Back to Journals » Advances and Applications in Bioinformatics and Chemistry » Volume 13
Structural and Molecular Docking Analytical Studies of the Predicted Ligand Binding Sites of Cadherin-1 in Cancer Prognostics
Authors Bakare OO , Fadaka AO , Keyster M , Pretorius A
Received 23 March 2020
Accepted for publication 5 June 2020
Published 28 July 2020 Volume 2020:13 Pages 1—9
DOI https://doi.org/10.2147/AABC.S253851
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Juan Fernandez-Recio
Olalekan Olanrewaju Bakare,1 Adewale Oluwaseun Fadaka,1 Marshall Keyster,2 Ashley Pretorius1
1Bioinformatics Research Group, University of the Western Cape, Cape Town 7535, South Africa; 2Biotechnology Department, University of the Western Cape, Cape Town 7535, South Africa
Correspondence: Olalekan Olanrewaju Bakare Tel +27 603112776
Email [email protected]
Introduction: Several studies have explored the design of antimicrobial peptides (AMPs) for the development of therapeutic and diagnostic molecules for the treatment and identification of pathogenic diseases as well as cancer. Human cadherin-1 protein has been identified to be involved in adhesion-mediated signalling pathways in normal cells and its loss through genetic and epigenetic alterations can result in an enhanced invasion and metastasis of malignancy in tumours. Therefore, the identification of cadherin during treatment of cancer can be used as prognostic biomarker to establish the responsiveness of patients to treatment regimen. Antimicrobial peptides (AMPs) offer several compensatory advantages in biomedical applications and have been used for treatment of diseases, dietary supplements and diagnosis of diseases. The aim of this research work was to use in silico approaches to analyse retrieved human cadherin-1 as prognostic targets in cancer treatments using modelled putative anticancer AMPs.
Methods: The structures of the putative AMPs and cadherin-1 were modelled using I-TASSER server and the protein overall quality was validated using PROCHECK. Thereafter, the protein motifs were predicted and the molecular interaction between the putative anticancer AMPs and protein was carried out using PatchDock.
Results: The results revealed that all the AMPs were good prognostic molecules for cancer with BOO1 having the highest binding affinity of 15,874.
Conclusion: This study revealed that all the generated AMPs have good prognostic value for monitoring the progress of cancer treatment using human cadherin-1 as receptor. This is the first report where AMPs were used in prognostics of cancer using human cadherin-1.
Keywords: cancer, antimicrobial peptides, prognosis, homology, modelling, docking, physicochemical, binding, validation, cadherin-1
Introduction
According to the WHO statistics, cancer is rated the second leading cause of death in the world with about 9.6 million death in 2018.1 This means one in six death cases is due to cancer with the 70% of these happening in low and middle income country.2 It has led to major economic losses in which International Health communities and Research institutes have spent a lot of money to combat its menace with a total economic cost of 1.16 trillion dollars spent in 2010 alone.3 One major factor leading to the enormous mortality rate in the low income country is the late stage presentation, inaccessible diagnosis, lack of precision to monitor treatment efficacy and overwhelming lack of data to drive policy.
Cancer is caused by many factors characterized by an abnormal growth of cells in the body with the tendency to grow rapidly, divide and proliferate.2 Environmental and lifestyle factors worsen the incidence of cancer as a major public health problem posing a global menace associated with metabolic and genetic abnormalities.4 One evidence linking cancer to abnormal gene regulation is the disturbance of cell–cell and cell extracellular matrix adhesion with the subsequent changes in adhesion-mediated signalling pathways of cadherins which induce malignant phenotypes in normal cells.5,6
Calcium independent adhesions (cadherins) are a class of type-1 transmembrane proteins found in tumors and normal cells which are highly dependent on Ca2+ ions to function and use cell adhesion molecule (adherens junctions) for binding cells together.7 Cadherin-mediated adhesion through the formation of cadherin–catenin complex regulates cell growth and differentiation, normal cell–cell adhesion, and maintain homeostatic tissue architecture. Disruption of signalling and altered expression of cadherins causes the disaggregation of tumors which can be temporal or permanent to promote the metastasis of such cells.8 This makes cadherins a major target in cancer therapy. Over a hundred subtypes of this superfamily have been identified from different species of organisms.7 Much research work has been conducted on the relationship between cadherins and tumorigenesis as well as drug-targeting cadherins which have undergone clinical trials.9 The loss of cadherin in tumorigenesis due to genetic and epigenetic alterations has been implicated in increased abnormal growth and metastasis in cancer cells, thus preventing its regulatory role in malignancy as a cancer cell suppressor. Identification of cadherin after treatment could be a potential approach in cancer prognostics for enhanced treatment.
