Back to Journals » Infection and Drug Resistance » Volume 16
Study on Virulence Genes, Drug Resistance and Molecular Epidemiology of Klebsiella pneumoniae with High Virulence in Inner Mongolia, China
Authors Li HF, Zhang LX, Zhang WL, Li J, Li YQ, Hu TP
Received 27 September 2022
Accepted for publication 16 December 2022
Published 23 February 2023 Volume 2023:16 Pages 1133—1144
DOI https://doi.org/10.2147/IDR.S391468
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Hong-Fu Li,1,2,* Li-Xia Zhang,2,* Wen-Lan Zhang,2,* Jing Li,2,* Ya-Qian Li,2,* Tong-Ping Hu2
1Department of Clinical Laboratory, Zhuhai Third People’s Hospital, Zhuhai, People’s Republic of China; 2Department of Clinical laboratory, the First Affiliated Hospital of Baotou Medical College, Inner Mongolia University of Science and Technology, Inner Mongolia, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tong-Ping Hu, Department of Clinical laboratory, the First Affiliated Hospital of Baotou Medical College, Inner Mongolia University of Science and Technology, No. 41, of Linyin Road, Kundulun District, Baotou City, Baotou, 014000, People’s Republic of China, Tel +86 13296922365, Email [email protected]
Objective: The purpose of this study was to analyse the clinical, microbiological and molecular epidemiological characteristics of patients with pyogenic liver abscess (PLA) caused by Klebsiella pneumoniae (KPN) in Inner Mongolia, China.
Methods: The KPN isolates from 78 KPN-PLA cases admitted to a tertiary teaching hospital in Baotou, Inner Mongolia, from 2016 to 2019 were studied systematically and described comprehensively. The virulence factors, drug resistance and sequence types of KPN in different samples were identified by a wire-drawing test, polymerase chain reaction, a drug susceptibility test and multi-site sequence typing.
Results: There were more male than female KPN-PLA patients (P< 0.05). The mortality rate was 2.5%, and KPN-PLA was significantly associated with diabetes mellitus (P< 0.05). Most of the KPN isolates in the puncture fluid of patients with KPN-PLA were hypervirulent KPN (HvKP). The positive rate of the KPN-PLA specimens was higher than that of the blood and urine specimens. The KPN isolates of the urine specimens had higher drug resistance than the other two (P< 0.05). The hypermucoviscous KPN, aerobic actin (aero) (+), K1 and K2 serotypes accounted for 80.8%, 89.7%, 56.4% and 26.9%, respectively. In addition to ironB (3.8%), the detection rates of virulence factors rmpA, irp2, entB, iucD, aero, wcaG, iutA, kfu, ybtA, iron, fimH and mrkD were higher (69.2%– 100.0%). The positive rate of KPN isolates of the KPN-PLA puncture fluid was higher than that of the blood and urine samples (P< 0.05). In addition, ST23 was found to be the dominant ST (32.1%) of KPN-PLA in the Baotou region.
Conclusion: In the KPN-PLA specimens, the KPN isolates were more virulent than those in the blood and urine specimens, and a carbapenem-resistant HvKP strain emerged. This research will help improve the understanding of HvKP and provide useful suggestions for KPN-PLA treatments.
Keywords: Klebsiella pneumoniae, virulence factor, capsular serotype, bacterial drug resistance, multilocus sequence typing
Introduction
Klebsiella pneumoniae (KPN) is a common gram-negative bacterium that causes clinical infection, and studies have shown that it has the second-highest detection rate among all bacterial infections.1,2 Compared with common KPN (ie classic KPN [cKP]), hypervirulent KPN (HvKP) has the following characteristics: (1) Usually, HvKP colonies display a high-mucus type – typically, a laboratory wire-drawing test is used to determine whether strains are of a high-viscosity type. (2) There are more virulence factors in HvKP than in cKP, such as rmpA and siderophores. (3) In most cases, HvKP infection is manifested by a primary bacterial liver abscess, which can cause infection foci that spread in the bloodstream.3–5
The virulence factor of HvKP is the key to distinguishing HvKP from cKP, and its virulence level is also related to the expression of virulence factors, indicating that the virulence molecule is an important breakthrough in studying the molecular characteristics of the HvKP gene.6–8 In a recent study, 90.9% of the pathogens that caused pyogenic liver abscess (PLA) were found to be HvKP, and the incidence of PLA was associated with a high prevalence of HvKP strains.7,8 Bacterial liver abscess caused by KPN (KPN-PLA) has become a global disease. Therefore, it is necessary to systematically study the clinical and microbiological characteristics of patients with KPN-PLA and compare them with KPN from blood and urinary system infections. Only through a detailed comparative study can we determine a more accurate site for HvKP detection in patients with KPN, provide more scientific and accurate test methods for future clinical treatments and provide a more effective test reference for clinicians’ treatment work.9,10
In recent years, the prevalence of KPN-PLA has been increasing. However, there are limited reports on the virulence and drug resistance of KPN-PLA in Mongolia.11,12 In this study, we explored the virulence, drug resistance and molecular epidemiology of KPN in KPN-PLA specimens and compared these characteristics in the systems of patients. This research will help improve the understanding of HvKP and provide valuable recommendations for KPN-PLA treatments.
