Back to Journals » International Journal of Nanomedicine » Volume 16
The Effects of Luminescent CdSe Quantum Dot-Functionalized Antimicrobial Peptides Nanoparticles on Antibacterial Activity and Molecular Mechanism
Authors Li W, Song P , Xin Y, Kuang Z, Liu Q, Ge F, Zhu L, Zhang X , Tao Y, zhang W
Received 9 December 2020
Accepted for publication 15 February 2021
Published 5 March 2021 Volume 2021:16 Pages 1849—1867
DOI https://doi.org/10.2147/IJN.S295928
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yan Shen
Wanzhen Li,1,* Ping Song,1,* Ying Xin,1 Zhao Kuang,1 Qin Liu,1 Fei Ge,1 Longbao Zhu,1 Xuguang Zhang,2 Yugui Tao,1 weiwei zhang1,2
1School of Biological and Food Engineering, Anhui Polytechnic University, Wuhu, Anhui, 241000, People’s Republic of China; 2Bankpeptide Biological Technology Company, Hefei, Anhui, 230031, People’s Republic of China
*These authors contributed equally to this work
Correspondence: weiwei zhang Email [email protected]
Background: With the development of bacterial resistance, the range of effective antibiotics is increasingly becoming more limited. The effective use of nanoscale antimicrobial peptides (AP) in therapeutic and diagnostic methods is a strategy for new antibiotics.
Methods: Combining both AP and cadmium selenide (CdSe) into a composite material may result in a reagent with novel properties, such as enhanced antibacterial activity, fluorescence and favorable stability in aqueous solution.
Results: AP-loaded CdSe NPs (AP-CdSe NPs) showed strong antibacterial activity against multidrug-resistant (MDR) Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus) in vitro and in vivo. Colony-forming unit (CFU) and minimum inhibitory concentration (MIC) assays showed that AP-CdSe NPs have highly effective antibacterial activity. The quantitative analysis of apoptosis by flow cytometry analysis further confirmed that MDR E. coli and S. aureus treated with AP-CdSe NPs had death rates of 98.76% and 99.13%, respectively. Also, AP-CdSe NPs was found to inhibit bacterial activity in an in vivo bacteremia model in mice infected with S. aureus. In addition, the antibacterial mechanism of AP-CdSe NPs was determined by RNA sequencing analysis. Gene ontology (GO) analysis and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis revealed the molecular mechanism of the antibacterial effect of AP-CdSe NPs. Importantly, histopathology analysis, and hematological toxicity analysis indicated that AP-CdSe NPs had few side effects.
Conclusion: These results demonstrate that AP loaded on CdSe NPs had a higher water solubility, bioavailability and antibacterial effect compared with raw AP. This study reports findings that are helpful for the design and development of antibacterial treatment strategies based on AP.
Keywords: antibacterial peptides, CdSe QD, antibacterial mechanism, RNASeq, in vitro and in vivo
Introduction
Bacterial infections have become a global public health problem, and their mortality, morbidity and treatment costs during hospitalization increase every year.1–3 With the rapid emergence of multidrug-resistant (MDR) “superbug” strains and the lack of new antibiotics, modern society is facing the post-antibiotic era.4–7 Therefore, there is an urgent need to develop alternatives to antibiotics.
Antimicrobial peptides (AP) have become attractive potential alternative therapeutic compounds.8 AP have a wide range of activities, and a broad-spectrum antibacterial activity against Gram-negative and Gram-positive bacteria.9,10 AP kill or suppress benign and pathogenic organisms, thereby disrupting the homeostasis between a healthy microbiota and immune system.11 The amino acid sequence and structure of different AP are obviously different, but they all have an amphipathic structure and can be combined with other compounds.12,13 Therefore, they can easily interact with the negatively charged components on the bacterial cell membrane and integrate into the lipid bilayer. However, their poor biocompatibility is one of the main obstacles to the clinical development of a large number of common AP.14,15 Target delivery of AP is necessary due to their short half-life and relative toxicity against healthy cells.16 The effective use of nanoscale proteins and peptides in therapeutic and diagnostic methods is a strategy to induce and maintain their functional conformation.
Research on nano-modification based on AP has shown that they can be combined with nanoparticles (NPs) to form nanomaterials with novel properties, due to their advantages in molecular recognition and antibacterial activity.15,17,18 Currently, quantum dots (QDs) not only have very important applications in solar cells and phototransistor components, but also in biological applications including biosensors,19,20 bioimaging21 and biomedical research.22 Cadmium selenide (CdSe) QDs with a size of 1–20 nm are currently used for in vivo imaging and diagnostic treatment.23 Growing an outer shell of AP on the outside of the QDs can greatly improve its stability.24 Therefore, the combined use of an AP and CdSe QDs (such as conventional antibiotics) definitely has the potential to improve the effectiveness of the two groups of compounds.25–27
In this study, an AP, used as a stabilizer and active group, was modified on the surface of CdSe QDs to form fluorescent AP NPs (AP-CdSe NPs) with high antibacterial activity.28 The AP-CdSe NPs showed strong antibacterial activity against MDR Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus) in vitro and in vivo. The results of colony-forming unit (CFU) and minimum inhibitory concentration (MIC) assays showed that AP-CdSe NPs have a broad-spectrum antibacterial activity, and displayed highly effective antibacterial activity against MDR E. coli and S. aureus. Fluorescence microscopy analysis revealed that AP-CdSe NPs have a high cellular uptake rate and killing rate for E. coli and S. aureus. The quantitative analysis of live/dead cells by flow cytometry analysis further confirmed the antibacterial activity of AP-CdSe NPs. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) were used to examine the morphological changes of bacteria after being destroyed by AP-CdSe NPs. In addition, through the use of a bacteremia model in mice infected with S. aureus to analyze the biodistribution of bacteria in mice and perform histological evaluation, it was determined that AP-CdSe NPs can inhibit bacterial activity and heal tissue damage through. Furthermore, gene expression profiling by RNA sequencing (RNASeq) analysis was used to determine the mechanism of antibacterial activity of AP-CdSe NPs.29 Concurrently, we prepared peptide nanoparticles using traditional modification methods, and their effects were better than those of AP alone.
Methods and Materials
Materials
Selenium (Se) powder (99.99%), 11-Mercaptoundecanoic acid (99%), CdCl2·2.5H2O (99%), NaBH4 (≥99%) were purchased from Aladdin Chemical Reagent Co., Ltd. (Shanghai, China). The beef extract peptone medium agar powder used to prepare the bacterial culture plates was obtained from the School of Biochemical Engineering, Anhui Polytechnic University (Wuhu, China).
