Back to Journals » Infection and Drug Resistance » Volume 11
Systematic review in South Africa reveals antibiotic resistance genes shared between clinical and environmental settings
Authors Ekwanzala MD, Dewar JB, Kamika I , Momba MNB
Received 10 April 2018
Accepted for publication 17 June 2018
Published 17 October 2018 Volume 2018:11 Pages 1907—1920
DOI https://doi.org/10.2147/IDR.S170715
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eric Nulens
Mutshiene Deogratias Ekwanzala,1 John Barr Dewar,2 Ilunga Kamika,3 Maggy Ndombo Benteke Momba1
1Department of Environmental, Water and Earth Sciences, Tshwane University of Technology, Pretoria, South Africa; 2Department of Life and Consumer Sciences, University of South Africa, Johannesburg, South Africa; 3Department of Environmental Sciences, University of South Africa, Johannesburg, South Africa
Abstract: A systematic review was conducted to determine the distribution and prevalence of antibiotic-resistant bacteria (ARB), antimicrobial-resistant genes (ARGs), and antimicrobial-resistant gene determinants (ARGDs) in clinical, environmental, and farm settings and to identify key knowledge gaps in a bid to contain their spread. Fifty-three articles were included. The prevalence of a wide range of antimicrobial-resistant bacteria and their genes was reviewed. Based on the studies reviewed in this systematic review, mutation was found to be the main genetic element investigated. All settings shared 39 ARGs and ARGDs. Despite the fact that ARGs found in clinical settings are present in the environment, in reviewed articles only 12 were found to be shared between environmental and clinical settings; the inclusion of farm settings with these two settings increased this figure to 32. Data extracted from this review revealed farm settings to be one of the main contributors of antibiotic resistance in healthcare settings. ARB, ARGs, and ARGDs were found to be ubiquitous in all settings examined.
Keywords: systematic review, ARB, clinical ARGs, environmental ARGs, ARGDs, South Africa
Introduction
The emergence and spread of antibiotic-resistant bacteria (ARB), antimicrobial-resistant genes (ARGs), and antimicrobial-resistant gene determinants (ARGDs) have been portrayed as one of the leading challenges of the 21st century and a health issue of concern that is rapidly expanding worldwide.1 Each year in Europe, 400,000 patients experience ill effects due to infection by antibiotic-resistant microorganisms, with an associated mortality of 25,000 patients.2 More and more enteric bacteria are being reported as being drug resistant in the USA,3 where antibiotic resistance (AR) is accountable for >2 million hospitalizations and at least 23,000 deaths annually.4 In addition to the direct cost of hospital services, diseases caused by antimicrobial-resistant microorganisms result in an individual and societal economic burden. In Europe only, reports by the European Centre for Disease Prevention and Control and European Medicines Agency evaluated the general expense of AR to society at €1.5 billion per year.2 It has been reported that accessible data are lacking to evaluate the financial ramifications regionally or nationally at the point when viable treatment for an infection is totally lost as an aftereffect of such resistance.1
The African region has shown negative trends in multiple resistance among key enteric pathogens such as Escherichia coli, Klebsiella, Salmonella spp., Vibrio cholerae, and Shigella spp. to nearly all commonly available antibiotics.5–7 Issues related to AR and its magnitude in Africa are hampered by lack of a surveillance system.8 Many African reviews suggest the need for a proper continental surveillance system to gather complete and adequate data on the true extent of the AR problem.9–11 It is therefore imperative that this rising trend should be controlled, as enteric bacterial infections pose a heavy challenge to human populations, particularly among children and immune-suppressed individuals in developing countries, where malnutrition, HIV/AIDS, and poor sanitation abound.8 In South Africa (SA), four major outbreaks of AR have been identified at the national level. The National Institute for Communicable Disease12 reported a methicillin-resistant Staphylococcus aureus in 2010;13 an extended-spectrum β-lactamase (ESBL) producing Klebsiella pneumoniae from 2010 to 2012;14 vancomycin-resistant Enterococci in 2012, and carbapenemase producing Enterobacteriaceae. In response to the outbreaks, the South African government established the South African Antimicrobial Resistance Strategy framework to combat the spread of AR.15 Its strategies focus mainly on ARB in clinical and healthcare settings. Currently, there is no strategy in place to contain and track the movement of ARB, ARGs, and ARGDs of environmental isolates in an attempt to prevent them from reaching clinics and healthcare settings. Although ARGs are regarded as emerging environmental contaminants, environments have globally been portrayed as one of the main contributors to the burden of AR.16
Several environmental ARB and ARGs resistomes, otherwise referred to as AR hotspots, have been identified. These include the following aquatic environments: surface water bodies (rivers, lakes, and streams),16,17 groundwater,18 effluents of hospitals wastewater,19 and municipal wastewater.20,21 In terrestrial environments, river sediments22–24 and antibiotic-treated manure soils25 have also been identified as contributing to the spread of ARB, ARGs, and ARGDs. In addition to these resistomes, food26 and drinking water27 have been found to be direct key reservoirs of ARB, ARGs, and ARGDs associated with human infection. As a result, the presence and dissemination of ARB, ARGs, and ARGDs in environmental settings present a profound threat to public health and highlight the need to be assessed against clinical settings in the South African context, so that the findings can be applied worldwide.
This quinquennial systematic review focused on the genetic epidemiology of ARB, ARGs, and ARGDs in SA from January 1, 2011 to December 31, 2016. The main objective of the current study was to elucidate the distribution and prevalence of ARB, ARGs, and ARGDs in different settings (eg, clinical, environmental, and farm settings) to identify key knowledge gaps in a bid to contain their spread and establish shared genes between the environmental and clinical settings.
Methodology
This quinquennial systematic review on ARGs was compiled using the PRISMA guidelines.28
Literature review
The literature search was performed using six online databases: PubMed Database, EBSCOhost Online Research Databases, MEDLINE, ISI Web of Knowledge, African Journals Online, and Scopus in January 2017 by three reviewers (MDE, PB, and VKTP – acknowledged under the “Acknowledgments” section). Predefined terms such as (Antibiotic OR Resistance OR Bacteria OR ARB OR Gene* OR ARG* OR Determinant* OR ARGD*) AND (South Africa OR SA) were used to retrieve relevant articles published from January 1, 2011 to December 31, 2016. No limiters or refiners were applied so to include gray literature in different databases.