Materials and Methods
The flowchart (Figure 1) simplifies our description of the entire approach.13
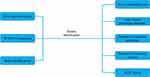 |
Figure 1 The flowchart of the entire approach. |
Research Subjects and Grouping of Strain-Screening Criteria
Research Subjects
Information and strains were collected from 78 patients with KPN from the First Affiliated Hospital of Baotou Medical College, Inner Mongolia University of Science and Technology, between 2016 and 2019. These comprised 46 males and 32 females, aged 24–81 years, with an average age of 60.6 years.
Grouping of Strain-Screening Criteria
The screening criteria were as follows: (1) Each patient met the diagnostic criteria for bacterial liver abscess, having (a) chills, fever, nausea and liver tenderness or percussive pain, (b) confirmation via radiological imaging and having undergone either percutaneous aspiration/drainage or surgical drainage and (c) positive bacterial culture or effective antibiotic treatment.14 (2) Each patient had no underlying diseases related to the liver or gallbladder. (3) Klebsiella pneumoniae was the only pathogenic bacteria.
Regarding the uri ne-specimen group (positive KPN strains in the same period), the screening criteria were as follows: (1) Patients who met the diagnostic criteria for bacterial urinary tract infection, having (a) bladder-irritation symptoms, such as frequent urination, urgency and dysuria, with possible accompanying systemic infection symptoms, such as elevated body temperature, chills and chills, (b) urine leukocytes exceeding the normal range and (c) a positive urine bacterial culture or effective antibiotic treatment. Patients who met (1), (2) and (3) at the same time were included in the study.14 (2) The only pathogenic bacterium was KPN.
Regarding the blood-sample group (selected blood-culture-positive KPN strains in the same period), the screening criteria were as follows: (1) Patients met the clinical diagnostic criteria for bloodstream infection (fever >38°C or hypothermia <36°C), which may have been accompanied by chills and one of the following conditions: (a) invasion portals or migration lesions, (b) toxemia or even bacteremia or sepsis, (c) rashes or bleeding spots, hepatosplenomegaly, blood neutropenia with a left shift of the nucleus and no other explanations or (d) systolic blood pressure <12 kPa (90 mmHg) or >5.3 kPa (40 mmHg) lower than the original systolic blood pressure.14 (2) Based on clinical diagnosis, the etiological diagnostic criteria of bloodstream infection were met, ie KPN was isolated from the blood culture and was the only pathogenic bacterium.
Identification of Strains
All strains were identified using instruments according to the identification method in National Clinical Inspection Procedures (fourth edition).15 The instruments used were a VITEK® 2 compact microbial-identification system and supporting reagents from bioMérieux (France).
Instruments and Reagents
Instruments: Polymerase chain-reaction (PCR) amplification instrument, constant-temperature incubator, gel-electrophoresis instrument and gel-imaging analyser.
Reagents: Columbia blood agar, Chinese blue agar, MH agar and drug-sensitive paper (OXOID, UK); imipenem and meropenem E-test strips (Zhengzhou Antu Bioengineering Co., Ltd); DNA extraction kit (Gen Biochemical Technology Co., Ltd); PCR amplification reagent (Hangzhou Baosai Biotechnology Co., Ltd); 2 × Taq PCR MasterMix (Beijing Baosai Biotechnology Co., Ltd) solutions; 50 × TAE buffers, EB nucleic-acid staining solution (10 mg/mL) and 100 bp DNA maker (all from Beijing Soleibo Technology Co., Ltd).
Drug Susceptibility Test
The K-B method was used in accordance with the regulations of the American Clinical Laboratory Standardization Institute (CLSI) with reference to the CLSI M100-S29 standard.16 In an extended-spectrum β-lactamases (ESBL) confirmation test, ESBLs were detected (SYA050, Biorab, Beijing, China) according to a CLSI-recommended confirmation test.17
The KB method used the following drug-sensitive paper chips (Oxoid Corporation, USA): ampicillin susceptibility assay paper chip (diffusion method) amp 10 ug, cefazolin susceptibility assay paper chip (diffusion method) KZ 30 ug, cefoperazone susceptibility test paper chip (diffusion method) CFP 75 ug, ampicillin/sulbactam susceptibility assay paper chip Sam 20 ug, 10 ug of MEM on a meropenem susceptibility test paper chip (diffusion method), cefotaxime susceptibility assay paper chip (diffusion method) CTX 30 ug, gentamicin susceptibility assay paper chip (diffusion method) CN 10 ug, tobramycin susceptibility assay paper chip (diffusion method) tob 10 ug, ciprofloxacin susceptibility assay paper chip (diffusion method) CIP 5 ug, tigecycline susceptibility assay paper chip (diffusion method) TGC 15 ug.
High-Mucus Phenotype Detection
The stored bacteria were removed from a −80°C refrigerator, and the bacteria were inoculated on a Columbia blood plate by a four-zone streak after being thawed at room temperature. The bacteria were inoculated on the Columbia blood plate after being incubated at 35°C for 24 h. A single colony was selected with an inoculation loop. A “drawing” length of >5 mm was considered to be “colony thread-drawing” positive, ie a high-yielding mucus phenotype strain; conversely, a length of ≤5 mm was considered “colony thread-drawing” negative.