Antimicrobial peptides (AMPs) have recently been explored as a source of therapy for many diseases due to their compensatory multifunctional attributes. The advantages of these vital molecules include signalling molecule, drug delivery vector, immune-modulatory agent, antitumor and mitogenic agents.10 The aim of this research work is to use an in silico approach to identify AMPs modelled from HMMER as selective inhibitors for more potent anti-cancer therapy with high effectiveness, short-lived side effects and high tolerability.
Materials and Methods
Retrieval of Cadherin-1 Sequences of Homo sapiens and Computing of the Physicochemical Parameters Analysis
The protein, cadherin-1, (P12830) amino acid sequences were retrieved from the Universal Protein Databases (UniProt) (https://www.uniprot.org/uniprot/P12830).11 Thereafter, the protein physicochemical parameters were generated from the ProtParam tools (http://web.expasy.org/protparam/) using exPAsy Server to reveal the protein structure and function with the computation of molecular weight, extinction coefficient, amino acid composition, estimated half-life, hydropathicity, theoretical PI, instability index and aliphatic index.12
Prediction of Secondary Structure
Secondary structure prediction such as alpha-helices, beta-sheets and random coils of cadherin was predicted using the PSIPRED secondary structure prediction server (http://bioinf.cs.ucl.ac.uk/psipred).13
Assessment of Experimentally Validated Antimicrobial Peptides (AMPs) Data
Experimentally validated anticancer AMPs from various databases such as the Collection of Anti-microbial Databases (CAMP) (http://www.camp.bicnirrh.res.in),14 Anti-microbial Peptides Database (APD) (http://aps.unmc.edu/AP/main.php)15 and Data Repository of Antimicrobial Peptides (DRAMP) (http://dramp.cpu-bioinfor.org).16 Thereafter, arrangement of these data and curation were performed to validate the authenticity of the anticancer AMPs. Subsequently, duplicate anticancer AMPs were removed using Cluster Density at High Intensity with Tolerance (CD-HIT) (http://www.bioinformatics.org/cd-hit).14
Hidden Markov Models (HMMER) Profile Construction
The latest version of Hidden Markov Models (HMMER) algorithm version 3.817 was used to construct specific anticancer AMPs profiles using the respective training datasets, having divided the retrieved experimentally validated anticancer AMPs into three quarters (training dataset) and one quarter (testing dataset). Query of the training dataset was carried out using the positive dataset (testing dataset) and negative dataset (neuropeptides). The HMMER profile was built on the Ubuntu 12.04 LTS operating system. The task was accomplished on a terminal and the command lines used to build the profile was written in accordance with the corresponding algorithm and the steps involved in their construction are shown in Figure S1.
Discovery of Novel Anticancer AMPs
Proteome sequences were retrieved from the Ensembl database (http://www.ensembl.org/index.html) and the UniProt database (http://www.uniprot.org/) and were used to query the profile with the list of all proteome sequences (in the FASTA format) retrieved. A cut-off E-value was set to be 0.05 for the search of putative anticancer AMPs. This was accomplished using “hmmsearch” module of the HMMER algorithm with the command line employed as stated in Figure S2.