Study selection criteria
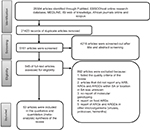
Figure 1 summarizes the steps taken to conduct the literature search and selection. The first step entailed removing duplicate articles that were found in the six databases. Subsequent to the removal of duplicate articles, the remaining articles were screened based on their title and abstract screening. Full-text articles were read and screened. Studies conducted outside SA were removed, as were those not reporting molecular detection of ARGs’ encoding gene. The remaining 53 full-text articles were read and included in the review. Only articles that contained information on the detection of AR encoding genes in clinical, environmental, and farm settings were included in this review. Food ARGs were also excluded to account only for direct environmental ARG input. Articles referring to ARGs detected from viruses and other microorganisms other than bacteria were also excluded.
In this review, ARGs and ARGDs were classified under clinical settings when isolated from hospital inpatient, outpatient, and community settings. Environmental settings constituted all the ARGs and ARGDs that occurred in the natural environments or entered the natural environments (studies reporting food ARGs were not included based on this classification). These environments included aquaculture, freshwater, groundwater, hospital, and municipal wastewater. ARGs and ARGDs isolated from agricultural environments (animal farms, rhizospheres, crop areas, and animal stool) were classified under farm settings.
Data extraction
Relevant data extracted from each of the full-text articles are shown in Table 1. Additionally, three others (MDE, PB, and VKTP – acknowledged under the “Acknowledgments” section) also extracted the same data independently to ensure reliability. When variance occurred, various articles were checked to reach a final decision. Relevant data extracted included first author’s name, year of sample collection, location of samples collected, isolated bacteria, AR class targeted, method used for molecular isolation and characterization of the ARGs, samples matrix and settings, and finally the results obtained. The results obtained included bacterial species and strain isolated, number of isolates tested for AR, specific antibiotics tested for resistance, percentage of resistant bacteria, and genetic epidemiology of ARGs detected.
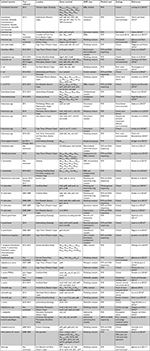  | Table 1 Characteristics of eligible articles Abbreviations: ESBL, extended-spectrum β-lactamase; AMR, antimicrobial resistance; MRSA, methicillin-resistant S. aureus. |
Quality assessment of studies
The quality of the reviewed studies was assessed using the checklist provided by Joanna Briggs Institute.28 A quality score was obtained from answering each of the 10 questions in the checklist. Consequently, each YES answer gave a point ranging from 0 to 10. A study that scored between 6 and 10 was included in this review.
Results
General overview
This quinquennial (2011–2016) systematic review search retrieved 26,584 combined articles from different online research databases (Figure 1). The six databases were combined, and 21,423 duplicate articles were removed. Of the 5,161 remaining articles, 4,216 articles were removed after title and abstract screening to leave 945 articles. Based on the eligibility criteria, 892 articles were removed based on unspecified locations, lack of molecular or genetic detection of an AR encoding gene, reports on food ARGs, and reports of ARGs and ARGDs in other microorganisms such as viruses, protozoa, and helminths. The remaining 53 articles were included in this qualitative systematic review. Studies reviewed represented seven South African provinces. Figure 2 shows the distribution of resistance patterns by province. No study describing detection and characterization of ARGs and ARGDs in ARB was found for Mpumalanga and the Northern Cape Province. The Eastern Cape Province accounted for 35.8% (n=19), followed by the Western Cape Province with 18.9% (n=10). Gauteng Province and KwaZulu-Natal Province had 17% (n=9) each. North-West represented 3.7% (n=2) and Limpopo Province had 1.9% (n=1). Three articles (5.7%) had data on AR for two or more provinces. Thirty-four articles included in this review were from clinical settings (64.1%) and 10 from environmental settings (18.9%). ARGs isolated from farm settings accounted for 15.1% (n=8). Only one study detected ARGs in food (1.9%) and was excluded from further analysis. Four articles (7.5%) reported AR patterns from two or more sources.29–32
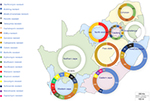  | Figure 2 Distribution of genotypic AMR studies in SA: the initial South African map was created using an open source GIS software (QGIS 2.18 – http://www.qgis.org). Subsequent chart was added using PowerPoint® 2016 (Microsoft Corporation, Redmond, WA USA). Abbreviations: ESBL, extended-spectrum β-lactamase; AMR, antimicrobial resistance; SA, South Africa. |
Methods used for ARGs detection and characterization
Genetic detection of ARGs in different matrixes was performed using principally three methods: PCR only, PCR coupled with DNA sequencing, and whole genome sequencing (WGS). The PCR technique appeared to be the gold standard involved in 70% (n=37) of detection for ARGs across all sample types, be it clinical, environmental, or from farm settings. DNA sequencing of the detected ARG was performed as a complementary test to the PCR to provide in-depth analysis in 28.1% (n=15) of the reviewed articles.33–45 The WGS technique was performed in only a single clinical study (1.9%).46
ARB distribution
A wide range of ARB was studied for the detection and characterization of ARGs. The most frequently cultured bacterium was S. aureus, which accounted for 13.2% (n=7) of all the studies.31,45,47–51 This was followed by Mycobacterium tuberculosis accounting for 11.3% (n=6) of the reviewed articles.33,34,37,41,42,46 Five reviewed articles (9.4%) studied E. coli40,52,53 and Enterococcus spp.30,54–56 Two different strains of E. coli were also studied, namely E. coli ST13135 and O157:H7.57 Aeromonas spp. represented 7.5% (n=4),58–61 while K. pneumoniae,62–64 Neisseria gonorrhoeae,39,65,66 and Salmonella spp.67–69 each accounted for 5.7% (n=3) in the reviewed articles. Two reviewed articles studied Vibrio spp.,70 of which one article specifically studied Vibrio cholera O1 strains29 and Pseudomonas spp.71 The other sources studied Pseudomonas aeruginosa.36 Enterobacteriaceae as a group was also studied in two articles.72,73
