Back to Journals » Journal of Inflammation Research » Volume 14
Stable Cells with NF-κB-ZsGreen Fused Genes Created by TALEN Editing and Homology Directed Repair for Screening Anti-inflammation Drugs
Authors Zhang S , Luo T, Wang J
Received 29 December 2020
Accepted for publication 22 February 2021
Published 17 March 2021 Volume 2021:14 Pages 917—928
DOI https://doi.org/10.2147/JIR.S298938
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Shuyan Zhang, Tao Luo, Jinke Wang
State Key Laboratory of Bioelectronics, Southeast University, Nanjing, 210096, People’s Republic of China
Correspondence: Jinke Wang
State Key Laboratory of Bioelectronics, Southeast University, Nanjing, 210096, People’s Republic of China
Email [email protected]
Background: NF-κB is a sequence-specific DNA-binding transcription factor that plays key roles in inflammation and cancer. It is well known that NF-κB is over-activated in these diseases. NF-κB inhibitors are therefore developed as promising drugs for these diseases. However, finding NF-κB inhibitors is dependent on effective screening platforms.
Methods: For providing an easy and visualizable tool for screening NF-κB inhibitors, and other NF-κB-related studies, this study edited all five genes of NF-κB family (RELA, RELB, CREL, NF-κB1, NF-κB2) in three different cell lines (293T, HepG2, and PANC1) with both TALEN and CRISPR. The edited NF-κB genes were repaired by homology-dependent repair using a linear homologous donor containing ZsGreen coding sequence. The edit efficiency was thus directly evaluated by detecting cellular fluorescence. The editing efficiency was also confirmed by PCR detection of NF-κB-ZsGreen fused genes.
Results: It was found that all genes were more efficiently edited by TALEN in all cells than CRISPR. The positive cells were then isolated from the TALEN-edited cell pool by flow cytometry. The purified positive cells were finally evaluated by regulating NF-κB activity with a known NF-κB inhibitor, BAY 11– 7082, and an NF-κB-targeting artificial microRNA, miR533. The results revealed that all the labeled NF-κB genes responded well to the two kinds of NF-κB activity regulators in all cell lines.
Conclusion: This study thus obtained 15 cell lines with NF-κB-ZsGreen fused genes, which provide an easy and visualizable tool for screening NF-κB inhibitors and other NF-κB-related studies.
Keywords: NF-κB, TALEN, edit, HDR, ZsGreen
Introduction
Targeted integration of transgenes is usually achieved by a homologous recombination (HR)-mediated method.1,2 It requires a repair template that harbors left and right homology arms (HAs) (500–3000 bp), thus allowing precise insertion of large DNA fragments. To facilitate such targeted integration of transgenes, a targeted DNA double-strand break (DSB) has to be produced in the genome by using custom-designed nucleases, such as zinc-finger nuclease (ZFN),3–6 transcription activator-like effector nucleases (TALEN),2,7 and the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 nuclease (Cas9).1,8 However, DSBs are lethal to cells so that it must be repaired by the cell’s DNA repair machinery. DSBs can be repaired via imprecise nonhomology-based repair mechanisms, such as nonhomologous end-joining (NHEJ), or by precise homology-dependent repair (HDR).9 HDR utilizes a template DNA that contains sequences homologous to the flanking sequences of the DSB.10 The homologous sequences are termed as homology arms. The HDR can use donors of variant forms including single-stranded DNA oligonucleotides (ssODNs) and double-stranded DNA (PCR fragments).11 The PCR fragments with 35-bp homology arms can function as efficient donors for genome editing in mouse embryos and human cells.12 The genome editing and HDR are now widely used to produce various chimeric antigen receptor (CAR) immunotherapeutic cells such as CAR-T. Additionally, other cell therapies also rely on gene modification to provide cells with particular functions.