Synthesis of AP, CdSe and AP-CdSe Nanoparticles
Peptides were synthesized by the fluorenylmethoxycarbonyl (Fmoc) solid-phase synthesis method. The 2-Cl-resin was selected as the carrier for amino acid binding. A mixture of N,N-diisopropylethylamine (DIEA), 1-hydroxybenzotriazole (HOBT), and N,N’-diisopropylcarbodiimide (DIC) were used as condensation reagents. The AP periplanetasin-4 (Leu-Arg-His-Lys-Val-Tyr-Gly-Tyr-Cys-Val-Leu-Gly-Pro-NH2) was synthesized from the carboxyl end to the amino end in the indicated order.30,31 The structure of the AP is shown in Supplementary Figure S1.
The CdSe NPs were prepared as previously reported by Sun et al.32 The AP-CdSe NPs were prepared with mini revise. Briefly, the AP (0.6 mM) and CdCl2 (0.5 mM) were added into 60 mL of Milli-Q water. The pH of the mixture solution was adjusted to 11. Also, NaBH4 (1 mM) was dissolved in 1 mL H2O, and Se powder (0.5 mM) was added and mixed until the mixture was clear and transparent. The precursor is then quickly added to the above solution under N2 flow. The nanomaterial (AP-CdSe NPs) was obtained after stirring for 2 h with a magnetic stirrer at 60°C. The AP-CdSe NPs were ultimately collected by centrifugation at 15,000 rpm, for 5 min, and washed twice with Milli-Q water. Amount of AP in the solutions was determined by measuring the absorbance at 234 nm after centrifugation. The Table S1 showed the drug loading rate of AP-CdSe NPs was up to 85%.
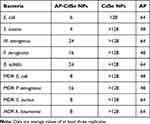 |
Table 1 MICs Values Test (μg/mL) |
Characterization
The purity of the AP was determined by mass spectrometry (MS) on a Q Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, USA), and nuclear magnetic resonance (NMR) spectroscopy on a picoSpin 80 NMR spectrometer (Thermo Fisher Scientific Inc.). The ultraviolet-visible (UV-vis) absorption spectra were recorded by UV-vis spectroscopy using a Shimadzu UV-2550 spectrometer (Shimadzu Corp., Kyoto, Japan). Fourier transform infrared (FT-IR) spectroscopy was performed on a IR Prestige-21 FT-IR spectrometer (Shimadzu Corp.). Fluorescence spectra of the AP-CdSe NPs were recorded by fluorescence spectroscopy using an RF-5301PC fluorescence spectrophotometer (Shimadzu Corp.). The morphology of the CdSe NPs and AP-CdSe NPs were examined by TEM on a JEOL 6300 F transmission electron microscope (JEOL Ltd., Tokyo Japan). The dynamic light scattering (DLS) of the NPs was measured using a Brookhaven Zeta PALS instrument (Brookhaven Instruments Corporation, Brookhaven, Holtsville, NY, USA).
Bacterial Culture
Escherichia coli ATCC 11303 (E. coli, Gram-negative), Staphylococcus aureus ATCC 25923 (S. aureus, Gram-positive), Bacillus subtilis ATCC 2233 (B. subtilis, Gram-positive), Micrococcus tetragenus ATCC 2835 (M. tetragenus, Gram-positive) and Pseudomonas aeruginosa ATCC 27853 (P. aeruginosa, Gram-negative) were obtained from Anhui Polytechnic University. MDR E. coli E76227, MDR P. aeruginosa BJ915, MDR S. aureus ATCC 25213, MDR Acinetobacter baumannii GIM (A. baumannii, Gram-negative) 1.650 were obtained from Jinan University (Guangzhou, China).
Minimum Inhibitory Concentration and Counting Colony-Forming Unit Test
The 96-well microtiter plate dilution method was used to determine the MIC of the AP-CdSe NPs. Logarithmic-phase bacteria were diluted with fresh medium and then two aliquots of 190 μL of the bacterial culture suspension were separately mixed with 10 μL of two different concentrations of AP-CdSe NPs followed by incubation at 37°C for 12 h (the final density of bacteria was 1.0 ×106 CFU/mL). Then, 200 μL of medium was added to each sample and in turn aliquots of 200 μL were drawn from each sample and further diluted by double dilution, and ultimately the concentration of each sample set was determined in triplicate by measuring three parallel samples.33
Log-phase MDR E. coli and MDR S. aureus were inoculated (1:40) in fresh medium containing AP-CdSe NPs solution at the corresponding concentrations (10, 20, 40 µg/mL), or AP solution (40 µg/mL), or CdSe NPs solution (40 µg/mL) and subsequently incubated in a shaking incubator at 37°C, for 12 h. The bacterial culture treated with phosphate-buffered saline (PBS, pH 7.5, 0.1 M) were used as blank. The numbers of viable bacterial cells on solid medium were counted using the CFU counting method. The statistical analysis of the CFU assay data for each group was performed with the Origin software (OriginLab Corp., Northampton, MA, USA).
Cellular Uptake Assays
The cellular uptake assay was conducted using a staining test. Log-phase MDR E. coli and S. aureus were incubated with AP-CdSe NPs solution at the corresponding concentrations (10, 20, 40 µg/mL) for 12h. The PBS-treated cells were used as the blank group. The CdSe NPs (40 µg/mL) was set as the control group. Cells were collected and stained with 5 µg/mL of 4ʹ-6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific Inc.). Bacterial culture suspension were also washed twice with PBS. Then, a fluorescence microscope was used to observe the cells in the red and blue channels.