Extraction of Bacterial DNA
The stored strains were removed from the −80°C refrigerator, thawed at room temperature and inoculated on Columbia blood plates by a four-zone streak; then, they were cultured at 35°C for 24 h, and KPN was isolated. A single strain was selected and inoculated in a 5 mL broth medium at 37°C and shaken at a constant temperature at 100 r/min for 24 h. It was centrifuged, and the supernatant was discarded to produce 1 mL of bacterial liquid. A DNA extraction kit was used to extract the DNA of the strain (for the specific method, please refer to the kit instructions). The obtained DNA template was stored in a −20°C refrigerator.
Detection of Capsular Serotypes and Virulence Factors
All strains were tested for capsular serotypes (K1, K2, K5, K20 and K57) and 13 virulence factors (rmpA, irp2, entB, iucD, aero, wcaG, iutA, kfu, ybtA, iron, ironB, fimH and mrkD) using the PCR method, typing and detection.13 Capsule serotype and virulence-factor gene primers were synthesised by Beijing Qingke Biotechnology Co., Ltd (Supplementary Table 1), and PCR reagents and a PCR instrument were used for amplification. The positive products were sent to the sequencing department of Ruiboxingke (Beijing) for sequencing, and the results were uploaded to http://blast.ncbi.nlm.nih.gov/ for comparison to confirm the existence of virulent genes.
MLST Typing
We used the Pasteur Institute’s MLST website (http://bigsdb.pasteur.fr/) for the DNA sequence analysis of seven housekeeping genes. The reaction system and reaction conditions for the PCRphoE amplification of these genes were as follows. The seven housekeeping gene primers of KPN (gapA, infB, rpoB, phoE, mdh, pgi and tonB) were synthesised by Beijing Qingke Biotechnology Co., Ltd (Supplementary Table 2) using an amplification reagent (Hangzhou Baosai Biotechnology Co., Ltd) and a PCR instrument for amplification. The PCR products were purified and sequenced in two directions by the sequencing department of Ruiboxingke (Beijing). The MLST database of the Pasteur Institute (https://bigsdb.pasteur.fr/klebsiella/primers-used/) was used to compare the sequences and identify the ST of the tested strain.
Statistical Analysis
Using SPSS 25 software for statistical analysis, enumeration data were described by the number of cases or rate using a χ2 test or Fisher’s exact test; a value of P<0.05 was considered statistically significant. A χ2 test was used to evaluate the relationship between hepatic abscess and diabetes mellitus (DM).
Results
Clinical Characteristics of Patients with KPN-PLA
A total of 78 cases that conformed to KPN-PLA were collected, all of which were community-acquired infections. There were 42 patients with underlying DM and 20 patients with hypertension. Diabetes was associated with mortality in KPN-PLA patients (P<0.05) (Table 1). There were 69 patients who received surgical drainage. The other 9 cases received percutaneous puncture without indwelling drainage tubes. The reasons were as follows: 5 patients chose conservative treatment, 3 patients or their families refused drainage and 1 patient was restless, and staff were unable to place the drainage tube. In 66 cases (95.7%), there was an improved prognosis. After treatment, these patients were re-examined. An imaging examination showed that the purulent cavity of the liver had shrunk or disappeared. Laboratory examination showed that WBC, PCT and other infection indicators had decreased or returned to normal. Also, 1 case (1.4%) was transferred to hospital without cure, and 2 cases (2.9%) died (2 diabetic patients). A total of 9 cases did not undergo liver-abscess puncture and drainage during hospitalisation, 5 cases (55.6%) had an improved prognosis and 4 cases (44.4%) were transferred to hospital without cure. Regarding the indication for drainage vs conservative treatment, please read.16,18
 |
Table 1 Relationship Between Diabetes Mellitus and Cure Rate of Abscess |
There was CRP <100 mg/L in 15 patients, 100–200 mg/L in 42 patients, 200–300 mg/L in 12 patients and >300 mg/L in 9 patients. There was WBC 4–10×109/L in 26 patients, WBC 10–15×109/L in 32 patients, WBC 16–20×109/L in 14 patients, WBC 21–25×109/L in 4 patients and WBC 40–50×109/L in 2 patients. The NEUs of 4 patients were 50.0%–70.0%; in 19 patients, 70.0%–80%; in 41 patients, 80%–90% and in 14 patients, >90%.
Detection Rate of HvKP
Of the 78 strains of KPN-PLA specimens, 78 strains of blood specimen KPN and 78 strains of urine specimen KPN were determined to be HvKP or cKP according to whether aero was positive or not. Seventy strains of HvKP (89.7%) were detected in KPN-PLA specimen KPN, which was significantly higher than the detection rate for the 49 strains of HvKP (62.82%) in blood KPN and 20 strains of HvKP (25.64%) in urine KPN (P<0.05) (Table 2, Supplementary Figure 1). The KPN-PLA specimens’ KPN drawing test of 63 strains (positive rate: 80.8%) was significantly higher than the 39 KPN strains from blood samples (positive rate: 50%) and the 16 KPN strains from urine samples (positive rate: 20.5%) (P<0.05).