Determination of the Physicochemical Parameters of the Putative Anticancer AMPs
3-D Structure Prediction of the Putative Anticancer AMPs and Cadherin
The anticancer AMPs and cadherin (PDB ID: 1FF5) (E-Cadherin double domain) 3-D structures were predicted using ITASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) and visualized using PyMol.19 The cadherin, protein model structure was utilized for the de novo modelling of the cadherin protein from the LOMETS threading programs.20
Model Evaluation and Prediction of Functional Motif
The quality of the generated model was checked using PROCHECK to compute parameters such as side chain conformations of the protein using atomic resolutions, lengths, geometry of the hydrogen bond, angles and planarity of the peptide bonds.21 The MOTIF (http://www.genome.jp/tools/motif/) finder was used to find the motifs present in the protein using the amino acid sequences.22
In Protein-Ligand Interaction Study
The docking of the anticancer AMPs to the cadherin protein was carried out using PatchDock 1.3 version, a free online web-server that enables the docking of the protein-small ligand molecule, available at http://bioinfo3d.cs.tau.ac.il/PatchDock/.23 In brief, the 3-D structures of the putative anticancer AMPs and the cadherin protein receptor PDB files from I-TASSER were uploaded onto the PatchDock server. Interaction analysis of the complex formation between the putative anticancer AMPs and the cadherin protein receptor was achieved using RasMol 2.7.5 software.24
Results
Physicochemical Properties of Cadherin-1
The result from ExPAsy server (Table 1) indicated that cadherin-1 from Homo sapiens has 882 amino acids with each residue contributing to give the protein an average molecular weight of 97.456kDa. The result also indicated that the most abundant amino acids are threonine, leucine, valine, aspartate, proline, and glutamate, respectively. The physicochemical parameters predicted the protein to be negatively charged because of the abundance of glutamic acid (6.8%) and aspartic acid (7.7%) over arginine (4.4%) and lysine (4.0%) since these four amino acids contribute the overall charge of the protein at neutral Ph.25 Also the protein has 4327 carbon, 6749 hydrogen, 1151 nitrogen, 1374 oxygen and 18 sulphurs adding up to 13,619 atoms.
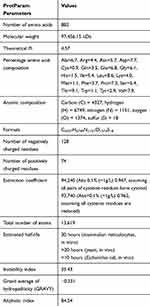 |
Table 1 Physicochemical Properties of Cadherin-1 |
The protein is also acidic (4.57) with an half-life of 30 hours in mammals, greater than 20 hours in yeast and greater than 10 hours in Escherichia coli indicating the number of hours that it can stay intact in these organisms without being degraded. The quantity of light that can be absorbed by the protein is indicated by the extinction coefficient was computed using tyrosine, tryptophan and cysteine while the aliphatic index was 84.54 with −0.351 GRAVY and instability index of 35.43.
Secondary Structure Prediction of Cadherin-1
The result in Figure 1 shows the predicted 2-D structure of cadherin from the PSIPRED server where one alpha helix, 29 beta strand, and 32 random coils were recorded. Cadherin is a transmembrane protein with an abundance of beta strands just like human retinol binding protein and porins.26,27 The abundance of beta strands in cadherin allows it to perform its binding function through cell adhesion.28
 |
Figure 1 Secondary structure prediction of cadherin 1 from PSIPRED. Cadherin 1 is predicted to have 29 beta strands and one alpha helix. |
Putative AMPs Discovery and Their Physicochemical Characterization
Retrieval of experimentally validated anticancer AMPs from CAMP, APD and DRAMP databases was carried out. It was revealed that APD, CAMP and DRAMP had a total of 231 anticancer AMPs which were either validated, inferred or synthetic. Only experimentally validated AMPs from the databases were retrieved for this study. After duplicate removal, 56 experimentally validated anticancer AMPs were generated. The peptides were divided into three quarters (training set) and one quarter (testing set). HMMER was used to build single profile of the anticancer AMPs using the training set. The training profile was used to scan the testing set (positive dataset) which showed no discrimination for a high specificity, accuracy and sensitivity. For improved sensitivity, accuracy and specificity, 17,234 neuropeptides in FASTA format were retrieved from the UNIPROT database (http://www.uniprot.org) and ENSEMBL server (http://www.ensembl.org/index.html) to test the discriminatory tendency of the profile constructed by HMMER, called “the query of profile”. The HMMER showed motif discrimination for the negative dataset (neuropeptides) since they have no evolutionary relationship.