Nuclear factor κB (NF-κB) is a family of DNA-binding transcription factors. The NF-κB family consists of five members, RelA/p65, RelB, c-Rel, p50, and p52, which are coded by the genes of RELA, RELB, CREL, NF-κB1, and NF-κB2, respectively. These members are classified into two groups: a group of p50 and p52, and the other group of RelA, RelB, and c-Rel.13 The former has the DNA-binding domain (DBD) but no transcription activation domain (TAD); however, the latter has both DBD and TAD. The main functional and canonical dimer of NF-κB family is p50·p65 heterodimer.13 NF-κB plays various key roles in many physiological and pathological processes, such as immunity,13–15 cell proliferation,16 apoptosis, inflammation,17,18 and especially oncogenesis.19 Some studies indicate that many diseases are associated with the aberrant activation of NF-κB and its signaling pathway.18,20 For instance, NF-κB is abnormally activated in many human cancers to promote survival and malignancy by upregulating antiapoptotic genes.21 Therefore, many pharmaceutical companies and scientists have been committing to develop NF-κB inhibitors for inflammation and cancer therapy.22
Chemicals are important fields to develop inhibitors of NF-κB activity. For instance, BAY 11–7082 is a widely used NF-κB inhibitor that inhibits NF-κB activity by inhibiting the phosphorylation of IκBα.23 In addition, microRNA (miRNA) is a kind of natural small RNA (~22 nt), which can regulate gene expression by blocking target mRNA translation or inducing target mRNA degradation.24–26 For finding the NF-κB inhibitors, an effective cellular platform is essential, in which the change of NF-κB activity under the treatment of various potential drugs should be better facilely and quantitative monitored. For this purpose, various NF-κB reporter stable cell lines with NF-κB reporter construct that contains several repeats of NF-κB binding sites, a minimal promoter upstream of the green fluorescent protein (GFP) and/or luciferase coding region were developed,27–33 which allows an assessment of NF-κB activity via fluorescence microscopy, flow cytometry, and/or luminometry. However, these constructs are independent on the natural promoters of NF-κB genes. They report a signal resulted from NF-κB activity but not NF-κB itself. In addition, the current platforms are only developed for NF-κB RelA; however, NF-κB family consists of five genes. RelA just forms the canonical NF-κB signaling pathway, while others such as p52·RelB consist of a noncanonical NF-κB signaling pathway.15,34 These different pathways were activated by different sets of stimulators and activate different sets of target genes.15,34 Therefore, more simple and comprehensive cell platforms are still in demand in the industry.
This study systematically edited all five genes of NF-κB family in three different cell lines (293T, HepG2, PANC1) with both TALEN and CRISPR. The linear HDR donor containing ZsGreen coding sequence was used to repair the TALEN/CRISPR-produced DBSs. In this way, a ZsGreen coding sequence was fused to the ends of NF-κB genes, which would produce ZsGreen-labeled NF-κB proteins under the control of natural promoters of NF-κB genes. The editing results were characterized by the fluorescent imaging of cells, quantitative analysis of cell fluorescence with flow cytometry, and quantitative PCR (qPCR) detection of NF-κB-ZsGreen fused genes. The editing results were also evaluated by treating cells with an NF-κB stimulator, TNFα. Finally, the positive cells were isolated from the edited cells with flow cytometry according to cellular fluorescence and further validated by treating cells with two kinds of NF-κB inhibitors, chemical and microRNA inhibitors. This study thus obtained 15 cell lines with NF-κB-ZsGreen fused genes, which provide an easy and visualizable tool for screening NF-κB inhibitors and other NF-κB-related studies.
Methods
TALEN and CRISPR Vectors
The TALEN and CRISPR expression plasmids that express TALEN and CRISPR/Cas9-sgRNA targeting five genes of NF-κB family came from our recent study.35 All vectors used for cell transfection were purified from DH5α using EndoFree Plasmid kits (QIAGEN, Germany) and quantified with a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific, USA).
HDR Donor Preparation
The linear HDR donors that were used to fusing ZsGreen coding sequences to NF-κB family genes were prepared by PCR amplification. Oligonucleotide ending with 35-nt sequences that were used as homology arms targeting NF-κB family genes were used as primers (Table S1). A linear DNA fragment containing the Gslink-ZsGreen coding sequences was used as a template. The PCR reaction (30 μL) contained 1 ng Gslink-ZsGreen DNA fragment (template), 10 μM hom-F (Table S1), 10 μM hom-R (Table S1), and 1×PrimeSTAR®HS DNA Polymerase (Takara, Japan). PCR program was 96°C 3 min, 28 cycles of 96°C 20 s, 58°C 20 s, and 72°C 1 min, and 72°C 5 min. The PCR products were purified with a Gel Extraction Kit (QIAGEN).