 |
Table 2 HvKP Detection Rate and Positive Rate of Puncture Fluid, Blood and Urine Samples of Liver Abscess |
Detection of Capsular Serotypes
In the KPN-PLA KPN samples, there were 44 K1 strains in KPN-PLA samples, which was significantly higher than those in blood samples (n=13) and urine samples (n=0) (P<0.05). In the KPN-PLA specimens, the KPN contained 21 strains of the K2 type, which accounted for 26.9% – higher than the 10 strains detected in the blood samples (12.8%) and the 3 strains detected in the urine samples (3.9%) (P<0.05). In the KPN-PLA samples, KPN was detected in 11 other serotypes (14.1%) compared with 48 detections in the blood samples (61.5%) and 72 detections in the urine samples (92.2%) (P<0.05). The K20, K54 and K57 capsular serotypes were not detected (Table 3, Supplementary Figure 1).
 |
Table 3 The Detection Rate of Capsular Serum Type of Puncture Fluid, Blood and Urine of Liver Abscess |
Detection of Virulence Factors
Thirteen virulence genes of 234 KPN strains were detected in the KPN-PLA specimens, blood specimens and urine specimens. Among them, the differences in the detection rates of rmpA, aero and iutA in the three specimens were all higher in the KPN-PLA specimens (87.2%, 89.7% and 93.3%, respectively) than in the blood specimens (53.8%, 62.8% and 53.8%, respectively) and the urine specimens (16.7%, 25.6% and 16.7%, respectively), which was a statistically significant difference (P<0.05). The detection rates of the three virulence genes irp2, wcaG and ybtA in the three specimens were all higher in the KPN-PLA specimens (79.5%, 69.2% and 79.5%, respectively) than in the blood specimens (46.2%, 25.6% and 46.2%, respectively) and urine samples (46.2%, 16.7% and 46.2%, respectively), while the detection rate of iucD in the KPN-PLA samples (83.3%) was lower than that in the blood samples (100.0%) and urine samples (100.0%) (P<0.05). The detection rate of mrkD in the KPN-PLA specimens (96.2%) and urine specimens (100.0%) was higher than that in the blood specimens (87.2%) (P<0.05). The detection rates of kfu and iron in the KPN-PLA specimens (73.1% and 70.5%, respectively) and the blood specimens (70.5% and 75.6%, respectively) were higher than those in the urine specimens (25.6% and 42.3%, respectively) (P<0.05) (Table 4, Supplementary Figure 1). Irp2, wacg, kfu, ybta and iron were more frequently detected in type K1 KPN than in type K2 (P<0.05) (Table 5).
 |
Table 4 Detection Rate of Virulence Factors in Puncture Fluid, Blood and Urine Samples of Liver Abscess |
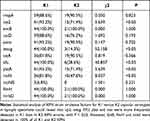 |
Table 5 Detection Rates of Virulence Factors of K1 and K2 Strains in KPN-PLA Specimens |
KPN MLST Results
A total of 234 strains of KPN from the KPN-PLA, blood and urine samples were typed by MLST. In this study, 17 MLST types were identified, of which the main types in Baotou were ST23 (16.2%, 38,234), ST86 (5.1%, 12,234), ST65 (4.7%, 11,234) and ST374 (4.3%, 10,234). In all strains, K1 was the main type of ST23 (92.1%, 35/38). The main K2 types in KPN isolated from the KPN-PLA samples were ST86 (38.1%, 8/21) and ST65 (28.6%, 6/21). In the blood and urine samples, ST374 (76.9%, 10/13) was the main K2 capsular serotype, and ST86, ST65 and ST374 were K2 capsular serotypes (Table 6).
 |
Table 6 MLST Result |
Drug Susceptibility Results of KPN in KPN-PLA, Blood and Urine Samples
The drug susceptibility results showed that KPN in the KPN-PLA specimens was highly sensitive to commonly used antibiotics; the resistance rate to all antibiotics except cefazolin (10.3%) was less than 10%, and the resistance rate to cefoperazone/sulbactam was less than 10%. The resistance rates to piperacillin/tazobactam, amikacin, ertapenem and meropenem were all 0.0%, but the resistance rate to imipenem was 1.3%. The resistance rate of KPN from the blood samples to all antibiotics except cefazolin (12.5%) and co-trimoxazole (15.6%) did not exceed 10%, among which the resistance rate to carbapenems and piperacillin/tazobactam was 0%. Compared with the KPN-PLA specimens and the blood samples, the resistance rate of KPN in the urine specimens to commonly used antibiotics was relatively high. The resistance rates to cefoperazone, cefotaxime, cefazolin, co-trimoxazole and ampicillin/sulbactam were all higher than 40%. The lowest resistance rate was to meropenem and imipenem (0.0%), but the resistance rate to ertapenem was 1.2%.
In the comparison of drug susceptibility results of KPN in the KPN-PLA samples, blood samples and urine samples, in addition to the antibiotics tigecycline, amikacin and carbapenem, all other antibiotics showed that the resistance rate of the urine specimens was higher than that of both the KPN-PLA specimens and the blood specimens (P<0.05). There was no significant difference in the resistance rates of the KPN-PLA specimens and the blood specimens to all antimicrobial agents (Table 7).