Subsequently, discovery of putative anticancer AMPs was carried out using the training profile constructed by HMMER against several proteomes of organisms downloaded from the ENSEMBL database to discover putative anticancer AMPs, where the lowest E values less than the default 0.05 were considered for further study.Five putative anticancer AMPs were identified and ranked according to their E values from lowest to the highest (Table S1).
Subsequently, the physicochemical parameters of the AMPs were computed to determine their characteristics required for the interaction study with the cadherin-1. For the five putative anticancer AMPs generated (Table 2), none have 100% similarity to the existing anticancer AMPs or experimentally validated ones from which they were discovered and as such can be considered novel. All the anticancer AMPs had positive charge in conformity with an ideal AMP. Also the AMPs had an abundance of lysine and arginine strengthening further the contribution of these amino acids to the positive charge of the AMPs and their isoelectric point at alkaline pH.
 |
Table 2 Physicochemical Parameters of the Putative Anticancer AMPs |
The predicted hydrophobic value of the AMPs was greater than 30% as anticipated for an ideal AMP and these values contributed significantly to their high binding potential to receptors and cells.29 This work is in accordance with the work of Aruleba et al, 30 where AMPs were used to investigate the structural and predicted binding sites of Slc2a4 as a therapeutic target in the treatment of cancer.
3-D Homology Prediction of the Anticancer Putative AMPs and Cadherin-1 Using I-TASSER
The prediction of the 3-D homology structure of the putative anticancer AMPs and the cadherin-1 was carried out using ITASSER and visualized using PyMol. As indicated in Figure S3, the result from the ITASSER gave rise to beta strands of all the putative anticancer AMPs. This means that the putative anticancer AMPs can modulate their interaction, specificity and binding affinity to a receptor target. In addition, the 3-D ITASSER result is in agreement with the 2-D result as indicated from the quality assessment scores such as the confidence scores, template modelling scores and the root mean square deviation. As indicated in Table 3, the confidence scores (C scores), the template modelling scores (TM scores) and RMSD of the putative anticancer AMPs and the cadherin 3-D homology models were of good model quality. A C score of a quality structure prediction model ranges between −5 and 2,31 TM score >0.5 indicates a model of correct topology32 whereas RMSD only indicates atomic deviation of the predicted 3-D homology of the putative anticancer AMPs and cadherin-1 from the templates available in ITASSER.33 The models were of good quality because the C score ranges between −5 and 2, TM scores were all above the limiting value of >0.5 indicating models of correct topology and RMSD values were between 4.4Å and 15.3Å for all models of the anticancer AMPs and cadherin-1.
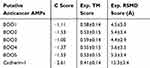 |
Table 3 Quality Assessment Scores of the Putative Anticancer AMPs and Cadherin-1 |
3-D Modelled Structure Validation of Cadherin-1
The quality of the model structure of cadherin1 was carried out using the evaluation and validation of the PROCHECK (https://servicesn.mbi.ucla.edu/PROCHECK/). As indicated in Figure 2, the result from the PROCHECK analysis indicates that the model of cadherin1 has 87.2% residues in the most favored regions, 12.8% in the additional allowed regions, 0.0% in the generously allowed regions and 0.0% at the disallowed regions. This model is said to be of high quality because of the distribution of the amino acid residues. The G factor was also determined using this program.
Probable Motifs Functional Analysis of Cadherin-1
Table S2 shows the investigation of the probable functional motif of cadherin-1 where the protein was said to contain five functional motifs. Cadherin, the first functional motif is located at position 161.253 in the amino acid sequence with an E value of 4.1e-3, cadherin C is located at position 734.878 in the amino acid sequence withE value 6.7e-49, Cadherin pro located at position 27.116 in the amino acid sequence with E value 5e-31, RET_CLD1 located at position 521.559 in the amino acid sequence with E value 0.017 and cadherin-3 located at position 357.428 in the amino acid sequence with E value 0.39.
Protein-peptide Interaction of the Putative Anticancer AMPs and Cadherin-1
Table 4 shows online protein–protein interaction between the cadherin-1 (receptor) and the putative anticancer AMPs (ligands) using PatchDock. All results show a high binding affinity score greater than 8741, indicating a high binding solution between the putative anticancer AMPs and cadherin-1. The result from this experiment can be pursued for rational prediction of cadherin inhibitors in the development of an efficacious anticancer therapy.