Cell Culture and Transfection
293T, HepG2, and PANC1 cells were obtained from the cell resource center of Shanghai institute for biological sciences, Chinese Academy of Sciences; China. 293T is a human embryonic kidney cell with stable gene sequence coding SV40 large T antigen, HepG2 is a human hepatoma cell, and PANC1 is a human pancreatic carcinoma cell. 293T is widely used in gene editing studies due to its high transfection efficiency. Hepatoma is a cancer that most seriously threatens the health and life of the Chinese. The pancreatic carcinoma is recognized as the king of carcinoma due to very limited therapy and high mortality. Therefore, the three cell lines were selected to use in this study. Cells were cultured with DMEM (HyClone, USA) containing 10% fetal bovine serum (FBS) (HyClone), 100 U/mL penicillin, and 100 μg/mL streptomycin in a humidified incubator with 5% CO2 for 37°C. Cells were seeded in a 24-well plate at a density of 5×104 cells/well and incubated for 24 h. The cells were co-transfected with 400 ng of TALEN vector (200 ng left TALEN and 200 ng right TALEN) or CRISPR/Cas9-sgRNA vector and 400 ng of HDR donor. The cell transfection was performed with lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instruction. The transfected cells were cultured for an additional 72 h. The cells were rinsed with PBS and imaged with a fluorescent microscope, IX51 with DP71 (Olympus, Japan), and quantitatively analyzed with a flow cytometer, BD Calibur (BD, USA). Ten thousand cells were analyzed in flow cytometry.
PCR Detection of Fused Genes
The genomic DNA (gDNA) was extracted from the edited cells by using a gDNA extraction kit (Tiangen, China). The gDNA was detected by quantitative PCR (qPCR) using a StepOnePlusTM Real-Time PCR system (Applied Biosystems, USA). The fused NF-κB genes were detected with forward primers (RELA-F, RELB-F, CREL-F, NFKB1-F, or NFKB2-F; Table S2) and a reverse primer (ZsGreen-R; Table S2). GAPDH gene was also detected as an internal control with primers GAPDH-F and GAPDH-R (Table S2). The PCR reaction (20 μL) consisted of 100 ng of gDNA, 10 μM forward primer, 10 μM reverse primer, and 2×SYBR Mix (Roche, Switzerland). PCR program was 95°C 3 min, 40 cycles of 95°C 15 s, and 60°C 1 min. The PCR products were detected by agarose gel electrophoresis.
TNFα Stimulation of NF-κB Fused Gene
The TALEN-edited cells were seeded in 24-well plates and incubated for 24 h at 37°C in 5% CO2. The cells were then stimulated with TNFα (Sigma, USA) at a final concentration of 10 ng/mL for 1 h. The control cells were not treated by TNFα. The cells were then washed with cold PBS and collected by trypsinization. The collected cells were washed with cold PBS. The cells were fixed in formaldehyde (Sigma) at a final concentration of 1% (w/v) for 10 min at room temperature. The crosslinking was terminated by adding glycine at the final concentration of 0.125 M. The cells were precipitated by centrifugation at 1500 rpm for 10 min at 4°C. The cells were lysed with 500 μL of lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.1% IGEPAL CA-630). The nuclei were collected by centrifugation at 1500 rpm for 10 min at 4°C. The nuclei were washed twice with PBS. Finally, the nuclei were imaged with fluorescent microscope IX51 with DP71 (Olympus) and analyzed with flow cytometry (Calibur).
Screening of Positive Cells
The TALEN-edited cells were collected by trypsinization. The cells were resuspended in 200 μL DMEM (HyClone) containing 2% fetal bovine serum (FBS) (HyClone), 100 U/mL penicillin, and 100 μg/mL streptomycin. The cells were used to screen positive cells with green fluorescence through a sorter of flow cytometry (BD FACSAriaTM III, USA). The fluorescent cells were purified from one million edited cells. The screened cells (about 100–200 thousand) were cultured at 37°C for 48 h in a 24-well plate.