 |
Table 7 Drug Sensitive Results of Liver Abscess Puncture Fluid KPN, Blood KPN and Urine KPN |
Discussion
Throughout the demographic analysis, we found that men were more likely to develop KPN-PLA than women. KPN-PLA prevalence increases with age. Diabetes was associated with mortality in KPN-PLA patients, which was consistent with the results of others.19,20 This may be related to the mechanism by which glucose can lead to increased CPS production of KPN by reducing cAMP level.21 Despite the majority of isolates being hvKP in KPN-PLA, mortality remained low. The possible reason is that although hvKP is more virulent, it is still highly sensitive to commonly used antibiotics.22
For patients with a definitive diagnosis of KPN-PLA, ultrasound- or computed-tomography-guided percutaneous hepatic pus drainage with appropriate antimicrobial therapy has been considered the treatment standard for KPN-PLA.17 In this study, the prognosis-improvement rate of patients who underwent percutaneous hepatic pus drainage was 95.7%, while that of patients who did not undergo the procedure was 55.6%. Essentially, drainage helps to better control the source of infection, accurately identify pathogens and select appropriate antimicrobial agents. The mortality rate of patients with KPN-PLA in this study was 2.6% lower than that reported in a previous study (5.0%), which could be due to the implementation of accurate and timely interventions in patients with KPN-PLA.23
In addition, a small proportion of severely ill patients chose to be transferred to hospital, potentially leading to an underestimation of mortality. The prevalence of DM in patients with KPN-PLA in this study was 53.8%, which was consistent with previous research.24
DM is considered to be an important risk factor for KPN-PLA, since poor glycaemic control impairs neutrophil phagocytosis and promotes the growth of pathogens in tissues, and metabolic disturbances can negatively affect the liver.13 Related studies have found that HvKP strains can produce more capsular polysaccharides in a high-sugar environment. Furthermore, the virulence of HvKP increases with elevated blood-glucose levels in patients, allowing HvKP to evade the immune response of the body and survive longer, ultimately leading to a poor prognosis.20,25 Therefore, the blood-glucose level should be strictly monitored and controlled in patients with KPN-PLA.
The microbiological characterisation of KPN-PLA isolates indicates that the virulence and drug resistance of KPN strains play an important role in bacterial pathogenicity. Previous studies have suggested that high viscosity is an important hallmark of HvKP and also causes invasive KPN – an important reason for the high virulence of PLA-infecting strains. The colony thread-drawing test is usually used to determine whether the KPN strain has high viscosity. When the inoculation loop or needle can stretch out sticky filaments of ≥5 mm in length from the colony on an agar plate, the string test is determined to be positive.26 Many reports define this as hypermucoviscous KPN (HmKP). A total of 50.4% of KPN strains in this study were identified as HmKP, which is higher than another study in East China (about 33%) but lower than that reported in other Asian countries.27
However, an increasing number of recent studies have shown that HmKP and HvKP are two distinct phenotypes of KPN.28 These strains of HmKP are not necessarily highly virulent, and the strains that are negative in drawing tests are not completely without high virulence. Therefore, cKP and HvKP cannot be determined only by the colony thread-drawing test, and they should be evaluated and identified by combining genotypical and clinical characteristics.29
Current research shows that KPN strains have at least 78 capsular serotypes. The most common HvKP capsular types are K1, K2, K5, K20, K54 and K57, of which K1 and K2 are associated with HvKP, which accounts for about 70% of HvKP strains in humans. It is highly pathogenic.30 In this study, the detection rate of the K1 capsular serotype in KPN-PLA puncture fluid was 56.4%. The detection rate of the K2 capsular serotype was 26.9%, which was consistent with the above conclusions and with other studies (K1: 40.5%–63.4%; K2: 14.2%–20.5%).31,32 The detection rates of the K1 and K2 capsular serotypes in KPN-PLA puncture-fluid isolates were higher than those in blood- and urine-specimen isolates.
The above results indicated that KPN strains derived from KPN-PLA puncture-fluid specimens were more likely to have higher virulence than KPN strains derived from blood and urine specimens.
Previous studies have shown that rmpA modulates mucus phenotypes by acting on capsular polysaccharide synthesis. It has been reported that 87.5% of PLA-derived KPN isolates carry this gene.33,34 In addition, in KPN strains, common virulence factors are aerobic actin (aero), irp2, iucD, iutA, iron, kfu, ybtA, entB (encoding siderophore), wcaG, fimH (encoding type-1 bacteria [hair]) and mrkD (encoding type-3 fimbriae).35–40
Aero is the main siderophore produced by HvKP strains and accounts for 80%–90% of total siderophores. It contributes to the high virulence of HvKP expression both in vivo and in vitro, so it is more likely to detect the aero virulence gene in KPN strains to determine whether it is HvKP.38 In this study, a total of 139 (70+49+20=139) strains of HvKP were identified by detecting aero virulence factors, which accounted for 59% (139/234) of all isolated strains. The proportion of HvKP in the KPN of KPN-PLA specimens was 89.7%. This indicates that the results obtained by the two methods to determine whether the KPN strain is HvKP (ie based on clinical manifestations and the detection of the aero gene) are similar.28
To exclude the one-sidedness of assessing whether KPN isolates are HvKP based on whether the aero gene is positive or not and the clinical manifestations of patients, we tested all 234 isolates for virulence genes. The rmpA gene is the most important gene, other than aero, that causes HvKP hypervirulence;41–43 rmpA is involved in regulating the synthesis of the exopolysaccharide capsule and is associated with a hyperviscous phenotype. The loss of rmpA may lead to the loss or thinning of the capsule, thereby impairing the ability to evade immune responses and significantly reducing the virulence of KPN strains.36 In the present study, the positive rate of rmpA in KPN-PLA puncture-fluid specimens was 87.2%, which was higher than that in blood specimens (53.8%) and urine specimens (16.7%). This difference was statistically significant (P<0.05). A total of 81.3% of the KPN isolates in the KPN-PLA puncture fluid carried both aero and rmpA genes, which were more likely to be HvKP strains – a result consistent with previous reports.9,44,45 Irp2, wacg, kfu, ybta and iron were more frequently detected in type K1 KPN than in type K2 (P<0.05). This result suggests, perhaps, that the K1 type KPN strain is the most virulent of all serotypes.