LIVE-DEAD Assays
Log-phase MDR E. coli and MDR S. aureus were treated with AP-CdSe NPs (10, 20, 40 µg/mL). AP solution (40 µg/mL) and CdSe NPs solution (40 µg/mL) were set as control group. The PBS was set as blank group. Cells were collected and stained with the reagent test kit (SYTO9 and propodium iodide; Thermo Fisher Scientific) for 30 min in the dark. Then, Used FL microscope observed the cell at red and green channels. The apoptotic cell death statistical test was analyzed by a CytoFLEX (Beckman Coulter).34
Cell Membrane and Wall Integrity Studies
Log-phase MDR E. coli and MDR S. aureus were treated with AP-CdSe NPs solution at the corresponding concentrations (10, 20, 40 µg/mL). The groups treated with AP solution (40 µg/mL) and CdSe NPs solution (40 µg/mL) were used as control groups. The PBS-treated group was used as blank group. All bacterial cells were collected by centrifugation at 3000 rpm, for 15 min and fixed by dehydrating in an ethanol gradient at 4°C, and then postfixed in a mixture of glutaraldehyde (2.5%) and paraformaldehyde (2%) for 12 h. Eventually, dried samples were examined by SEM using a Hitachi S-4800 scanning electron microscope (Hitachi Ltd., Tokyo, Japan).35
Log-phase MDR E. coli and MDR S. aureus were treated with AP-CdSe NPs solution at the corresponding concentrations (10, 20, 40 µg/mL). The groups treated with AP solution (40 µg/mL) and CdSe NPs solution (40 µg/mL) were used as control groups. The PBS-treated group was used as blank group. All bacterial cells were collected by centrifugation at 3000 rpm, for 15 min, and fixed with a mixture of glutaraldehyde (2.5%) and paraformaldehyde (2%) for 24 h. Then, the MDR E. coli and MDR S. aureus cells were subjected to a series of biological dehydration tests, embedded, sectioned, mounted on 200-mesh copper grids and examined by TEM.36
RNA Extraction and Quantification of Gene Expression Level
Log phase MDR E. coli cells were exposed to AP-CdSe NPs (40 µg/mL) for 12 h at 37◦C in the shaking incubator. The PBS-treated cells were used as control. Total cellular RNA was extracted. After the library was constructed, Qubit3.0 was used for preliminary quantification, and then the Qsep 100 BioFragment Analyzer (BiOptic Inc., New Taipei City, Taiwan) was used to detect the insert size of the library. Once the insert size met expectations, the quantitative real-time polymerase chain reaction (qPCR) method was used to accurately quantify the effective concentration of the library in order to ensure the quality of the library.
Before differential gene expression analysis, for each sequenced library, the read counts were adjusted using a scaling normalization factor through the edge R package. Then, the DEGSeq (differentially expressed genes Seq) R package (1.20.0) was used to analyze the differential gene expression in the two conditions. The Benjamini & Hochberg method was used to adjust the P value. A corrected P value of 0.005 and a log2 (fold change) of 1 were set as the threshold for significant differential expression.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis of Differentially Expressed Genes
The off-machine data was filtered to obtain clean reads. The clean reads were compared with the reference genome sequence, and the number of reads contained in each gene were counted, that is, the expression level, according to the genome annotation file and the position of the reads, and then statistical testing was performed. The differentially expressed genes (DEGs) were obtained and functionally annotated and ultimately Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) enrichment analyses were performed on the DEGs to obtain the GO and KEGG sets enriched by the DEGs. Concurrently, according to reads and reference base due to group differences, together with statistical inference, single nucleotide variations, such as SNP and Indel, were found.
Animal Bacteremia Model
Six-week-old female SKH1 nude mice (~25 g) were obtained from the Model Animal Research Center of Nanjing University (Nanjing, China). All animals were maintained in accordance with the Animal Welfare Act and the Guide for the Care and Use of Laboratory Animals protocols. All animal studies were approved by the Suzhou University (Suzhou, China) Animal Care (Approval Number: 202010A415). The SKH1 nude mice were randomly and equally divided into 4 groups (n=21), on the 7th day of intravenous tail injection. The four groups of mice were separated into cages: blank, infection group (MDR S. aureus), and treatment groups (MDR S. aureus). At 24 h after the injection, the mice of the treatment groups were intravenous tail injections with a dose of 2 mg/kg AP or AP-CdSe NPs for treatment groups.
The mice of each group were sacrificed at 1 d, 3 d, 5 d and 7 d after treatment, and blood and organs (heart, liver, kidneys, spleen and lungs) were collected. The organs were harvested, weighed, and homogenized in sterile water. The number of surviving bacteria was counted by the CFU method. The weight and survival rate of the mice in each group were monitored daily.
Histological Analysis
The following organs (heart, liver, kidneys, spleen and lungs) were collected in each group after all treatment. The harvested organs were fixed in 4% formaldehyde for 24 h. Afterwards organ samples were embedded in paraffin, sectioned into 4-μm thick slices, placed on slides and stained with hematoxylin and eosin (H&E). The stained slides were examined by light microscopy.
Safety Evaluation
To evaluate the biosafety of the AP-CdSe NPs, mice (Blank and AP-CdSe NPs treatment groups) were sacrificed after 30 days, and major organs (liver, spleen, kidneys, heart, and lungs) were collected and stained with H&E to evaluate histopathological changes in vivo. Blood samples were also collected and hematological parameters, including uric acid, blood glucose, cholesterol, and alanine aminotransferase, were measured. Organs from healthy nude mice were used as blank.37
Results and Discussion
Characterization of AP-CdSe NPs
AP-CdSe NPs were synthesized using a simple method as described above. The AP is mainly modified as a stabilizer active agent of CdSe NPs. The structure of the AP was also confirmed by MS analysis and NMR spectroscopy. The MS chromatogram showed the molecular weight of the AP was 1505.8, which is consistent with the design results (Supplementary Figure S2). The NMR spectroscopy results showed that the AP was highly pure (Supplementary Figure S3 and S4). The characterization of AP-CdSe NPs is shown in Figure 1. The extinction spectrum of CdSe NPs (red line) shows peaks at around 281 nm, and all shifted to the shorter wavelength after being modified by AP (Figure 1A). For the AP-CdSe NPs, the peak at 246 nm is blue-shifted, indicating the successful modification by AP on the CdSe NPs. Their structures were also confirmed by FT-IR spectroscopy, as shown by the difference changes observed in the peak of the fingerprint spectrum (Figure 1B). The fluorescence excitation spectra of AP-CdSe NPs at different concentrations, shown in Figure 1C, reveal that the fluorescence intensity of the solution became increasingly stronger with the increase of the concentration. The successful conjugation of the AP on the surface of the CdSe NPs also revealed the fluorescence property of QDs. AP is modified on the QDs without altering its fluorescence properties. The fluorescence property was confirmed in the image displayed in Figure 1F, which shows the fluorescence signal of CdSe NPs (I) and AP-CdSe NPs (II). The morphology and size distribution of the NPs are shown in Figure 1D and E, respectively. The average size of the obtained CdSe NPs and AP-CdSe NPs were 9 nm and 14 nm, respectively (Figure S7). Amount of AP released into the solutions was determined by measuring the absorbance at 234 nm using UV-vis spectroscopy at different time points. The Figure S5 showed the initial rate of release was much faster, the amount of release was much larger, and the time to reach equilibrium was at 12 h. This experiment demonstrated that the release rate of AP from nanoparticles was up to 82%. The stability of AP-CdSe NPs in bacterial culture suspension was evaluated by examining the size of NPs (Figure S6) in vitro. When the NPs were dispersed in PBS and bacterial culture medium for more than 196 h, no obvious size change was observed, indicating good stability in biological media. Taken together, these results suggest that APs were successfully decorated on the surface of CdSe NPs and exhibited fluorescence properties.
