epigenetic epidemiology of chronic widespread pain. J Pain Res. 2017;10:2021–2029. doi:10.2147/JPR.S143869
6. Fitzcharles MA, Perrot S, Häuser W. Comorbid fibromyalgia: a qualitative review of prevalence and importance. Eur J Pain. 2018;22(9):1565–1576. doi:10.1002/ejp.1252
7. O’Brien AT, Deitos A, Triñanes Pego Y, et al. Defective endogenous pain modulation in fibromyalgia: a meta-analysis of temporal summation and conditioned pain modulation paradigms. J Pain. 2018;19(8):819–836. doi:10.1016/j.jpain.2018.01.010
8. Bains A, Kohrman S, Punko D, et al. A link between inflammatory mechanisms and fibromyalgia. Adv Exp Med Biol. 2023;1411:357–378.
9. Mastrangelo F, Frydas I, Ronconi G, et al. Low-grade chronic inflammation mediated by mast cells in fibromyalgia: role of IL-37. J Biol Regul Homeost Agents. 2018;32(2):195–198.
10. Gregory NS, Brito RG, Fusaro MCGO, et al. ASIC3 is required for development of fatigue-induced hyperalgesia. Mol Neurobiol. 2016;53(2):1020–1030. doi:10.1007/s12035-014-9055-4
11. Coskun Benlidayi I. Role of inflammation in the pathogenesis and treatment of fibromyalgia. Rheumatol Int. 2019;39(5):781–791. doi:10.1007/s00296-019-04251-6
12. Smart PA, Waylonis GW, Hackshaw KV. Immunologic profile of patients with fibromyalgia. Am J Phys Med Rehabil. 1997;76(3):231–234. doi:10.1097/00002060-199705000-00014
13. Staud R, Vierck CJ, Robinson ME, et al. Effects of the N-methyl-D-aspartate receptor antagonist dextromethorphan on temporal summation of pain are similar in fibromyalgia patients and normal control subjects. J Pain. 2005;6(5):323–332. doi:10.1016/j.jpain.2005.01.357
14. Holton KF, Taren DL, Thomson CA, et al. The effect of dietary glutamate on fibromyalgia and irritable bowel symptoms. Clin Exp Rheumatol. 2012;30(6 Suppl 74):10–17.
15. Olivan-Blázquez B, Herrera-Mercadal P, Puebla-Guedea M, et al. Efficacy of memantine in the treatment of fibromyalgia: a double-blind, randomised, controlled trial with 6-month follow-up. Pain. 2014;155(12):2517–2525. doi:10.1016/j.pain.2014.09.004
16. Yang Y, Li Y, Qi R, et al. Constructe a novel 5 hypoxia genes signature for cervical cancer. Cancer Cell Int. 2021;21(1):345. doi:10.1186/s12935-021-02050-3
17. Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
18. Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–2914. doi:10.1093/bioinformatics/btv300
19. Lancichinetti A, Fortunato S. Consensus clustering in complex networks. Sci Rep. 2012;2:336. doi:10.1038/srep00336
20. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi:10.1186/1471-2105-9-559
21. Li M, Li X, Guo Y, et al. Development and assessment of an individualized nomogram to predict colorectal cancer liver metastases. Quant Imaging Med Surg. 2020;10(2):397–414. doi:10.21037/qims.2019.12.16
22. Bejaoui M, Villareal MO, Isoda H. 3,4,5-Tri-O-caffeoylquinic acid promoted hair pigmentation through β-catenin and its target genes. Front Cell Dev Biol. 2020;8:175. doi:10.3389/fcell.2020.00175
23. Han XY, Wang -Y-Y, Wei H-Q, et al. Multifocal neuroendocrine cell hyperplasia accompanied by tumorlet formation and pulmonary sclerosing pneumocytoma: a case report. World J Clin Cases. 2020;8(16):3583–3590. doi:10.12998/wjcc.v8.i16.3583
24. Li J, Ren W, Huang X-J, et al. Bullatine A, a diterpenoid alkaloid of the genus Aconitum, could attenuate ATP-induced BV-2 microglia death/apoptosis via P2X receptor pathways. Brain Res Bull. 2013;97:81–85. doi:10.1016/j.brainresbull.2013.05.015
25. Ibáñez-Vera AJ, García-Romero JC, Alvero-Cruz JR, et al. Effects of monopolar dielectric radiofrequency signals on the symptoms of fibromyalgia: a single-blind randomized controlled trial. Int J Environ Res Public Health. 2020;17(7):2465. doi:10.3390/ijerph17072465
26. Nugraha B, Anwar SL, Gutenbrunner C, et al. Polymorphisms of brain-derived neurotrophic factor genes are associated with anxiety and body mass index in fibromyalgia syndrome patients. BMC Res Notes. 2020;13(1):402. doi:10.1186/s13104-020-05226-8