One article each was studied for the presence of ARGs in Acinetobacter baumanii,74 Stenotrophomonas maltophilia,75 Ureaplasma parvum,76 Clostridium difficile,38 Bacteroides fragilis,77 Campylobacter spp.,78 and Helicobacter pylori.44 Four articles investigated several bacteria in their studies60,79,80 of which one investigated bacteria as colony forming bacteria assemblages.43
Characteristics of ARGs in SA
Studies that only used culture-independent methods reported mostly on the prevalence of ARGs and ARGDs in isolated ARB. The gene aac (6¢)-aph (2≤) was assessed through PCR for gentamicin resistance in S. aureus51 and it was found that 92.6% of strains harbored the gentamicin resistance gene. The aac (6′)-lb-cr gene was studied in E. coli ST131 for ESBLs resistance.35 The aac (6′)-lb-cr gene was not detected in negative ESBLs resistance and was found to be carried by most isolates positive for ESBLs resistance. The aacA-aphD gene responsible for the aminoglycoside resistance determinant was studied in Staphylococcus spp.47 The authors found a total of nine human isolates that carried the aacA-aphD gene, comprising four Staphylococcus epidermidis and five Staphylococcus haemolyticus.47 The aadA gene conferring resistance to streptomycin (aminoglycosides) was studied in E. coli and K. pneumoniae.52,53,81 It was found that six E. coli isolates belonging to different pathotypes carried the aadA gene,52 with one reference53 showing that all the screened E. coli colonies were positive for the aadA gene. Peirano et al81 found that all the EBSLs-resistant K. pneumoniae positives also carried a plasmid-encoded aadA gene cassette. As it confers resistance to gentamycin, the ant (3″)-la gene was studied in Salmonella spp., shown in 32% of bacterial isolates and detected in 80% of clinical specimens.69
In some studies, ARGs were not detected, while phenotypic resistance was observed. In one study, the tat gene was not detected although phenotypic results showed high tetracycline resistance.51 In another study,52 cat II, ampC, and blaZ genes were not detected even when the phenotypic pattern exhibited resistance.
Mutations were mainly assessed by DNA sequencing. The 23S-rRNA gene was assessed for resistance to clarithromycin in H. pylori.44 Two-point mutations were detected in the 23S-rRNA gene and assigned as conferring clarithromycin resistance, since isolates showed phenotypic resistance to this antibiotic. A study by Bhembe et al33 found isoniazid-resistant M. tuberculosis strains in 71.4% (100/140) isolates with mutations at codon 315. For the katG gene region, seven different mutations were observed: two-point mutation was found in 35.7% (50/140) and 14.3% (20/140) and five-point mutation in 7.1% (10/140). Mutations among the rifamycin-resistant isolates were located at codon 42 (21.4%) followed by 14.3% of mutations on codon 52, codon 87, and codon 531, which is known to be a hotspot for rpoB gene mutations. In addition, four types of mutation patterns were observed in the rrs gene region; S2170A, R2201G, K2202E, and a deletion in position 2207.33
rrs A1401G was further investigated to reveal the extent of capreomycin resistance in M. tuberculosis.41 It was found that 58% of isolates with an rrs A1401G mutation showed a high level of resistance to amikacin and decreased phenotypic susceptibility to capreomycin.41 Only one study made use of WGS technology46 to study a collection of M. tuberculosis isolates from KwaZulu-Natal. These authors could identify the order of acquisition of katG, rpoB, gidB, pncA, rrs, and gyrA genes to the emergence of extensively resistant M. tuberculosis. Table 1 lists details of the characteristics of ARGs and ARGDs in isolated ARB.
ARGs clinical and environmental nexus
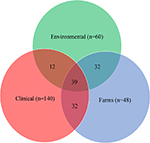
Shared ARGs in environmental, clinical, and farm settings are presented in Figure 3. Numbers inside shared intersecting circles represent the number of shared ARGs in environmental, clinical, and farm settings. All three settings shared 39 ARGs and ARGDs. Twelve ARGs and ARGDs were shared between environmental and clinical settings. Farm settings shared 32 ARGs and ARGDs in a clinical setting and the same number of ARGs and ARGDs was shared between environments and farms (n=32). Forty-eight out of 140 ARGs and ARGDs were only found in clinical settings. This was followed by environmental ARGs and ARGDs found only in environmental settings, where there were 22 out 60 ARGs and ARGDs. In farm settings, there were six out of 48 ARGs in total. Table 2 shows the shared ARGs and ARGDs per setting.
  | Table 2 Shared ARGs and ARGDs per setting Abbreviations: ARGs, antimicrobial-resistant genes; ARGDs, antimicrobial-resistant gene determinants. |
Discussion
In this systematic review, the findings of 53 published articles that reported detection and characterization of ARB, ARGs, and ARGDs in different matrixes were summarized. As data from abstracts and full text were screened and extracted by three reviewers, the authors believe this process reduced the possibility of bias. This review was not designed to review phenotypic characteristics or gene sequences of isolates, but rather collected data on the genetic epidemiology of ARB, ARGs, and ARGDs. The Northern Cape and Mpumalanga Provinces were not represented in the data sets and this may be due, in these provinces, to a paucity of research facilities and/or logistical problems regarding transport of specimens to such research facilities. The Eastern Cape was found to be the leading province in the number of articles published reporting clinical investigations. Most studies were conducted in clinical settings compared to environmental settings. There is a dearth of information on the link between laboratory and clinical data systems and it is thus difficult to identify patterns of environmental and community-acquired ARGs compared to hospital-acquired bacterial resistance.15
Three main molecular biology methods were used to detect and characterize ARGs in the different settings, namely PCR, DNA sequencing, and WGS. The PCR technique was found to be the most commonly published technique in articles, probably because of relatively easy access to PCR cyclers and decreasing costs associated with PCR. A study by Ali et al82 indicated that PCR was the gold standard for identification of ARGs in methicillin-resistant S. aureus. Microarray technologies have been used to detect multiple ARGs and ARGDs simultaneously in ARB,83–85 but this method was not used in any of the reviewed studies where most characterization of specific ARG or ARGD made use of DNA sequencing of ARG and ARGD amplicons.