Antibacterial Activity of AP-CdSe NPs
AP have been reported as novel and effective antibacterial active short peptide fragments. However, since some APs have low water solubility, their bioavailability is extremely low.38,39 In this study, the polarity of an AP was successfully changed to improve its antibacterial activity, as indicated by the results shown in Figure 2 and Table 1. For instance, the MIC values (Table 1) for the AP against regular E. coli, S. aureus, M. tetragenus, P. aeruginosa and B. subtilis were relatively high at 48.0, 48.0, 64.0, 48.0 and 64.0 μg/mL, respectively. Also, there was only a little difference in the MIC values for the AP against MDR bacteria. On the other hand, AP-CdSe NPs not only showed relatively low MIC values against these regular bacterial cells at 6.0, 4.0, 24.0, 16.0 and 24.0 μg/mL, but also against MDR bacteria at 16.0, 24.0, 8.0 and 8.0 μg/mL, respectively. By penetrating the bacterial cell membrane, the AP-conjugated CdSe NPs had enhanced antibacterial activity according to the MIC assay results. AP-CdSe NPs had a more prominent effect on the activity of S. aureus (Gram-positive bacteria) and E. coli (Gram-negative bacteria) than on the other bacterial cells.
These results were also consistent with those obtained by the CFU assays, shown in Supplementary Figure S8 and Figure 2. As shown in the Supplementary Figure S8, photographs of the inhibition zone in cultures of MDR E. coil and S. aureus after treatment with AP-CdSe NPs (40 µg/mL) were qualitative test of antibacterial drugs and measured. The diameter of the inhibition zone of AP-CdSe NPs can reach 27.5 mm for MDR S. aureus and 16.5 mm for MDR E. coil. The results showed that both the MDR E. coil and S. aureus are sensitive to AP-CdSe NPs, and the sensitivity of S. aureus is stronger than that of E. coil. The sensitivity of raw AP was bad. The CFU assays were used to evaluate the antibacterial activity of AP-CdSe NPs against MDR E. coli and S. aureus. The results showed that AP-CdSe NPs at 40 μg/mL against MDR E. coli and S. aureus, resulted in survival rates of 10.3 and 3.8%, respectively, whereas the survival rates of AP at 40 μg/mL were 43.7% and 22.3%, and those of CdSe NPs at 40 μg/mL were 96.7% and 95.3%, respectively (Figure 2). These results revealed that CdSe NPs alone had no antibacterial activity. In addition, the inhibitory effect of AP-CdSe NPs was higher than that of AP and CdSe NPs. Besides showing that AP-CdSe NPs have high antibacterial activity, these results also proved that AP-CdSe NPs exhibit a synergistically enhanced antibacterial performance against MDRs bacteria.
Cellular Uptake and Antibacterial Activity Assays
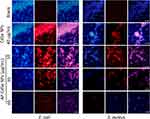
The drug delivery efficacy and antibacterial activity were investigated by fluorescence microscopy analysis. All MDR E. coli and MDR S. aureus cells were stained with DAPI, including the blank group, and with AP-CdSe NPs at various concentrations (10, 20, 40 µg/mL). The merged images indicated the drug delivery efficacy and antibacterial activity of AP-CdSe NPs. The results in Figure 3, where red cells are those stained with AP-CdSe NPs and blue cells are stained with DAPI, show that AP-CdSe NPs were taken up at the high rate of 100% in MDR E. coli and MDR S. aureus cells. Even at 10 µg/mL, AP-CdSe NPs could enter the cells, but the antibacterial effect is poor. At the concentration of 40 µg/mL of AP-CdSe NPs, the number of cells in the field of view is smaller, but shows high antibacterial activity. In addition, the sampling time in cellular uptake assays had been set at the shorter time when most cells are still alive. It is clear that in just 1 hour, AP-CdSe NPs also completely enter the inside of the cell (Figure S9). Above all, the results show that AP-CdSe NPs are easily taken up by bacterial cells and have a good antibacterial activity against MDR E. coli and MDR S. aureus.
Fluorescence Microscopy Analysis of Antibacterial Activity
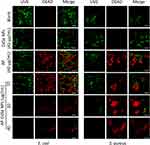
The CFU assay showed the remaining viable cells and evaluated antibacterial activity, but it did not reveal the dead cells. The uptake assay showed the decrease of cell number under the microscope and evaluated antibacterial activity.37,38 Thus, the results of these assays only Indirectly explain the antibacterial effect of the AP-CdSe NPs. Therefore, the live-dead assay was used to directly investigate the antibacterial activity of AP-CdSe NPs. In Figure 4, green fluorescent cells represented the living bacterial cells, while the red fluorescent cells represented dead bacterial cells. There were very few living MDR E. coli and MDR S. aureus cells (green fluorescent cell) in the blank group. Similar results were obtained for the group treated with CdSe NPs, which indicated that the NPs had no antibacterial activity against MDR E. coli and MDR S. aureus. There were some dead cells (red fluorescent cells) in the AP-treated groups, indicating that AP alone had antibacterial activity. However, the same concentration of AP-CdSe NPs displayed higher antibacterial activity compare with the AP-treated groups. All areas had dead cell (red fluorescent cells) and showed high killing rate. The reason might be that the AP loaded on CdSe NPs had a higher water solubility and bioavailability.
To further clarify that the AP-CdSe NPs reduce the survival rate of bacteria, a quantitative analysis of live/dead cells was performed by flow cytometry analysis using the SYTO 9-PI apoptosis detection kit in MDR E. coli and MDR S. aureus. The results shown in Figure 5A and B revealed that the apoptosis rates of the blank group were 3.16% and 11.5%, and those of the group treated with CdSe (40 µg/mL) were 12.89% and 12.65% respectively. The results also revealed that CdSe NPs alone had no antibacterial activity. On the other hand, the AP (40 µg/mL) exhibited antibacterial activity with apoptosis rates of 61.36% and 59.47%, respectively. The result is consistent with the conclusion reported by others.40–42 Also, as shown in Figure 5, after loading AP on CdSe NPs, the antibacterial activity of AP-CdSe NPs (40 µg/mL) was significantly higher than that of the AP-treated groups, with apoptosis rates of 98.76% and 99.13% for MDR E. coli and MDR S. aureus, respectively. The quantitative analysis of live/dead cells by flow cytometry analysis further confirmed the enhanced antibacterial activity of AP-CdSe NPs.