27. Hackshaw KV. The Search for Biomarkers in Fibromyalgia. Diagnostics. 2021;11:2.
28. Yang WJ, Wang H-B, Wang W-D, et al. A network-based predictive gene expression signature for recurrence risks in stage II colorectal cancer. Cancer Med. 2020;9(1):179–193. doi:10.1002/cam4.2642
29. Donadelli M. The cancer secretome and secreted biomarkers. Semin Cell Dev Biol. 2018;78:1–2. doi:10.1016/j.semcdb.2017.09.004
30. Paltridge JL, Belle L, Khew-Goodall Y. The secretome in cancer progression. Biochim Biophys Acta. 2013;1834(11):2233–2241. doi:10.1016/j.bbapap.2013.03.014
31. Rodriguez-Pintó I, Agmon-Levin N, Howard A, et al. Fibromyalgia and cytokines. Immunol Lett. 2014;161(2):200–203. doi:10.1016/j.imlet.2014.01.009
32. Sniderman AD, Cianflone K. The adipsin-ASP pathway and regulation of adipocyte function. Ann Med. 1994;26(6):388–393. doi:10.3109/07853899409148358
33. Tafere GG, Wondafrash DZ, Zewdie KA, et al. Plasma adipsin as a biomarker and its implication in type 2 diabetes mellitus. Diabetes Metab Syndr Obes. 2020;13:1855–1861. doi:10.2147/DMSO.S253967
34. Mathews JA, Wurmbrand AP, Ribeiro L, et al. Induction of IL-17A precedes development of airway hyperresponsiveness during diet-induced obesity and correlates with complement factor D. Front Immunol. 2014;5:440. doi:10.3389/fimmu.2014.00440
35. Hietaharju A, Kuusisto H, Nieminen R, et al. Elevated cerebrospinal fluid adiponectin and adipsin levels in patients with multiple sclerosis: a Finnish co-twin study. Eur J Neurol. 2010;17(2):332–334. doi:10.1111/j.1468-1331.2009.02701.x
36. Takeshita A, Kondo T, Okada T, et al. Elevation of adipsin, a complement activating factor, in the mouse placenta during spontaneous abortion. J Reprod Dev. 2010;56(5):508–514. Doi:10.1262/jrd.10-036K
37. Cicuttini FM, Wluka AE. Not just loading and age: the dynamics of osteoarthritis, obesity and inflammation. Med J Aust. 2016;204(2):47. doi:10.5694/mja15.01069
38. D’Onghia M, Ciaffi J, Lisi L, et al. Fibromyalgia and obesity: a comprehensive systematic review and meta-analysis. Semin Arthritis Rheum. 2021;51(2):409–424. doi:10.1016/j.semarthrit.2021.02.007
39. Schrepf A, Harte SE, Miller N, et al. Improvement in the spatial distribution of pain, somatic symptoms, and depression after a weight loss intervention. J Pain. 2017;18(12):1542–1550. doi:10.1016/j.jpain.2017.08.004
40. Kirnap M, Çolak R, Eser C, et al. A comparison between low-dose (1 µg), standard-dose (250 µg) ACTH stimulation tests and insulin tolerance test in the evaluation of hypothalamo–pituitary–adrenal axis in primary fibromyalgia syndrome. Clin Endocrinol. 2001;55(4):455–459. doi:10.1046/j.1365-2265.2001.01373.x
41. Tishler M, Smorodin T, Vazina-Amit M, et al. Fibromyalgia in diabetes mellitus. Rheumatol Int. 2003;23(4):171–173. doi:10.1007/s00296-002-0279-7
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.