Despite several advantages offered by WGS, it was used only once in all the reviewed articles. In recent years, WGS has been implemented in the USA by the Food and Drug Administration and the Centers for Disease Control and Prevention as a tool for outbreak source tracking and surveillance in clinical and environmental settings and ex post facto sentinel studies.86 The application of WGS allows the detection of all ARGs with no exception, including ARGDs such as integrons and plasmids.87 Analysis with WGS has shown the ability to resolve phenotypic and genotypic discrepancies88–90 and should be coupled with phenotypic tests to circumvent gene expression problems in obtaining a full AR profile of ARB. It is clear from the findings of this review that WGS is now needed in ARB studies in SA and worldwide.
This review shows that more attention is paid to clinical isolates compared to environmental isolates. The same trend is observed internationally, as studies show that ARB are mostly investigated in clinical compared to environmental settings. This is despite the fact that ARGs are thought to originate from environmental bacteria, as clinically relevant resistance genes have been detected in the genomes of environmental ARB.91 The use of high throughput functional metagenomics techniques92 showed evidence of recent exchange of ARGs between ARB of environmental and clinical origin. One study in this review demonstrated the line of transmission from clinical to environmental settings.31 The same study also revealed that water could be a transmission vector of staphylococcal urinary tract infection among HIV and AIDS patients in the Limpopo Province. Iweriebor et al93 detected the same ARB, ARGs, and ARGDs in hospital wastewater, sewage treatment plant, and receiving water bodies. High evidence of transmission from the environment to humans can only be established through genetic relatedness of ARGs using molecular typing techniques.94 In this review, a wide range of ARB were studied, and these bacteria also cover the most important clinical isolates worldwide.
A considerable number of ARGs and ARGDs were found to be shared across all three settings (n=39). Farm settings shared most ARGs with clinical and environmental settings (n=32) compared with clinical vs environmental settings (n=12). An analysis of 71 environmental specimens found soil to have the most diverse ARGs,95 concluding that soil is a major contributor of ARB, ARGs, and ARGDs. This agrees with the findings of this review, since the soil isolates were classified under farm settings to share ARGs and ARGDs with clinical settings as well. In the same article, it was found that soil and human feces shared more resistance classes with each other than other matrixes. A considerable number of ARGs and ARGDs were only found in clinical isolates (n=48). This indicates that a considerable number of ARGs and ARGDs are still isolated from clinical settings with unknown origins. There is a dire need for development of innovative approaches to track relevant clinically isolated ARB, ARGs, and ARGDs. Important ARB, ARGs, and ARGDs were also found only in environmental and farm settings. A study by Agga et al96 found that swine and cattle shared ARGs and ARGDs with wastewater settings. This agrees with the findings of this review that farm settings shared significant ARGs and ARGDs with environmental settings and indicates the need to contain these ARGs and ARGDs to limit their spread into clinical settings.
An approach to ARGs and ARGDs tracking and containment
Based on the current findings, there is a need for high throughput technologies such as WGS and shotgun metagenomics analysis as means for tracking and characterizing ARB, ARGs, and ARGDs better. The WGS sequencing technique is of paramount importance in solving the discrepancy between phenotypic and genotypic types of AR, as well as building an improved WGS database. This molecular approach has been widely used in other countries such as Denmark, where a study compared the use of WGS as a routine technique against phenotypic tests and reported 99.74% accuracy to suggest that WGS be used as an alternative method to phenotypic testing.97 Another study by Leekitcharoenphon et al98 used WGS to track the source of Salmonella Eko from Nigerian isolates and successfully identified the source of bacteria. A report by Berendonk et al99 has suggested a standardized system of core parameters to tackle AR and as the world is moving into WGS, SA should also advance in the same direction to track endemic AR patterns better. To obtain an in-depth view of the occurrence and distribution of ARGs and ARGDs, WGS should be complemented by shotgun metagenomics to observe routes of dissemination better and elucidate their distribution across various environments.
According to a study by Rowe et al,100 farms and wastewater treatment plants serve as an important reservoir of ARGs, mobile genetic elements, and pathogenic bacteria for river environments. The two metagenomic approaches, WGS and shotgun metagenomics, are now critical in tracking and characterizing AR in different environments and are also needed in SA to streamline individual investigations of ARB, ARGs, and ARGDs.
Furthermore, an AR genomic epidemiology application is needed in a bid to link environmental ARB, ARGs, and ARGDs with clinical ones. This tool should describe the genomics, laboratory, clinical, and epidemiological contextual information required to support data sharing and integration for AR surveillance and outbreak investigations. The application should geospatially map ARB, ARGs, and ARGDs between environmental and clinical settings and report on recent antibiogram data from different environments. The application should initially utilize recently published online studies to create an initial database of environmental, farm, and clinical ARB, ARGs, and ARGDs. In addition, geospatial mapping of ARGs and ARB would permit the identification of national AR hotspots and actions for containment. The application should also provide baseline information necessary to link the environmental spread of AR to routes of transmission. The development of this application would aid national clinicians, physicians, scientists, and policy makers to formulate appropriate strategies to combat AR.
Conclusion
Although data could not be found for the Northern Cape and Mpumalanga Provinces, genes conferring AR were found to be ubiquitous across SA. The available data present a broader scope on isolation and prevalence information on ARGs in ARB and emphasize the dire need for surveillance and documentation of ARGs, ARB, and ARGDs in all provinces. Owing to the lack of sequence analysis in the current review, future reviews will investigate shared ARB, ARGs, and ARGDs based on related sequence data. One of the main findings in this review was the discrepancy between phenotypic and genotypic patterns. This calls for a move toward complementing the gold standard PCR with the use of WGS in examining ARB as well as shotgun metagenomics technologies when dealing with a consortium of bacteria. In extending this tracking and genetic characterization of ARB, ARGs, and ARGDs, this study suggests the adoption of an AR genomic epidemiology application.