Cell Integrity Study
According to the results of the MIC assay, CFU assay, fluorescence microscopy analysis, and flow cytometry analysis, AP-CdSe NPs have highly effective antibacterial activity against MDR E. coli and MDR S. aureus. Changes in cell integrity may be the main mechanism of inducing bacterial cell apoptosis.43 Furthermore, the effects of the antibacterial activity of AP-CdSe NPs on the cell morphology and structure were examined by SEM (Figure 6) and subcellular ultrastructure analysis by TEM (Figure 7).
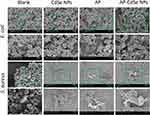
The integrity of the bacterial cell, including the cell membrane, was examined by SEM, as shown in the SEM images of MDR E. coli and MDR S. aureus treated with AP-CdSe NPs, displayed in Figure 6. The E. coli and S. aureus cells in the blank group maintained complete cell morphology and the integrity of the membrane structure. The enlarged region confirmed that cells were smooth and had an intact cell membrane. Similar integrity of the membrane structure was observed in bacterial culture treated with CdSe NPs. Also, the results revealed that CdSe NPs alone had no antibacterial activity. The E. coli and S. aureus cells in the AP-treated group showed damaged membrane structures, and the enlarged region revealed that cells had not intact cell membrane and leaked intracellular contents. The AP showed effective antibacterial activity against MDR E. coli and MDR S. aureus based on the damaged cell area. After the E. coli and S. aureus cells were treated with AP-CdSe NPs (40 µg/mL), the damaged cell area became larger in SEM images. The number of damaged cells increased significantly, and the results also indicated that the damage to the cells was more severe with increasing concentration of AP-CdSe NPs, indicating that the sensitivity of bacteria to AP-CdSe NPs is concentration-dependent. Taken together, these results indicated that AP-CdSe NPs had highly effective antibacterial activity compared with CdSe NPs and AP alone.
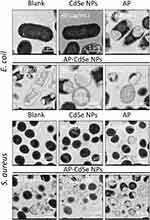
The evaluation of the antibacterial activity of AP-CdSe NPs through subcellular ultrastructure analysis by TEM revealed the changes in the internal structure of cells compared with the SEM images.44 As shown in the Figure 7, the internal structure of E. coli and S. aureus cells was intact in the blank group; the TEM image clearly show the bacterial nuclear and cytoplasmic areas inside the cell. Similarly, the internal structure of E. coli and S. aureus cells was also intact in bacterial culture treated with CdSe NPs. The subcellular ultrastructural analysis of bacterial culture treated with AP by TEM revealed that their internal morphology was significantly different from that of normal cells, as cells in the field of view appeared broken and the cytoplasm was missing. Moreover, in cells of the group treated with AP-CdSe NPs the cell damage was more pronounced than that in cells of the AP-treated group. Additionally, it was also found that the number of damaged cells in the AP-CdSe NPs group increased with the increase of the concentration of AP-CdSe NPs. The SEM and TEM images revealed that AP-CdSe NPs induce cell death combined with the destruction of the cell integrity, suggesting a possible mechanism of cell death that needs further verification.
Gene Expression Analysis
Gene expression profiling, such as by RNASeq analysis, can be used to study the metabolism at the cellular molecular level. cDNA from E. coli (control and AP-CdSe NPs groups) was sequenced in triplicate. Statistical analysis revealed a high total number of reads of the sequencing samples and a high ratio of high-quality reads as shown in the Table 2. The results suggested that the sequencing data were of good quality. In order to identify DEGs, the expression of each gene can be analyzed by FPKM (the expected number of fragments per kilobase of the transcript sequence per kilobase pair). The HTSeq program was used to analyze differential gene expression. The density estimation of gene expression levels in the AP-CdSe NPs and control groups is compared in Figure 8A. The results showed that the AP-CdSe NPs affected gene expression. To determine the differential gene expression in E. coli bacterial culture treated with AP-CdSe NPs, this study identified DEGs relative to control groups. As shown in Figure 8B, the number of DEGs was 373, including 216 upregulated genes and 157 downregulated genes. Gene expression cluster analysis was used to determine the clustering pattern of gene expression in the control group and AP-CdSe NPs group, and the results are shown in Figure 8C. We selected the top 50 DEGs with the largest expression variance for cluster analysis. These DEGs will be used for GO and KEGG enrichment analysis. The correlation analysis between samples is based on the expression levels of all genes or transcripts obtained by sequencing and is a way to analyze whether the expression profiles between two samples are correlated. As shown in Figure 8D, the results indicated a high degree of correlation between the control and AP-CdSe NPs. Also, the experiment with biological replicate samples is reliable and the sample selection is reasonable. The R2 value (0.859) < 0.9 indicated a poor-level of correlation between the control group with the AP-CdSe NPs group. Such result may be due to the treatment of E. coli cells with AP-CdSe NPs. Taken together, these results suggested that the E. coli bacterial culture treated with AP-CdSe NPs had shown differential expressions. This may be the antibacterial molecular mechanism of AP-CdSe NPs.
 |
Table 2 The RNASeq Outcomes of Control and AP-CdSe NPs Samples |
Antibacterial Pathway Analysis by GO and KEGG Enrichment Analysis
GO is often used to provide background knowledge of gene function classification labels and gene function research. The use of the GO database to search through species and gene information, thus provides the GO annotation information (functional information) of the DEGs. The distribution information and significance of the gene set in the GO category are obtained based on the GO annotation of the gene. GO analysis identified a total of 6 terms related to cellular components (CC), 15 terms for biological processes (BP), and 15 terms for molecular functions (MF) after E. coli treatment with AP-CdSe NPs (Figure 9). Regarding CC ontology, the major represented categories were “small molecule catabolic process” (GO:0044282) and “organic acid catabolic process” (GO:0016054). Regarding BP ontology, the major represented category was “periplasmic space” (GO:0042597). Regarding MF ontology, the major represented categories were “small molecule binding” (GO:0036094) and “bacterial-type proximal promoter sequence” (GO:0000986). The relative cDNA abundance of GO analysis terms was detected by RT-PCR analysis, and the results confirmed that AP-CdSe NPs significantly changed the cDNA abundance. All results showed that gene expression in E. coli bacterial culture treated with AP-CdSe NPs suggested a molecular mechanism for the antibacterial effect of AP-CdSe NPs.