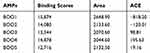 |
Table 4 Docking Interaction Analysis of the Putative Anticancer AMPs and Cadherin-1 |
The results of the docking interaction analysis of the cadherin-1 and the putative anticancer AMPs were downloaded in PDB files and visualized using RasMol software. This is displayed in Figure S4 with the blue colour indicating the protein, cadherin-1 and the red, putative anticancer AMPs.
Discussion
Detection of cadherin through a prognostic biomarker can give information about cancer patients' improvements toward treatment regimens.34 Cadherin mediate cell–cell adhesion to regulate epithelial monolayer behaviour and most human solid tumors are epithelial in origin, necessitating the attention of adhesion molecules at the epithelial cell junctions.9 Cadherins are well conserved proteins with their cytoplasmic domains interacting with beta catenins at its central region to perform regulatory and stability functions.35 One of the roles of cadherin in tumorigenesis is its loss, which contributes to enhanced invasion and metastasis in human cancer, thus preventing it from binding a newly synthesized beta catenin.36 The increased upregulation of human cadherin-1 in malignancy has been linked to the suppression of cancer cells.35 Therefore, detection of cadherin could be a potential approach in cancer prognostics for enhanced treatment. Numerous research works have used AMPs in the treatment of diseases.37,38 Recently, AMPs have been used for HIV detection using its P24 as receptor.39 The aim of this research work was to detect cadherin using putative AMPs as prognostic biomarkers for potential applications in monitoring cancer patients' improvements using in silico approaches. This study is novel and significant because it is the first report where AMPs were used as prognostic biomarkers for human cadherin-1 detection to monitor patients’ responsiveness during treatment of cancer. To this end, analysis of human cadherin-1 was carried out with the discovery of five putative anticancer AMPs and their interaction studies analysed using several types of bioinformatics tools for precision prognosis.
The cysteine residue of cadherin-1 contributed less stability due to its lower amount in the protein. Thus the stability of human cadherin-1 could be due to the exposure of the hydrophobic core, hydrogen bonding, net charge and the ionization state of the amino acid residues.40 The presence of noncovalent bonds between amino acids and chemical forces between protein and its immediate environment in human cadherin-1 could be another reason for its stability.41 AMPs have gained interest as novel sources of drugs and diagnostics of diseases. Its significance for these therapeutic and diagnostic roles have been attributed to small size and diverse activities triggered towards diseases.42 The five putative anticancer AMPs discovered from this research had a3> Boman index >1, an indicator that these peptides would make a good prognostic biomarker for the detection of cadherin-1 in humans. A Boman index of less than zero might have antibacterial activities and that greater than zero might have multifunctional and hormone-like activities.30,43 The alpha-helices and extended sheets of the putative anticancer AMPs 3-D structures can help in the folding complementation, insertion and intermolecular interaction.44 This is because alpha helices make efficient use of hydrogen bonds for binding between hydrogen of the amino group and the oxygen of the carboxyl group.30 Thus, the presence of alpha helices in the AMPs make them interact with another biomolecule for significant impact as targets in prognosis. The 3-D structure of the human cadherin-1 and the putative anticancer AMPs showed good quality as indicated with the C scores, TM-scores and RMSD values. The modelled 3-D structure of human cadherin-1 reliability was evaluated using PROCHECK which deemed it a high quality model because of the distribution of the amino acid residues in the highly favoured, allowed, disallowed and generously allowed regions.21 Subsequently, five major functional motifs were predicted and several ligand active sites in the human cadherin-1 protein were revealed.
Finally, the predicted putative anticancer AMPs were used in this study to identify human cadherin-1, an important protein for the cell–cell adherence and it can serve as prognostic biomarker for monitoring the progress of cancer patients during treatment.
Furthermore, the protein–protein interaction PatchDock server was used to dock the putative anticancer AMPs (BOO1–BOO5) with human cadherin-1 in which high binding affinity scores greater than 8741 were generated for the predicted 3-D structure of the AMPs when docked with cadherin-1 3-D structure. This is an indication of high binding solution for the AMPs with BOO1 having the highest score (Table 4). The results generated from this study can be pursued for the rational design of novel, selective and potent human cadherin-1 biomarker in the search for novel anticancer prognostic biomarker with high accuracy and specificity.