Acknowledgments
MDE would like to thank Mr P Budeli and Ms VKT Phetla for their initial input during data collection and equally thank Emmanuel Oladipo Babafemi and Hannah Lishman for their critical review of the manuscript. This systematic review received funding from the National Research Foundation (grant number: 112851) and South African Research Chairs Initiative in Water Quality and Wastewater Management. Opinions expressed and conclusions arrived at are those of the authors and are not necessarily to be attributed to the funders.
Disclosure
The authors report no conflicts of interest in this work.
References
World Health Organization (WHO). Antimicrobial Resistance: 2014 Global Report on Surveillance. Geneva: World Health Organization; 2014. | ||
Jung Y, Matthews KR. Antimicrobial Resistance and Food Safety. New York: Elsevier; 2015. | ||
Centers for Disease Control and Prevention (CDC). National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS).CDC; 2016. Available from: https://www.cdc.gov/narms/. Accessed September 24, 2018. | ||
Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States, 2013. CDC, US Department of Health and Human Services; 2013. Available from: http://www.cdc.gov/drugresistance/threat-report-2013. Accessed September 24, 2018. | ||
Nys S, Okeke IN, Kariuki S, Dinant GJ, Driessen C, Stobberingh EE. Antibiotic resistance of faecal Escherichia coli from healthy volunteers from eight developing countries. J Antimicrob Chemother. 2004;54(5):952–955. | ||
Berkley JA, Lowe BS, Mwangi I, et al. Bacteremia among Children Admitted to a Rural Hospital in Kenya. N Engl J Med. 2005;352(1):39–47. | ||
Kariuki S, Revathi G, Kariuki N, et al. Increasing prevalence of multidrug-resistant non-typhoidal salmonellae, Kenya, 1994–2003. Int J Antimicrob Agents. 2005;25(1):38–43. | ||
Ndihokubwayo JB, Yahaya AA, Desta AT, Ki-Zerbo G. Antimicrobial resistance in the African Region: Issues, challenges and actions proposed. Afr Health Monit. 2013;16:27–30. | ||
Abdulgader SM, Shittu AO, Nicol MP, Kaba M. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol. 2015;6:348. | ||
Ampaire L, Muhindo A, Orikiriza P, Mwanga-Amumpaire J, Bebell L, Boum Y. A review of antimicrobial resistance in East Africa. Afr J Lab Med. 2016;5(1):1–6. | ||
Sonda T, Kumburu H, van Zwetselaar M, et al. Meta-analysis of proportion estimates of Extended-Spectrum-Beta-Lactamase-producing Enterobacteriaceae in East Africa hospitals. Antimicrob Resist Infect Control. 2016;5(1):1–9. | ||
National Institute for Communicable Diseases (NICD); South Africa. Germs; South Africa; Annual Report; 2012. Available from: http://www.nicd.ac.za/assets/files/2012_GERMS-SA_Annual_Report.pdf. Accessed October 5, 2018. | ||
Naidoo R, Nuttall J, Whitelaw A, Eley B. Epidemiology of Staphylococcus aureus bacteraemia at a tertiary children’s hospital in Cape Town, South Africa. PLoS One. 2013;8(10):e78396. | ||
Bamford C, Bonorchis K, Ryan A, et al. Antimicrobial Susceptibility Patterns of Selected Bacteraemic Isolates from South African Public Sector Hospitals, 2010. Southern Afr J Epidemiol Infect. 2011;26(4):243–250. | ||
Mendelson M, Matsoso M. The South African antimicrobial resistance strategy framework. AMR Control. 2015:54–61. | ||
Czekalski N, Gascón Díez E, Bürgmann H. Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J. 2014;8(7):1381–1390. | ||
Marti E, Jofre J, Balcazar JL. Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS One. 2013;8(10):e78906–78908. | ||
Chee-Sanford JC, Amniov RI, Krapac IJ, Garrigues-Jeanjean N, Mackie RI, Aminov R. Occurrence and Diversity of Tetracycline Resistance Genes in Lagoons and Groundwater Underlying Two Swine Production Facilities Occurrence and Diversity of Tetracycline Resistance Genes in Lagoons and Groundwater Underlying Two Swine Production Facilities. Appl Environ Microbiol. 2001;67(4):1494–1502. | ||
Varela AR, Ferro G, Vredenburg J, et al. Vancomycin resistant enterococci: From the hospital effluent to the urban wastewater treatment plant. Sci Total Environ. 2013:450155–451161. | ||
Birošová L, Mackuľak T, Bodík I, Ryba J, Škubák J, Grabic R. Pilot study of seasonal occurrence and distribution of antibiotics and drug resistant bacteria in wastewater treatment plants in Slovakia. Sci Total Environ. 2014;490:440–444. | ||
Harris S, Morris C, Morris D, Cormican M, Cummins E. Antimicrobial resistant Escherichia coli in the municipal wastewater system: Effect of hospital effluent and environmental fate. Sci Total Environ. 2014;468–469:1078–1085. | ||
Kristiansson E, Fick J, Janzon A, et al. Sequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements Supporting Information Supporting materials and methods. Chem Anal. 2011;6(2):e17038. | ||
Maal-Bared R, Bartlett KH, Bowie WR, Hall ER. Phenotypic antibiotic resistance of Escherichia coli and E. coli O157 isolated from water, sediment and biofilms in an agricultural watershed in British Columbia. Sci Total Environ. 2013;443:315–323. | ||
Abia ALK, Ubomba-Jaswa E, Momba MNB. High prevalence of multiple-antibiotic-resistant (MAR) Escherichia coli in river bed sediments of the Apies River, South Africa. Environ Monit Assess. 2015;187(10):652. | ||