 |
Figure 9 Differentially expressed genes (DEGs) GO enrichment analysis results. |
KEGG is a database containing biochemical reactions, signaling pathways, metabolic pathways and biological processes.45 The biological pathway enrichment analysis based on the KEGG database is performed on the DEG set, and pathways with significantly enriched DEGs are extracted, which is useful for the development of downstream experiments. Through the KEGG annotation of genes, the hypergeometric distribution is used to calculate the P value, and a P <0.05 is used as the significance threshold to obtain statistically significant high-frequency annotations relative to the background, in order to obtain the distribution information and significant enrichment of genes in KEGG pathways (Figure 10). KEGG enrichment analyses of E. coli treated by AP-CdSe NPs transcriptome sequences revealed the presence of significant enrichment of DEGs in three pathways compared with the control group. The results showed that AP-CdSe NPs affected the bacterial metabolic pathways, thus inhibiting E. coli cell growth.
 |
Figure 10 Differentially expressed genes (DEGs) KEGG enrichment analysis results. |
Antibacterial Activity of AP-CdSe NPs in vivo
Through in vitro antibacterial test, we confirmed that AP-CdSe NPs have high antibacterial activity. At the same time, transcriptome analysis was performed to determine the antibacterial molecular mechanism of AP-CdSe NPs. Further evaluation of the antibacterial activity of AP-CdSe NPs in vivo will greatly increase their potential clinical application. Due to its drug resistance and high mortality, S. aureus infections have become a major problem in public health worldwide. Therefore, we explored and established a bacteremia model in mice infected with S. aureus.
The biodistribution of bacteria in mice of different treatment groups (blank, infection and treatment groups) was analyzed at different times to determine the antibacterial activity of AP-CdSe NPs in vivo. As shown in Figure 11A, the number of bacteria in blood and organs in the blank group was almost the same (~1.1×104 CFU/mL or g) and remained at such level for 7 days. After injecting bacteria into the mice through the tail vein, the number of bacteria in blood and organs was ~3.3×104 CFU/mL or g in the infected group at 7 days. These results indicated that the bacteremia model was successfully developed. The AP alone treated mice with bacteremia exhibited antibacterial effect, as indicated by the significant decrease in the number of bacteria in these mice, although the clinical symptoms of bacterial infection were still present. This finding showed that AP alone was not enough to cure mice with bacteremia. However, when AP-CdSe NPs was injected into S. aureus infected mice, the number of bacteria in the blood and organs of the mice gradually decreased over time. The number of bacteria was almost the same as that in the blank group at 7 days, indicating that AP-CdSe NPs could also inhibit bacterial activity in vivo and cure mice with bacteremia. The survival rate and weight of mice were monitored after tail vein injection, as shown in Figure 11B. The survival rate of the AP-CdSe NPs group was 83%, which was significantly higher than that (31%) of the non-treated infected group. The results showed that AP-CdSe NPs improved the survival rate of mice with bacteremia compared with the AP-treated S. aureus infected group. This finding further confirmed the antibacterial effect of AP-CdSe NPs in vivo.
Clinical evaluation of the treatment of mice with bacteremia can be achieved not only through the biological distribution of bacteria, but also by immunostaining of major organs. The mice in the blank group did not have any histological damage or bacteremia. However, the organs and tissues of the S. aureus infected mice presented obvious symptoms of clinical bacteremia. Meanwhile, in the AP treatment group, the organ tissue damage of the mice was repaired, but there were obvious symptoms of clinical bacteremia. On the other hand, in the AP-CdSe NPs treatment group, the injured tissue completely recovered and appeared to be in a normal tissue state (Figure 11C). These results indicated that AP-CdSe NPs can heal tissue damage caused by bacteremia in mice. This result is consistent with the result of the biodistribution of bacteria shown in Figure 11A, confirming the highly effective antibacterial effect of AP-CdSe NPs in vivo.
Safety Evaluation
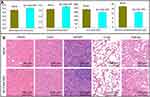
Side effects of the AP-CdSe NPs were examined to evaluate its safety. Blood biochemical assays were also performed to examine possible organ toxicity in mice after treatment with AP-CdSe NPs. As shown in Figure 12A, the levels of blood glucose, cholesterol, uric acid, and alanine aminotransferase (indexes for the function of kidney, liver, blood glucose, and blood lipids, respectively) all showed no significant changes, compared with the blank group, and were within normal range values. The assessment of histopathological changes by H&E staining of major mouse organs revealed that the AP-CdSe NPs treatment groups all showed similar histological features compared with the blank groups, and no obvious impairment or inflammation was observed in any of the groups (Figure 12B). These results confirmed that AP-CdSe NPs was non-toxic and had no side effects.
Conclusion
In this study, an AP was synthesized by the Fmoc solid-phase method and decorated on the CdSe NPs. The results of the characterization of the newly developed AP-CdSe NPs revealed that they had good monodispersity, size stability and consistent spectral characteristics. The AP-CdSe NPs had a diameter of approximately 14 nm and showed excellent fluorescence properties. In addition, CFU assays showed that AP-CdSe NPs display a broad-spectrum antibacterial activity, and had highly effective antibacterial activities against MDR E. coli and S. aureus. The MIC assays showed that the AP-CdSe NPs had highly effective antibacterial activities at low dosages of 8.0 μg/mL against MDR E. coli and MDR S. aureus. Fluorescence microscopy analyses revealed that AP-CdSe NPs had high cellular uptake rate and killing rate for E. coli and S. aureus. The quantitative analysis of live/dead cells by flow cytometry analysis further confirmed the antibacterial activity of AP-CdSe NPs. Moreover, SEM and TEM observations were performed to investigate the mechanisms of cell integrity by examining changes in cell membrane and wall. Gene expression analysis of E. coli bacterial culture treated with AP-CdSe NPs indicated a molecular mechanism for the antibacterial effect of AP-CdSe NPs. GO and KEGG pathway enrichment analyses revealed key biological pathways involved in the metabolic pathways of E. coli treated with AP-CdSe NPs. In addition, AP-CdSe NPs inhibited bacterial activity and healed tissue damage in a bacteremia model in mice infected with S. aureus. The evaluation of the biosafety of the AP-CdSe NPs showed that they do not have any side effects. This study also reports that the transcriptome analysis of the mechanism provides novel insights into the molecular mechanism of the antibacterial effects of AP-CdSe NPs. Both the in vitro and the in vivo studies suggested that AP-CdSe NPs can be a potential nanoscale antimicrobial agent against E. coli and S. aureus.