The wcaG gene is also involved in capsular polysaccharide biosynthesis and is also prevalent in KPN-PLA isolates.44,46 In this study, the detection rate of the wcaG gene in KPN-PLA puncture fluid was 69.2%, which was higher than in blood (25.6%) and urine (16.7%) (P<0.05).
The above results further confirm the conclusion that KPN strains derived from KPN-PLA puncture specimens are more likely to have a high-mucus phenotype and higher virulence than KPN strains derived from blood and urine specimens.
Although the virulence factor is not a separate factor for the determination of HvKP, we note an intrinsic correlation that needs to be confirmed by further studies. Since the detection of virulence factors has a warning effect on HvKP, in the clinical treatment of KPN-PLA, the treatment time should be appropriately extended, and long-term follow-ups should be performed to improve the possibility of prognosis improvement and minimise the recurrence rate.29 Unfortunately, the current study could not definitively determine the exact number of HvKP in the 234 KPN strains because the reference standard for HvKP has not yet been established. However, from the perspective of clinical laboratory diagnostics, it is recommended that clinicians should not ignore the diagnostic value of virulence factors in the identification of HvKP isolates. At the same time, the continued exploration of virulence factors may provide new therapeutic targets and treatment modalities for HvKP infection.47
In addition, KPN strains isolated from KPN-PLA puncture-fluid specimens are highly sensitive to almost all antibacterial drugs, such as β-lactamase inhibitors, cephalosporins, quinolones and carbapenems, which may be related to the fact that the KPN-PLA puncture-fluid isolates were almost all HvKP.47 However, one carbapenem-resistant strain was detected in our study, which was of the K1 capsular serotype and positive for all virulence factors except iron, which rarely occurs in HvKP.7 Relevant studies have found that this carbapenem-resistant strain is more likely to appear in patients with a history of hepatobiliary-related diseases; the KPN-PLA puncture-fluid specimens selected in our study excluded specimens from patients with a history of hepatobiliary-related diseases. A further comparison and analysis of the drug susceptibility results of the 78 KPN-PLA puncture-fluid samples selected in this study and in patients with KPN-PLA who had a history of hepatobiliary-related diseases showed that the drug-resistance rates of almost all the tested antimicrobials in patients with KPN-PLA who had a history of hepatobiliary disease were significantly higher than those in patients with KPN-PLA but without a history of hepatobiliary disease.
The results of the MLST analysis showed the molecular epidemiological characteristics of 234 KPN strains. The ST23 type was the main type in the KPN-PLA puncture-fluid isolates and accounted for 32.1%, which was similar to that in previous reports.48 The ST23 strain is one of the major clonal strains of HvKP, the founder strain of the clonal lineage of ST23 and a representative of a specific genetic background for high virulence.48 Although ST23 strains can spread not only locally but also worldwide, ST23 isolates from KPN-PLA puncture-fluid specimens were genotypically closely related.49 Notably, ST23 was closely related to the K1 capsular serotype among the KPN-PLA aspirate specimen isolates, whereas ST65- and ST86-like isolates were associated with the K2 capsular serotype, which is similar to other reports from Asia.50 In the KPN strains of blood and urine samples, the K2 capsular serotype was mainly ST374, but the specific reason for this phenomenon is still unclear. Among all 234 KPN strains, the ST23 type accounted for the highest proportion (16.2%), which proved that the K1 ST23 isolate was the dominant clone of HvKP in the Baotou area. In 75.6% and 76.9% of KPN isolates in blood and urine samples, respectively, the capsular serotype could not be identified, which was much higher than in KPN-PLA puncture-fluid isolates (47.4%) (P<0.05). The reason for this difference may be related to the genetic difference between HvKP and cKP, and the specific cause remains to be further explored. More research is needed to elucidate the capsular serotype, the virulence of KPN and the relationship between genotypes, drug resistance and MLST typing, especially for strains with simultaneous high virulence, high drug resistance and high pathogenicity.
Conclusion
This study suggests to clinicians that most of the KPN isolates in the puncture fluid of KPN-PLA patients in Inner Mongolia, China, are HvKP. The positive rate of KPN-PLA specimens was higher than in blood and urine specimens. The K1 and K2 types were the main capsular serotypes of HvKP, and the main ST types of HvKP were ST23, ST86 and ST65. The detection rates of the virulence genes rmpA, aero and iutA in KPN-PLA specimens were higher than those in blood and urine specimens, which indicated that the KPN-PLA specimens may have had stronger virulence. Although the KPN-PLA specimen isolates were highly sensitive to commonly used antibiotics, there remained some carbapenem-resistant HvKP isolates.
These results will provide an effective clinical reference for the detection methods and mechanisms of HvKP and will have guiding significance for its clinical medication.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval and Consent to Participate
This study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of The First Affiliated Hospital of Baotou Medical College. The sample collection were performed with written informed consent from all patients.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This study was not funded.