Joy SR, Li X, Snow DD, Gilley JE, Woodbury B, Bartelt-Hunt SL. Fate of antimicrobials and antimicrobial resistance genes in simulated swine manure storage. Sci Total Environ. 2014;481(1):69–74. | ||
Verraes C, Van Boxstael S, Van Meervenne E, et al. Antimicrobial Resistance in the Food Chain: A Review. Int J Environ Res Public Health. 2013;10(7):2643–2669. | ||
Bai X, Ma X, Xu F, Li J, Zhang H, Xiao X. The drinking water treatment process as a potential source of affecting the bacterial antibiotic resistance. Sci Total Environ. 2015;533:24–31. | ||
Moher D, Liberati A, Tetzlaff J, Altman DGPRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269. | ||
Ismail H, Smith AM, Tau NP, Sooka A, Keddy KH; Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa. Cholera outbreak in South Africa, 2008–2009: laboratory analysis of Vibrio cholerae O1 strains. J Infect Dis. 2013;208(Suppl 1):S39–S45. | ||
Iweriebor BC, Obi LC, Okoh AI. Virulence and antimicrobial resistance factors of Enterococcus spp. isolated from fecal samples from piggery farms in Eastern Cape, South Africa. BMC Microbiol. 2015;15(1):1–11. | ||
Samie A, Shivambu N. Molecular detection of methicillin resistance gene (mec A gene) and pathogenic genes among Staphylococcus aureus isolates from clinical and drinking water samples of HIV and AIDS patients in Limpopo. Afr J Microbiol Res. 2011;5(30):5498–5506. | ||
Schmidt T, Kock MM, Ehlers MM. Diversity and antimicrobial susceptibility profiling of staphylococci isolated from bovine mastitis cases and close human contacts. J Dairy Sci. 2015;98(9):6256–6269. | ||
Bhembe NL, Nwodo UU, Govender S, et al. Molecular detection and characterization of resistant genes in Mycobacterium tuberculosis complex from DNA isolated from tuberculosis patients in the Eastern Cape Province South Africa. BMC Infect Dis. 2014;14(1):479. | ||
Dookie N, Sturm AW, Moodley P. Mechanisms of first-line antimicrobial resistance in multi-drug and extensively drug resistant strains of Mycobacterium tuberculosis in KwaZulu-Natal, South Africa. BMC Infect Dis. 2016;16(1):609. | ||
Gqunta K, Govender S. Characterization of ESBL-producing Escherichia coli ST131 isolates from Port Elizabeth. Diagn Microbiol Infect Dis. 2015;81(1):44–46. | ||
Jacobson RK, Minenza N, Nicol M, Bamford C. VIM-2 metallo-β-lactamase-producing Pseudomonas aeruginosa causing an outbreak in South Africa. J Antimicrob Chemother. 2012;67(7):1797–1798. | ||
Klopper M, Warren RM, Hayes C, et al. Emergence and spread of extensively and totally drug-resistant tuberculosis, South Africa. Emerg Infect Dis. 2013;19(3):449–455. | ||
Kullin B, Brock T, Rajabally N, et al. Characterisation of Clostridium difficile strains isolated from Groote Schuur Hospital, Cape Town, South Africa. Eur J Clin Microbiol Infect Dis. 2016;35(10):1709–1718. | ||
Lewis DA, Sriruttan C, Müller EE, et al. Phenotypic and genetic characterization of the first two cases of extended-spectrum-cephalosporin-resistant Neisseria gonorrhoeae infection in South Africa and association with cefixime treatment failure. J Antimicrob Chemother. 2013;68(6):1267–1270. | ||
Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother. 2016;60(7):4394–4397. | ||
Sirgel FA, Tait M, Warren RM, et al. Mutations in the rrs A1401G gene and phenotypic resistance to amikacin and capreomycin in Mycobacterium tuberculosis. Microb Drug Resist. 2012;18(2):193–197. | ||
Sirgel FA, Warren RM, Streicher EM, Victor TC, van Helden PD, Böttger EC. gyrA mutations and phenotypic susceptibility levels to ofloxacin and moxifloxacin in clinical isolates of Mycobacterium tuberculosis. J Antimicrob Chemother. 2012;67(5):1088–1093. | ||
Suzuki S, Ogo M, Koike T, Takada H, Newman B. Sulfonamide and tetracycline resistance genes in total- and culturable-bacterial assemblages in South African aquatic environments. Front Microbiol. 2015;6(e1002158):796. | ||
Tanih NF, Ndip LM, Ndip RN. Characterisation of the genes encoding resistance to metronidazole (rdxA and frxA) and clarithromycin (the 23S-rRNA genes) in South African isolates of Helicobacter pylori. Ann Trop Med Parasitol. 2011;105(3):251–259. | ||
Jansen van Rensburg MJ, Whitelaw AC, Elisha BG. Genetic basis of rifampicin resistance in methicillin-resistant Staphylococcus aureus suggests clonal expansion in hospitals in Cape Town, South Africa. BMC Microbiol. 2012;12(1):46. | ||
Cohen KA, Abeel T, Manson Mcguire A, Mcguire AM, et al. Evolution of Extensively Drug-Resistant Tuberculosis over Four Decades: Whole Genome Sequencing and Dating Analysis of Mycobacterium tuberculosis Isolates from KwaZulu-Natal. PLoS Med. 2015;12(9): e1001880. | ||
Schmidt VM, Pinchbeck GL, Nuttall T, Mcewan N, Dawson S, Williams NJ. Antimicrobial resistance risk factors and characterisation of faecal E. coli isolated from healthy Labrador retrievers in the United Kingdom. Prev Vet Med. 2015;119(1–2):31–40. | ||
Adegoke AA, Okoh AI. Species diversity and antibiotic resistance properties of Staphylococcus of farm animal origin in Nkonkobe Municipality, South Africa. Folia Microbiol. 2014;59(2):133–140. | ||
Singh-Moodley A, Marais E, Perovic O. Discrepancies in the identification of methicillin-resistant Staphylococcus aureus and the absence of mecC in surveillance isolates in South Africa. South Afr J Epidemiol Infect. 2015;30(4):15–17. | ||