Validation of Screened Cells by NF-κB Regulators
The positive cells cultivated in 24-well plates were treated with 50 μM BAY 11–7082 (Abmole, USA) for 1 h. The cells were imaged with fluorescent microscope (IX51 with DP71) and analyzed with flow cytometry (Calibur). The positive 293T, HepG2, and PANC1 cells in 24-well plates were transfected with 800 ng of pDMP-miR533, respectively. The cells were cultured for an additional 72 h. The cells were imaged with a fluorescent microscope (IX51 with DP71) and analyzed with flow cytometry (Calibur).
PCR Detection of NF-κB Target Gene Expression
The total RNA was purified from cells using the Trizol® reagent (Invitrogen). Then, 500 ng of total RNA was reversely transcribed into cDNA using PrimeScript™ RT Master Mix (Takara) according to the manufacturer’s instructions in a total volume of 10 μL. Triplicate samples per treatment were evaluated on a ABI Step One Plus (Applied Biosystems). The reaction mixture contained 2 μL cDNA, 0.25 μM of primers, and 10 μL of Hieff qPCR SYBR Green Master Mix (Yeasen, China) in a total volume of 20 μL. The primers used in qPCR were list in Table S3. PCR program was 95°C 5 min, 40 cycles of 95°C 10 s, and 60°C 30 s. The melting curve analysis was performed. The threshold value Ct values were normalized by subtracting the Ct values obtained for GAPDH. The ΔCt values were then used to calculate relative changes of mRNA expression.
Results
Editing NF-κB Family Genes in Three Cell Lines with Both TALEN and CRISPR
Five pairs of TALEN and five CRISPR expression plasmids were prepared by our recent study that developed an easy ready-to-use TALEN plasmid preparation pipeline.35 Five pairs of TALEN expression plasmids were constructed to express the TALEN proteins that can produce double-strand breaks (DSBs) at the end of coding sequences of five NF-κB family genes (Figure 1A). Five CRISPR expression plasmids were constructed to express Cas9 protein and single-guide RNA (sgRNA) that can produce DSBs at the same sites as TALENs (Figure 1A). A linear HDR donor that contains a Gslink and ZsGreen coding sequences and ends with 35-nt homologous (hom) arms was used as a template for repairing the DSBs (Figure 1B).
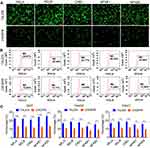
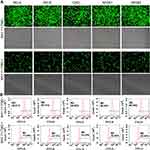
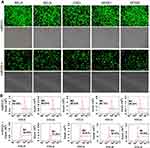
Five NF-κB family genes (RELA, RELB, CREL, NF-κB1, NF-κB2) were systematically edited in three different cell lines (293T, HepG2 and PANC1) by co-transfecting cells with TALEN/CRISPR and HDR donor. The successful editing and HDR would produce an NF-κB-ZsGreen fused gene in cells, which allows cells to express an NF-κB-ZsGreen fused protein under the control of natural promoter of NF-κB gene. By directly imaging cells with fluorescent microscope and quantitatively analyzing cells with flow cytometry, the editing efficiency can be evaluated. The results indicated that all five genes were successfully edited by both TALEN and CRISPR in all cells (Figure 2, Figure S1, and Figure S2). However, both fluorescent imaging (Figure 2A, Figure S1A, and Figure S2A) and flow cytometry quantitative assay (Figure 2B, Figure S1B, and Figure S2B) revealed that TALEN had higher editing efficiency than CRISPR. On average, TALEN edited 22.2%, 15.5%, and 17% cells in 293T, HepG2, and PANC1 cells, respectively, while CRISPR edited 13.2%, 9.7%, and 10.3% cells in 293T, HepG2, and PANC1 cells, respectively (Figure 2C). The editing success and efficiency were also confirmed by the subsequent qPCR detection of NF-κB-ZsGreen fused genes in gDNA (Figure 3).