Disclosure
The authors report no personal, financial, commercial, or academic conflicts of interest for this work.
References
1. Behzadi P, Elham B. The microbial agents of urinary tract infections at central laboratory of Dr. Shariati Hospital, Tehran, Iran. Turk Klin Tip Bilim. 2008;28(4):445–449.
2. Hu FP, Guo Y, Zhu DM, et al. Surveillance of bacterial resistance in CHINET tertiary hospitals in 2019. Chin J Infect Chemother. 2020;20(03):233–243.
3. Chen M, Li Y, Zhang J, Chen S, Zhang Y. Molecular and genetic characteristics of highly virulent Klebsiella pneumoniae in respiratory infection. Chin J Hosp Infect Dis. 2020;30(01):6–9.
4. Lin FJ, Xu ZC, Lin ZW, et al. Analysis of virulence factors and molecular typing characteristics of highly pathogenic Klebsiella pneumoniae in bloodstream infection. Chin J Pathogen Biol. 2018;13(04):364–367.
5. Liu YS, Gao Y. Clinical research progress of highly virulent Klebsiella pneumoniae liver abscess. Chin J Infect Chemother. 2020;20(01):95–101.
6. Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J Intern Med. 2020;287(3):283–300. doi:10.1111/joim.13007
7. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):e00001–19. doi:10.1128/CMR.00001-19
8. Behzadi P, Urbán E, Matuz M, et al. The role of gram-negative bacteria in urinary tract infections: current concepts and therapeutic options. Adv Exp Med Biol. 2021;1323:35–69.
9. Khonsari MS, Behzadi P, Foroohi F, et al. The prevalence of type 3 fimbriae in Uropathogenic Escherichia coli isolated from clinical urine samples. Meta Gene. 2021;28:100881. doi:10.1016/j.mgene.2021.100881
10. Issakhanian L, Behzadi P. Antimicrobial agents and urinary tract infections. Curr Pharm Des. 2019;25(12):1409–1423. doi:10.2174/1381612825999190619130216
11. Qu TT, Zhou JC, Jiang Y, et al. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect Dis. 2015;15:161. doi:10.1186/s12879-015-0899-7
12. Yang Y, Liu JH, Hu XX, et al. Clinical and microbiological characteristics of hypervirulent Klebsiella pneumoniae (hvKp) in a hospital from North China. J Infect Dev Ctries. 2020;14(6):606–613. doi:10.3855/jidc.12288
13. Behzadi P, Gajdács M. Writing a strong scientific paper in medicine and the biomedical sciences: a checklist and recommendations for early career researchers. Biol Futur. 2021;72(4):395–407. doi:10.1007/s42977-021-00095-z
14. Shang H, Wang YS, Shen ZY. National Clinical Laboratory Operation Regulations. People’s Health Publishing House; 2015.
15. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing[S]. Twentyninth Informational Supplement. Clinical and Laboratory Standards Institute; 2019.
16. Mezhir JJ, Fong Y, Jacks LM, et al. Current management of pyogenic liver abscess: surgery is now second-line treatment. J Am Coll Surg. 2010;210(6):975–983. doi:10.1016/j.jamcollsurg.2010.03.004
17. Chan KS, Shelat V. Pyogenic liver abscess. In: The IASGO Textbook of Multi-Disciplinary Management of Hepato-Pancreato-Biliary Diseases. Singapore: Springer; 2022. doi:10.1007/978-981-19-0063-1_66
18. Chan KS, Chia CTW, Shelat VG. Demographics, radiological findings, and clinical outcomes of Klebsiella pneumonia vs. non-Klebsiella pneumoniae pyogenic liver abscess: a systematic review and meta-analysis with trial sequential analysis. Pathogens. 2022;11(9):976. doi:10.3390/pathogens11090976
19. Lee JH, Jang YR, Ahn SJ, Choi SJ, Kim HS. A retrospective study of pyogenic liver abscess caused primarily by Klebsiella pneumoniae vs. non-Klebsiella pneumoniae: CT and clinical differentiation. Abdom Radiol. 2020;45(9):2669–2679. doi:10.1007/s00261-019-02389-2
20. Lin CT, Chen YC, Jinn TR, Wu CC, Hong YM, Wu WH. Role of the cAMP-dependent carbon catabolite repression in capsular polysaccharide biosynthesis in Klebsiella pneumoniae. PLoS One. 2013;8(2):e54430. doi:10.1371/journal.pone.0054430
21. Shelat VG, Chia CL, Yeo CS, Qiao W, Woon W, Junnarkar SP. Pyogenic liver abscess: does Escherichia coli cause more adverse outcomes than Klebsiella pneumoniae? World J Surg. 2015;39(10):2535–2542. doi:10.1007/s00268-015-3126-1
22. Siu LK, Yeh KM, Lin JC, et al. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12(11):881–887. doi:10.1016/S1473-3099(12)70205-0
23. Ma R, Wang X, Nie D. Clinical characteristics of bloodstream infection with highly virulent Klebsiella pneumoniae. China J Infect Control. 2018;17(01):26–30.