Wasserman E, Orth H, Senekal M, Harvey K. High prevalence of mupirocin resistance associated with resistance to other antimicrobial agents in Staphylococcus aureus isolated from patients in private health care, Western Cape. South Afr J Infect Dis. 2014;29(4):126–132. | ||
Amoako DG, Bester LA, Somboro AM, Baijnath S, Govind CN, Essack SY. Plasmid-mediated resistance and virulence mechanisms in the private health sector in KwaZulu-Natal, South Africa: An investigation of methicillin resistant Staphylococcus aureus (MRSA) clinical isolates collected during a three month period. Int J Infect Dis. 2016;46:38–41. | ||
Adefisoye MA, Okoh AI. Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa. Microbiologyopen. 2016;5(1):143–151. | ||
Nontongana N, Sibanda T, Ngwenya E, Okoh A. Prevalence and antibiogram profiling of Escherichia coli pathotypes isolated from the kat river and the fort beaufort abstraction water. Int J Environ Res Public Health. 2014;11(8):8213–8227. | ||
Ateba CN, Lekoma KP, Kawadza DT. Detection of vanA and vanB genes in vancomycin-resistant enterococci (VRE) from groundwater using multiplex PCR analysis. J Water Health. 2013;11(4):684–691. | ||
Molale GL, Bezuidenhout CC. Environmental Science and Pollution Research Antibiotic resistance, efflux pump genes and virulence determinants in Enterococcus spp. from surface water systems. Environ Sci Pollut Res. 2016;23(21):21501–21510. | ||
Lochan H, Moodley C, Rip D, Chb MB, Sa F, et al. Emergence of vancomycin-resistant Enterococcus at a tertiary paediatric hospital in South Africa. S Afr Med J. 2016;106(6):562–566. | ||
Iweriebor BC, Iwu CJ, Obi LC, Nwodo UU, Okoh AI. Multiple antibiotic resistances among Shiga toxin producing Escherichia coli O157 in feces of dairy cattle farms in Eastern Cape of South Africa. BMC Microbiol. 2015;15(1):213. | ||
Chenia HY. Prevalence and characterization of plasmid-mediated quinolone resistance genes in Aeromonas spp. isolated from South African freshwater fish. Int J Food Microbiol. 2016;231:26–32. | ||
Igbinosa IH, Igbinosa EO, Okoh AI. Detection of antibiotic resistance, virulence gene determinants and biofilm formation in Aeromonas species isolated from cattle. Environ Sci Pollut Res. 2015;22(22):17596–17605. | ||
Chenia HY, Vietze C. Tetracycline resistance determinants of heterotrophic bacteria isolated from a South African tilapia aquaculture system. Afr J Microbiol Res. 2012;6(39):6761–6768. | ||
Igbinosa IH, Okoh AI. Antibiotic Susceptibility Profile of Aeromonas Species Isolated from Wastewater Treatment Plant. ScientificWorldJournal. 2012;2012(1):1–6. | ||
Jacobson RK, Manesen MR, Moodley C, et al. Molecular characterisation and epidemiological investigation of an outbreak of blaOXA-181 carbapenemase-producing isolates of Klebsiella pneumoniae in South Africa. S Afr Med J. 2015;105(12):1030–1035. | ||
Peirano G, Moolman J, Pitondo-Silva A, Pitout JDD. The characteristics of VIM-1-producing Klebsiella pneumoniae from South Africa. Scand J Infect Dis. 2012;44(1):74–78. | ||
Perovic O, Singh-Moodley A, Dusé A, et al. National sentinel site surveillance for antimicrobial resistance in Klebsiella pneumoniae isolates in South Africa, 2010–2012. S Afr Med J. 2014;104(8):563–568. | ||
Fayemiwo SA, Müller EE, Gumede L, Lewis DA. Plasmid-Mediated Penicillin and Tetracycline Resistance Among Neisseria gonorrhoeae Isolates in South Africa: Prevalence, Detection and Typing Using a Novel Molecular Assay. Sex Transm Dis. 2011;38(4):329–333. | ||
Magooa MP, Müller EE, Gumede L, Lewis DA. Determination of Neisseria gonorrhoeae susceptibility to ciprofloxacin in clinical specimens from men using a real-time PCR assay. Int J Antimicrob Agents. 2013;42(1):63–67. | ||
Igbinosa IH. Prevalence and detection of antibiotic-resistant determinant in Salmonella isolated from food-producing animals. Trop Anim Health Prod. 2015;47(1):37–43. | ||
Iwu CJ, Iweriebor BC, Obi LC, Basson AK, Okoh AI. Multidrug-Resistant Salmonella Isolates from Swine in the Eastern Cape Province, South Africa. J Food Prot. 2016;79(7):1234–1239. | ||
Zishiri O, Mkhize N, Mukaratirwa S. Prevalence of virulence and antimicrobial resistance genes in Salmonella spp. isolated from commercial chickens and human clinical isolates from South Africa and Brazil. Onderstepoort J Vet Res. 2013;83(1):1–11. | ||
Igbinosa EO, Obi LC, Tom M, Anthony I. Detection of potential risk of wastewater effluents for transmission of antibiotic resistance from Vibrio species as a reservoir in a peri-urban community in South Africa. Int J Environ Health Res. 2015;2011(21):37–41. | ||
Igbinosa IH, Nwodo UU, Sosa A, Tom M, Okoh AI. Commensal Pseudomonas species isolated from wastewater and freshwater milieus in the Eastern Cape Province, South Africa, as reservoir of antibiotic resistant determinants. Int J Environ Res Public Health. 2012;9(7):2537–2549. | ||
Singh-Moodley A, Perovic O, et al. Antimicrobial susceptibility testing in predicting the presence of carbapenemase genes in Enterobacteriaceae in South Africa. BMC Infect Dis. 2016;16(1):536. | ||
Perovic O, Britz E, Chetty V, Singh-Moodley A. Molecular detection of carbapenemase-producing genes in referral Enterobacteriaceae in South Africa: A short report. S Afr Med J. 2016;106(10):975–977. | ||