Acknowledgments
Anhui Nature Science Foundation (1808085QC85 and 2008085QH397), Anhui University of Technology Introduced Talent Research Startup Fund (2019YQQ010), Anhui polytechnic university youth talent support program (2016BJRC006) and Science and Technology Innovation Training Program (201710363084).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors have declared that there are no competing interests in this work.
References
1. Almeida JR, Palacios ALV, Patiño RSP, et al. Harnessing snake venom phospholipases A2 to novel approaches for overcoming antibiotic resistance. Drug Develop Res. 2019;80(1):68–85. doi:10.1002/ddr.21456
2. Tan P, Lai Z, Zhu Y, Shao C, Shan A. Engineering. multiple strategy optimization of specifically targeted antimicrobial peptide based on structure–activity relationships to enhance bactericidal efficiency. ACS Biomater Sci Eng. 2019;6(1):398–414. doi:10.1021/acsbiomaterials.9b00937
3. Chen Y, Gao Y, Chen Y, Liu L, Peng Q, Peng Q. Nanomaterials-based photothermal therapy and its potentials in antibacterial treatment. J Control. Release. 2020;328:251–262. doi:10.1016/j.jconrel.2020.08.055
4. He B, Ma S, Peng G, He DJNNB. Medicine. TAT-modified self-assembled cationic peptide nanoparticles as an efficient antibacterial agent. Nanomed Nanotechnol. 2018;14(2):365. doi:10.1016/j.nano.2017.11.002
5. Raheem N, Straus S. mechanisms of action for antimicrobial peptides with antibacterial and antibiofilm functions. Front Microbiol. 2019;10:2866. doi:10.3389/fmicb.2019.02866
6. Bucki R, Niemirowicz-Laskowska K, Deptua P, Wilczewska AZ, Janmey P. Susceptibility of microbial cells to the modified PIP2-binding sequence of gelsolin anchored on the surface of magnetic nanoparticles. J Nanobiotechnol. 2019;17(1):81. doi:10.1186/s12951-019-0511-1
7. Li X, Bai H, Yang Y. Supramolecular antibacterial materials for combatting antibiotic resistance. Adv Mater. 2019;31(5):e1805092. doi:10.1002/adma.201805092
8. Zhou J, Liu Y, Shen T, Chen L, Wang CJMP. Antimicrobial activity of the antibacterial peptide PMAP-36 and its analogues. Adv Mater. 2019;136:103712. doi:10.1002/adma.201805092
9. Hou Z, Shankar YV, Liu Y, et al. Nanoparticles of short cationic peptidopolysaccharide self-assembled by hydrogen bonding with antibacterial effect against multidrug-resistant bacteria. ACS Appl Mater Inter. 2017;9(44):38288–38303. doi:10.1021/acsami.7b12120
10. Shuai J, Guan F, He B. Self-assembled nanoparticles of symmetrical cationic peptide against citrus pathogenic bacteria. J Agr Food Chem. 2019;67(20):5720–5727. doi:10.1021/acs.jafc.9b00820
11. Huang N, Chen X, Zhu X, Xu M, Liu JJB. Ruthenium complexes/polypeptide self-assembled nanoparticles for identification of bacterial infection and targeted antibacterial research. Biomaterials. 2017;141:296–313. doi:10.1016/j.biomaterials.2017.07.005
12. Tao B, Lin C, Deng Y, Zhang Y, Cai K. Copper-nanoparticle-embedded hydrogel for killing bacteria and promoting wound healing with photothermal therapy. J Mater Chem B. 2019;7(15):2534–2548. doi:10.1039/c8tb03272f
13. Richards SJ, Isulfi K, Wilkins LE, Lipecki J, Fullam E, Gibson MIJB. Multivalent antimicrobial polymer nanoparticles target mycobacteria and gram-negative bacteria by distinct mechanisms. BiomacromoleculesI. 2017;19(1):256–264. doi:10.1021/acs.biomac.7b01561
14. Zharkova MS, Orlov DS, Golubeva OY, et al. Application of antimicrobial peptides of the innate Immune system in combination with conventional antibiotics—a novel way to combat antibiotic resistance? Front Cell Infect Mi. 2019:9. doi:10.3389/fcimb.2019.00128
15. Torres LMFC, Almeida MT, Santos TL, et al. Antimicrobial alumina nanobiostructures of disulfide- and triazole-linked peptides: synthesis, characterization, membrane interactions and biological activity. Colloid Surface B. 2019;2017(177):94–104. doi:10.1016/j.colsurfb.2019.01.052
16. Subbalakshmi C, Nagaraj RJC. Characterization of silver and fluorescent dyes incorporated in self‐assembling peptides and their potential applications in biology. ChemPlusChem. 2014;79(9):1326–1333. doi:10.1002/cplu.201402077
17. Gao J, Na H, Zhong R, Yuan M, Zhang FJC. One step synthesis of antimicrobial peptide protected silver nanoparticles: the core-shell mutual enhancement of antibacterial activity. Colloid Surface B. 2019;186:110704. doi:10.1016/j.colsurfb.2019.110704
18. Mohtashamian S, Boddohi S. Nanostructured polysaccharide-based carriers for antimicrobial peptide delivery. J Pharm Innov. 2016;47(2):85–94. doi:10.1007/s40005-016-0289-1
19. Nasrin F, Chowdhury AD, Takemura K, et al. Single-step detection of norovirus tuning localized surface plasmon resonance-induced optical signal between gold nanoparticles and quantum dots. Biosens Bioelecteron. 2018;122:16–24. doi:10.1016/j.bios.2018.09.024
20. Sabek J, Torrijos-Moran L, Griol A, et al. Real time monitoring of a UV light-assisted biofunctionalization protocol using a nanophotonic biosensor. Biosensors-Basel. 2018;9(1):1. doi:10.3390/bios9010006
21. Maiti S, Kundu S, Roy CN, Das TK, Saha A. Synthesis of excitation independent highly luminescent graphene quantum dots through perchloric acid oxidation. Langmuir. 2017;33(51):14634–14642. doi:10.1021/acs.langmuir.7b02611