To further validate the edited NF-κB genes, the TALEN-edited cells were treated with an NF-κB stimulator, TNFα. Because the activated NF-κB proteins will translocate into nuclear, the nuclei were isolated from the TNFα-treated and -untreated cells. The isolated nuclei were analyzed by both fluorescent imaging and flow cytometry. The results indicated that the TNFα treatment induced a significant increase of fluorescent nuclei in all cells (Figure 4, Figure S3, and Figure S4).
Screening of Positive Cells and Treatment
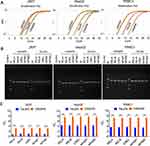
To obtain the pure positive cells, the TALEN-edited cells were sorted with flow cytometry according to ZsGreen fluorescence. The results indicated that the pure positive cells could be efficiently obtained from TALEN-edited cell lines (Figure S5 and Figure 5). Therefore, the pure positive cells were obtained for five NF-κB family genes from three cell lines (15 cell lines in total) (Figure 5).
To validate the potential application of these purified cell lines in screening of NF-κB inhibitors, all cell lines were treated with two kinds of representative NF-κB inhibitors, a small molecular chemical, BAY 11–7082, and a microRNA, miR533. The results indicated that both BAY 11–7082 and miR533 significantly decreased the number of fluorescent cells in all 15 cell lines (Figure 6, Figure 7, and Figure S6–S9).
To further validate the response of stable cells to the treatment of NF-κB inhibitors, the expression of some typical NF-κB target genes in the BAY 11–7082-treated and –untreated stable HepG2 and PANC1 cells with RELA-ZsGreen gene was detected by qPCR. The detected NF-κB target genes included CCL2, IL1A, IL1B, NFKB1, NFKBIA, and PTX3. These genes are all NF-κB direct target genes (NF-κB Target Genes, Boston University Biology, https://www.bu.edu/nf-kb/gene-resources/target-genes/). The results indicated that the BAY 11–7082 treatment to the stable cells leads to the significant downregulation of these genes (Figure S10).
Discussion
NF-κB is a key transcription factor involved in inflammation and cancer. To provide an easy and visualizable tool for screening NF-κB inhibitors and other NF-κB-related studies, this study edited all five genes of NF-κB family in three different cell lines with both TALEN and CRISPR. The edited NF-κB genes were repaired by HDR using a linear homologous donor containing ZsGreen coding sequence. In this way, the NF-κB proteins were labeled by fluorescent ZsGreen in cells. By detecting the edited cells with fluorescent imaging, flow cytometry, and qPCR, it was demonstrated that TALEN had higher editing efficiency than CRISPR in editing all genes in all cells. The positive cells were easily isolated from the TALEN-edited cells by flow cytometry. The purified positive cells responded well to two kinds of known NF-κB activity regulators in all cell lines. As a result, this study obtained 15 cell lines with five NF-κB-ZsGreen fusing genes, providing useful cellular platforms for screening potential NF-κB-target drugs.
In our recent study, we developed an easy and rapid pipeline for preparing ready-to-use TALEN expression plasmids.35 Using the pipeline, this study constructed the TALEN expression plasmids that can express the TALEN proteins targeting five genes of NF-κB family. By the quantitative analysis of cell fluorescence with flow cytometry and the edited genes with qPCR, this study demonstrated that all genes were more effectively edited by TALEN than CRISPR in all cells, indicating that TALEN is still a valuable genome editing tool with better performance than CRISPR. The high editing efficiency is very important for a gene editing tool. Additionally, TALEN can be used to edit any sequence in genome, whereas CRISPR is limited by the requirement of protospacer adjacent motif (PAM). Moreover, TALEN has high targeting performance due to cutting a target by a pair of TALEN; however, CRISPR is still challenged by its potential off-target in application. As previously reported, this study also demonstrated that a linear DNA fragment ended with short homology arm (35 nt) can be used as an efficient HDR donor in genome editing. This kind of linear HDR donor can be facilely prepared by PCR amplification using primers ended with 35-nt homology arms.