Future Work
The in vitro study of the prognostic activity of the putative anticancer AMPs will be carried out. In addition, the EC50 of all the AMPs and their prognostic or selective indices will be assessed for the optimized AMPs. The anticancer prognostic potential of these AMPs will be carried out on different pseudo types of the cancer cell lines to determine their broad spectrum of prognostic activities. Finally, the binding complex formed between the cancer cell lines and putative AMPs will be solved using structural biology techniques to validate the observations made by the in silico binding study.
Data Sharing Statement
All data generated or analysed during this study are included in this published article.
Acknowledgments
Special appreciation to National Research Foundation (NRF), South Africa and South African National Zakat Fund (SANZAF).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
3. Kim YA, Oh I-H, Yoon S-J, et al. The economic burden of breast cancer in Korea from 2007–2010. Cancer Res Treat. 2015;47(4):583. doi:10.4143/crt.2014.143
4. Kalyanaraman B. Teaching the basics of cancer metabolism: developing antitumor strategies by exploiting the differences between normal and cancer cell metabolism. Redox Biol. 2017;12:833–842. doi:10.1016/j.redox.2017.04.018
5. Kottke MD, Delva E, Kowalczyk AP. The desmosome: cell science lessons from human diseases. J Cell Sci. 2006;119(5):797–806. doi:10.1242/jcs.02888
6. Carvalho J, van Grieken NC, Pereira PM, et al. Lack of microRNA‐101 causes E‐cadherin functional deregulation through EZH2 up‐regulation in intestinal gastric cancer. J Pathol. 2012;228(1):31–44. doi:10.1002/path.4032
7. Goffinet AM, Tissir F. Seven pass cadherins CELSR1-3. In: Seminars in Cell & Developmental Biology. Elsevier; 2017.
8. Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. 2007;8(1):11. doi:10.1038/nrn2043
9. Labernadie A, Kato T, Brugués A, et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol. 2017;19(3):224. doi:10.1038/ncb3478
10. Tincho M, Gabere M, Pretorius A. In silico identification and molecular validation of putative antimicrobial peptides for HIV therapy. J AIDS Clin Res. 2016;7(9).
11. Pettitt J. The cadherin superfamily. In: WormBook: The Online Review of C. Elegans Biology [Internet]. WormBook; 2005.
12. Gill SC, Von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182(2):319–326. doi:10.1016/0003-2697(89)90602-7
13. McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. doi:10.1093/bioinformatics/16.4.404
14. Waghu FH, Gopi L, Barai RS, et al. CAMP: collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. 2014;42(D1):D1154–D1158. doi:10.1093/nar/gkt1157
15. Wang Z, Wang G. APD: the antimicrobial peptide database. Nucleic Acids Res. 2004;32(suppl_1):D590–D592. doi:10.1093/nar/gkh025
16. Fan L, Marchetti L, Parlanti P, et al. DRAMP: a comprehensive data repository of antimicrobial peptides. Sci Rep. 2016;6(1):1–7. doi:10.1038/s41598-016-0001-8
17. Roddy J. Reducing False Sequence Annotation Due to Alignment Overextension. 2018.
18. Fang Q. Predicting Functional Alterations Caused by Non-Synonymous Variants in CHO Using Models Based on Phylogenetic Tree and Evolutionary Preservation. University of Sheffield; 2018.