24. Lee CH, Chen IL, Chuah SK, et al. Impact of glycemic control on capsular polysaccharide biosynthesis and opsonophagocytosis of Klebsiella pneumoniae: implications for invasive syndrome in patients with diabetes mellitus. Virulence. 2016;7(7):770–778. doi:10.1080/21505594.2016.1186315
25. Wu H, Li D, Zhou H, Sun YF, Guo L, Shen DX. Bacteremia and other body site infection caused by hypervirulent and classic Klebsiella pneumoniae. Microb Pathog. 2017;104:254–262. doi:10.1016/j.micpath.2017.01.049
26. Kong H, Yu F, Zhang W, Li XF. Clinical and microbiological characteristics of pyogenic liver abscess in a tertiary hospital in East China. Medicine. 2017;96(37):e8050. doi:10.1097/MD.0000000000008050
27. Catalan-Najera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes. Virulence. 2017;8(7):1111–1123. doi:10.1080/21505594.2017.1317412
28. Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9):e00776–18. doi:10.1128/JCM.00776-18
29. Shi Q, Lan P, Huang D, et al. Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. 2018;18(1):94. doi:10.1186/s12866-018-1236-2
30. Fung CP, Chang FY, Lee SC, et al. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis. Gut. 2002;50(3):420–424. doi:10.1136/gut.50.3.420
31. Zhang S, Zhang X, Wu Q, et al. Clinical, microbiological, and molecular epidemiological characteristics of Klebsiella pneumoniae-induced pyogenic liver abscess in southeastern China. Antimicrob Resist Infect Control. 2019;8:166. doi:10.1186/s13756-019-0615-2
32. Siu LK, Lin JC, Gomez E, et al. Virulence and plasmid transferability of KPC Klebsiella pneumoniae at the veterans affairs healthcare system of New Jersey. Microb Drug Resist. 2012;18(4):380–384. doi:10.1089/mdr.2011.0241
33. Yu WL, Ko WC, Cheng KC, et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42(10):1351–1358. doi:10.1086/503420
34. Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun. 2015;83(8):3325–3333. doi:10.1128/IAI.00430-15
35. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
36. Candan ED, Aksoz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62(4):867–874. doi:10.18388/abp.2015_1148
37. Luo Y, Wang Y, Ye L, Yang J. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect. 2014;20(11):O818–O824. doi:10.1111/1469-0691.12664
38. Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;6:38929. doi:10.1038/srep38929
39. Zhang S, Yang G, Ye Q, Wu QP, Zhang JM, Huang YB. Phenotypic and genotypic characterization of Klebsiella pneumoniae isolated from retail foods in China. Front Microbiol. 2018;9:289. doi:10.3389/fmicb.2018.00289
40. Ahmadi M, Ranjbar R, Behzadi P, et al. Virulence factors, antibiotic resistance patterns, and molecular types of clinical isolates of Klebsiella Pneumoniae. Expert Rev Anti Infect Ther. 2022;20(3):463–472. doi:10.1080/14787210.2022.1990040
41. Hozzari A, Behzadi P, Khiabani PK, et al. Clinical cases, drug resistance, and virulence genes profiling in Uropathogenic Escherichia coli. J Appl Genet. 2020;61(2):265–273. doi:10.1007/s13353-020-00542-y
42. Yu WL, Lee MF, Chang MC, et al. Intrapersonal mutation of rmpA and rmpA2: a reason for negative hypermucoviscosity phenotype and low virulence of rmpA-positive Klebsiella pneumoniae isolates. J Glob Antimicrob Resist. 2015;3(2):137–141. doi:10.1016/j.jgar.2015.03.008
43. Tan TY, Ong M, Cheng Y, Ng LSY. Hypermucoviscosity, rmpA, and aerobactin are associated with community-acquired Klebsiella pneumoniae bacteremic isolates causing liver abscess in Singapore. J Microbiol Immunol Infect. 2019;52(1):30–34. doi:10.1016/j.jmii.2017.07.003
44. Ahmadi Z, Noormohammadi Z, Behzadi P, et al. Molecular detection of gyrA mutation in clinical strains of Klebsiella pneumoniae. Iran J Public Health. 2022;51(10):2334–2339. doi:10.18502/ijph.v51i10.10992
45. Ahmadi Z, Noormohammadi Z, Ranjbar R, et al. Prevalence of tetracycline resistance genes tet (A, B, C, 39) in Klebsiella pneumoniae isolated from Tehran, Iran. Iran J Med Microbiol. 2022;16(2):141–147. doi:10.30699/ijmm.16.2.141
46. Russo TA, Gulick AM. Aerobactin synthesis proteins as antivirulence targets in hypervirulent Klebsiella pneumoniae. ACS Infect Dis. 2019;5(7):1052–1054. doi:10.1021/acsinfecdis.9b00117
47. Moore R, O’Shea D, Geoghegan T, Mallon PWG, Sheehan G. Community-acquired Klebsiella pneumoniae liver abscess: an emerging infection in Ireland and Europe. Infection. 2013;41(3):681–686. doi:10.1007/s15010-013-0408-0
48. Lam M, Wyres KL, Duchene S, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9(1):2703. doi:10.1038/s41467-018-05114-7
49. Struve C, Roe CC, Stegger M, et al. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio. 2015;6(4):e00630. doi:10.1128/mBio.00630-15
50. Siu LK, Fung CP, Chang FY, et al. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol. 2011;49(11):3761–3765. doi:10.1128/JCM.00977-11
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