22. Rezaei H, Motovali-bashi M, Radfar S. An enzyme-free electrochemical biosensor for simultaneous detection of two hemophilia A biomarkers: combining target recycling with quantum dots-encapsulated metal-organic frameworks for signal amplification. Anal Chim Acta. 2019;1092:66–74. doi:10.1016/j.aca.2019.09.037
23. Dennis AM, Buck MR, Wang F, et al. Role of interface chemistry in opening new radiative pathways in InP/CdSe giant quantum dots with blinking‐suppressed two‐color emission. Adv Funct Mater. 2019;29(37):1809111. doi:10.1002/adfm.201809111
24. Wg A, Yw B, Yx B, et al. Size-dependent anti-inflammatory activity of a peptide-gold nanoparticle hybrid in vitro and in a mouse model of acute lung injury. Acta Biomater. 2019;85:203–217. doi:10.1016/j.actbio.2018.12.046
25. Zhou J, Liu Y, Shen T, Chen L, Wang CJCB, Design D. Enhancing the antibacterial activity of PMAP‐37 by increasing its hydrophobicity. Chem Biol Drug Des. 2019;94(4):1986–1999. doi:10.1111/cbdd.13601
26. Pham T-N, Loupias P, Dassonville‐Klimpt A, Sonnet P. Drug delivery systems designed to overcome antimicrobial resistance. Med Res Rev. 2019;39(6):2343–2396. doi:10.1002/med.21588
27. Sun Z, Zheng W, Zhu G, Lian J, Jiang XJAAM. Interfaces. albumin broadens the antibacterial capabilities of nonantibiotic small molecule-capped gold nanoparticles. ACS Appl Mater Inter. 2019;11(49):45381–45389. doi:10.1021/acsami.9b15107
28. Makowski M, Silva TC, Amaral CPD, Goncalves S, Santos NCJP. Advances in Lipid and Metal Nanoparticles for Antimicrobial Peptide Delivery. Pharmaceutics. 2019;11(11):588. doi:10.3390/pharmaceutics11110588
29. Sun D, Zhang W, Mou Z, et al. Transcriptome analysis reveals silver nanoparticle-decorated quercetin antibacterial molecular mechanism. ACS Appl Mater Inter. 2017;9(11):10047–10060. doi:10.1021/acsami.7b02380
30. Kuang H, Ku SH, Kokkoli EJADDR. The design of peptide-amphiphiles as functional ligands for liposomal anticancer drug and gene delivery. Adv Drug Deliver Rev. 2017;110:80–101. doi:10.1016/j.addr.2016.08.005
31. Mihklepp K, Kivirand K, Nikopensius M, et al. Design and Production of Antibodies for the Detection of Streptococcus uberis. Enzyme Microb Tech. 2017;96:135–142. doi:10.1016/j.enzmictec.2016.10.009
32. Yang X, Zhang W, Zhao Z. Quercetin loading CdSe/ZnS nanoparticles as efficient antibacterial and anticancer materials. J Inorg Biochem. 2017;167:36–48. doi:10.1016/j.jinorgbio.2016.11.023
33. Sun D, Zhang W, Li N, et al. Silver nanoparticles-quercetin conjugation to siRNA against drug-resistant Bacillus subtilis for effective gene silencing: in vitro and in vivo. Mat Sci Eng C-Mater. 2016;63:522–534. doi:10.1016/j.msec.2016.03.024
34. Rekha R, Vaseeharan B, Vijayakumar S, et al. Crustin-capped selenium nanowires against microbial pathogens and Japanese encephalitis mosquito vectors – insights on their toxicity and internalization. J Trace Elem Med Bio. 2019;51:191–203. doi:10.1016/j.jtemb.2018.10.017
35. Lu B, Zhu G, Yu C, et al. Functionalized graphene oxide nanosheets with unique three-in-one properties for efficient and tunable antibacterial applications. Nano Res. 2021;14(1):185–190. doi:10.1007/s12274-020-3064-6
36. Yga B, Yca B, Yc C, Am B, Qp A. Potentials of nanotechnology in treatment of methicillin-resistant Staphylococcus aureus. Eur J Med Chem. 2020;113056.
37. Obrecht D, Robinson J, Bernardini F. Recent progress in the discovery of macrocyclic compounds as potential anti-infective therapeutics. Curr Med Chem. 2009;16(1):42–65. doi:10.2174/092986709787002844
38. Splith K, Neundorf IJEBJ. Antimicrobial peptides with cell-penetrating peptide properties and vice versa. Eur Biophys J Biophy. 2011;40(4):387–397. doi:10.1007/s00249-011-0682-7
39. Zhang W, Yu W, Ding X, et al. Self-assembled thermal gold nanorod-loaded thermosensitive liposome-encapsulated ganoderic acid for antibacterial and cancer photochemotherapy. Artif Cell Nanomed B. 2019;47(1):406–419. doi:10.1080/21691401.2018.1559177
40. Yoon IN, Lu LF, Hong J, et al. The American cockroach peptide periplanetasin‐4 inhibits Clostridium difficile toxin A‐induced cell toxicities and inflammatory responses in the mouse gut. J Pept Sci. 2017;23(11):833–839. doi:10.1002/psc.3046
41. Lee H, Hwang JS, Dong GLJBJ. Periplanetasin-4, a novel antimicrobial peptide from the cockroach, inhibits communications between mitochondria and vacuoles. Biochem J. 2019;476(8):1267–1284. doi:10.1042/BCJ20180933
42. Kim IW, Lee JH, Seo M, Lee HJ, JSJJoM H. Biotechnology. anti-inflammatory activity of antimicrobial peptide periplanetasin-5 derived from the cockroach periplaneta americana. J Microbiol Biotechn. 2020;30(9):1282–1289. doi:10.4014/jmb.2004.04046
43. Yu CH, Chen GY, Xia MY, Xie Y, Peng Q. Understanding the sheet size-antibacterial activity relationship of graphene oxide and the nano-bio interaction-based physical mechanisms. Colloid Surfaces B. 2020;191:111009. doi:10.1016/j.colsurfb.2020.111009
44. Xia MY, Xie Y, Yu CH, Chen GY, Peng Q. Graphene-based nanomaterials: the promising active agents for antibiotics-independent antibacterial applications. J Control Release. 2019;307:16–31. doi:10.1016/j.jconrel.2019.06.011
45. Li J, Wang Z, Qiu W, et al. The effect of interaction between EtOH dosage and exposure time on gene expression in DPSC. Genomics. 2019;111(3):500–507. doi:10.1016/j.ygeno.2018.03.009
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.