19. DeLano WL. The PyMOL molecular graphics system; 2002. Available from: http://www.pymol.org.
20. Yang J, Yan R, Roy A, et al. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12(1):7. doi:10.1038/nmeth.3213
21. Laskowski RA, MacArthur MW, Moss DS, et al. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–291. doi:10.1107/S0021889892009944
22. Chhabra G, Sharma P, Anant A, et al. Identification and modeling of a drug target for clostridium perfringens SM101. Bioinformation. 2010;4(7):278. doi:10.6026/97320630004278
23. Schneidman-Duhovny D, Inbar Y, Nussinov R, et al. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(suppl_2):W363–W367. doi:10.1093/nar/gki481
24. Goodsell DS. Representing structural information with RasMol. Curr Protoc Bioinformatics. 2005;11(1):
25. Li R, Sonik A, Stindl R, et al. Aneuploidy vs. gene mutation hypothesis of cancer: recent study claims mutation but is found to support aneuploidy. Proc Natl Acad Sci. 2000;97(7):3236–3241. doi:10.1073/pnas.97.7.3236
26. Roche DB, Viet PD, Bakulina A, et al. Classification of β-hairpin repeat proteins. J Struct Biol. 2018;201(2):130–138. doi:10.1016/j.jsb.2017.10.001
27. Slusky JS. Outer membrane protein design. Curr Opin Struct Biol. 2017;45:45–52. doi:10.1016/j.sbi.2016.11.003
28. Bardag-Gorce F, Hoft RH, Wood A, et al. The role of E-cadherin in maintaining the barrier function of corneal epithelium after treatment with cultured autologous oral mucosa epithelial cell sheet grafts for limbal stem deficiency. J Ophthalmol. 2016;2016:1–13. doi:10.1155/2016/4805986
29. Giuliani A, Pirri G, Nicoletto S. Antimicrobial peptides: an overview of a promising class of therapeutics. Open Life Sci. 2007;2(1):1–33. doi:10.2478/s11535-007-0010-5
30. Aruleba RT, Adekiya T, Oyinloye B, et al. Structural studies of predicted ligand binding sites and molecular docking analysis of Slc2a4 as a therapeutic target for the treatment of cancer. Int J Mol Sci. 2018;19(2):386. doi:10.3390/ijms19020386
31. Yang J, Zhang Y. Protein structure and function prediction using I‐TASSER. Curr Protoc Bioinformatics. 2015;52(1):
32. Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725. doi:10.1038/nprot.2010.5
33. Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008;9(1):40. doi:10.1186/1471-2105-9-40
34. Christou N, Perraud A, Blondy S, et al. E‑cadherin: a potential biomarker of colorectal cancer prognosis. Oncol Lett. 2017;13(6):4571–4576. doi:10.3892/ol.2017.6063
35. Kourtidis A, Lu R, Pence LJ, et al. A central role for cadherin signaling in cancer. Exp Cell Res. 2017;358(1):78–85. doi:10.1016/j.yexcr.2017.04.006
36. Bertocchi C, Wang Y, Ravasio A, et al. Nanoscale architecture of cadherin-based cell adhesions. Nat Cell Biol. 2017;19(1):28–37. doi:10.1038/ncb3456
37. Wang S, Zeng X, Yang Q, et al. Antimicrobial peptides as potential alternatives to antibiotics in food animal industry. Int J Mol Sci. 2016;17(5):603. doi:10.3390/ijms17050603
38. Mor A. Antimicrobial Peptides. Kirk‐Othmer Encyclopedia of Chemical Technology; 2000.
39. Williams M, Tincho MB, Gabere M, Uys A, Meyer M, Pretorius A. Molecular validation of putative antimicrobial peptides for improved human immunodeficiency virus diagnostics via HIV protein p24. J AIDS Clin Res. 2016;7:571.
40. Pace CN, Fu H, Lee Fryar K, et al. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014;23(5):652–661. doi:10.1002/pro.2449
41. Pace CN, Grimsley GR, Scholtz JM. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem. 2009;284(20):13285–13289. doi:10.1074/jbc.R800080200
42. Ageitos J, Sánchez-Pérez A, Calo-Mata P, et al. Antimicrobial peptides (AMPs): ancient compounds that represent novel weapons in the fight against bacteria. Biochem Pharmacol. 2017;133:117–138. doi:10.1016/j.bcp.2016.09.018
43. Gómez EA, Giraldo P, Orduz S. InverPep: a database of invertebrate antimicrobial peptides. J Glob Antimicrob Resist. 2017;8:13–17. doi:10.1016/j.jgar.2016.10.003
44. Kumar P, Kizhakkedathu JN, Straus SK. Antimicrobial peptides: diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules. 2018;8(1):4. doi:10.3390/biom8010004
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