Lowings M, Ehlers MM, Dreyer AW, Kock MM. High prevalence of oxacillinases in clinical multidrug-resistant Acinetobacter baumannii isolates from the Tshwane region, South Africa – an update. BMC Infect Dis. 2015;15(1):521. | ||
Adegoke AA, Okoh AI. Antibiogram of Stenotrophomonas maltophilia Isolated From Nkonkobe Municipality, Eastern Cape Province, South Africa. Jundishapur J Microbiol. 2015;8(1):e13975. | ||
Govender S, Gqunta K, Le Roux M, de Villiers B, Chalkley LJ. Antibiotic susceptibilities and resistance genes of Ureaplasma parvum isolated in South Africa. J Antimicrob Chemother. 2012;67(12):2821–2824. | ||
Meggersee R, Abratt V. The occurrence of antibiotic resistance genes in drug resistant Bacteroides fragilis isolates from Groote Schuur Hospital, South Africa. Anaerobe. 2015;32:1–6. | ||
Shobo CO, Bester LA, Baijnath S, Somboro AM, Peer AKC, Essack SY. Antibiotic resistance profiles of Campylobacter species in the South Africa private health care sector. J Infect Dev Ctries. 2016;10(11):1214–1221. | ||
de Jager P, Chirwa T, Naidoo S, Perovic O, Thomas J. Nosocomial Outbreak of New Delhi Metallo-β-Lactamase-1-Producing Gram-Negative Bacteria in South Africa: A Case-Control Study. PLoS One. 2015;10(4):e0123337–13. | ||
Mhlongo N, Essack S, Govinden U. NDM-1, novel TEM-205, novel TEM-213 and other extended-spectrum β-lactamases co-expressed in isolates from cystic fibrosis patients from South Africa. S Afr J Infect Dis. 2015;30(3):103–107. | ||
Peirano G, van der Bij AK, Freeman JL, et al. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum β-lactamases: global distribution of the H30-Rx sublineage. Antimicrob Agents Chemother. 2014;58(7):3762–3767. | ||
Ali R, Al-Achkar K, Al-Mariri A, Safi M. Role of Polymerase Chain Reaction (PCR) in the detection of antibiotic-resistant Staphylococcus aureus. Egypt J Med Hum Genet. 2014;15(3):293–298. | ||
Perreten V, Vorlet-Fawer L, Slickers P, Ehricht R, Kuhnert P, Frey J. Microarray-Based Detection of 90 Antibiotic Resistance Genes of Gram-Positive Bacteria Microarray-Based Detection of 90 Antibiotic Resistance Genes of Gram-Positive Bacteria. J Clin Microbiol. 2005;43(5):2291–2302. | ||
Frye JG, Lindsey RL, Rondeau G, et al. Development of a DNA microarray to detect antimicrobial resistance genes identified in the National Center for Biotechnology Information database. Microb Drug Resist. 2010;16(1):9–19. | ||
Naas T, Cuzon G, Bogaerts P, Glupczynski Y, Nordmann P. Evaluation of a DNA microarray (Check-MDR CT102) for rapid detection of TEM, SHV, and CTX-M extended-spectrum β-lactamases and of KPC, OXA-48, VIM, IMP, and NDM-1 carbapenemases. J Clin Microbiol. 2011;49(4):1608–1613. | ||
Butaye P, Má AR, Threlfall J. Introduction to antimicrobial-resistant foodborne pathogens. Antimicrobial Resistance and Food Safety: Methods and Techniques. New York: Elsevier; 2015. | ||
Lupo A, Papp-Wallace KM, Bonomo RA, Endimiani A. Non-phenotypic tests to detect and characterize antibiotic resistance mechanisms in Enterobacteriaceae. Antimicrobial Resistance and Food Safety: Methods and Techniques. New York: Elsevier; 2015:233–257. | ||
Köser CU, Ellington MJ, Peacock SJ. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 2014;30(9):401–407. | ||
Ellington MJ, Ekelund O, Aarestrup FM, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect. 2017;23(1):2–22. | ||
Zhao S, Tyson GH, Chen Y, et al. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol. 2016;82(2):459–466. | ||
Cantas L, Shah SQA, Cavaco LM, et al. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front Microbiol. 2013;4:96. | ||
Rizzo L, Manaia C, Merlin C, et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ. 2013;447(3):345–360. | ||
Iweriebor BC, Gaqavu S, Obi LC, Nwodo UU, Okoh AI. Antibiotic susceptibilities of enterococcus species isolated from hospital and domestic wastewater effluents in alice, eastern cape province of South Africa. Int J Environ Res Public Health. 2015;12(4):4231–4246. | ||
Huijbers PMC, Blaak H, de Jong MCM, Graat EAM, Vandenbroucke-Grauls CMJE, de Roda Husman AM. Role of the Environment in the Transmission of Antimicrobial Resistance to Humans: A Review. Environ Sci Technol. 2015;49(20):11993–12004. | ||
Nesme J, Cécillon S, Delmont TO, Monier JM, Vogel TM, Simonet P. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr Biol. 2014;24(10):1096–1100. | ||
Agga GE, Arthur TM, Durso LM, Harhay DM, Schmidt JW. Antimicrobial-resistant bacterial populations and antimicrobial resistance genes obtained from environments impacted by livestock and municipal waste. PLoS One. 2015;10(7):e0132586. | ||
Zankari E, Hasman H, Kaas RS, et al. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother. 2013;68(4):771–777. | ||
Leekitcharoenphon P, Hendriksen RS, Le Hello S, et al. Global Genomic Epidemiology of Salmonella enterica Serovar Typhimurium DT104. Appl Environ Microbiol. 2016;82(8):2516–2526. | ||
Berendonk TU, Manaia CM, Merlin C, et al. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. 2015;13(5):310–317. | ||
Rowe W, Verner-Jeffreys DW, Baker-Austin C, Ryan JJ, Maskell DJ, Pearce GP. Comparative metagenomics reveals a diverse range of antimicrobial resistance genes in effluents entering a river catchment. Water Sci Technol. 2016;73(7):1541–1549. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.