This study used miR533 to represent an NF-κB inhibitor of microRNA. This microRNA is an artificial microRNA, which was designed and verified by our previous work.36 In this study, this microRNA was expressed by a plasmid that contains an NF-κB-specific promoter and a downstream microRNA expression sequence. The NF-κB-specific promoter consists of an NF-κB decoy sequence and minimal promoter. The microRNA expression sequence can transcribe a microRNA targeting RELA coding sequence. This study showed that the expression of other four NF-κB genes other than RELA could be also inhibited by this microRNA. The reason is that the NF-κB genes RELB,37 CREL,38,39 NF-κB1,40 and NF-κB241 are the direct target genes of RELA (NF-κB Target Genes, Boston University Biology, https://www.bu.edu/nf-kb/gene-resources/target-genes/).
Conclusions
This study edited five NF-κB family genes in three cell lines with TALEN and CRISPR. The edited genes were fused to the ZsGreen coding sequences by HDR. The results indicated that TALEN had higher editing efficiency than CRISPR in editing all genes in all cells. The screened positive cells formed 15 cell lines with five NF-κB-ZsGreen fusing genes. These cell lines could respond to variant NF-κB inhibitors and thus become useful platforms for screening potential NF-κB-target drugs.
Abbreviations
TALE, transcription activator-like effector; TALEN, TALE nuclease; DBD, DNA-binding domain; TAD, transcription activation domain; HR, homologous recombination; HAs, homology arms; ZFN, zinc-finger nuclease; CRISPR, clustered regularly interspaced short palindromic repeats; sgRNA, single-guided RNA; ssODNs, single-stranded DNA oligonucleotides; CAR, chimeric antigen receptor; PCR, polymerase chain reaction; qPCR, quantitative PCR; gDNA, genomic DNA; NF-κB, nuclear factor kappa B; NHEJ, non-homologous end joining; HDR, homology-dependent repair; DSB, double-strand break; TF, transcription factor; FBS, fetal bovine serum.
Funding
This work was supported by the National Natural Science Foundation of China (61971122).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yang H, Wang HY, Shivalila CS, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–1379. doi:10.1016/j.cell.2013.08.022
2. Hockemeyer D, Wang HY, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. doi:10.1038/nbt.1927
3. Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. P Natl Acad Sci USA. 1996;93:1156–1160. doi:10.1073/pnas.93.3.1156
4. Bibikova M, Beumer K, Trautman JK, et al. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300(5620):764. doi:10.1126/science.1079512
5. Moehle EA, Rock JM, Lee YL, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases (vol 104, pg 3055, 2007). P Natl Acad Sci USA. 2007;104:6090. doi:10.1073/pnas.0611478104
6. Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300(5620):763. doi:10.1126/science.1078395
7. Christian M, Cermak T, Doyle EL, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–U476. doi:10.1534/genetics.110.120717
8. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas Systems. Science. 2013;339(6121):819–823. doi:10.1126/science.1231143
9. Danner E, Bashir S, Yumlu S, et al. Control of gene editing by manipulation of DNA repair mechanisms. Mamm Genome. 2017;28(7–8):262–274. doi:10.1007/s00335-017-9688-5
10. Jasin M, Haber JE. The democratization of gene editing: insights from site-specific cleavage and double-strand break repair. DNA Repair. 2016;44:6–16. doi:10.1016/j.dnarep.2016.05.001
11. Paix A, Schmidt H, Seydoux G. Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs. Nucleic Acids Res. 2016;44(15):e128. doi:10.1093/nar/gkw502
12. Paix A, Folkmann A, Goldman DH, et al. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. P Natl Acad Sci USA. 2017;114(50):E10745–E10754. doi:10.1073/pnas.1711979114
13. Ghosh S, May MJ, Kopp EB. NF-κB and rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16(1):225–260. doi:10.1146/annurev.immunol.16.1.225
14. Baldwin AS. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14(1):649–683. doi:10.1146/annurev.immunol.14.1.649
15. Bonizzi G, Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25(6):280–288. doi:10.1016/j.it.2004.03.008
16. Biswas DK, Cruz AP, Gansberger E, et al. Epidermal growth factor-induced NF-κB activation: a major pathway of cell-cycle progression in estrogen-receptor negative breast cancer cells. P Natl Acad Sci USA. 2000;97(15):8542–8547. doi:10.1073/pnas.97.15.8542
17. Lane DP, Midgley CA, Hupp TR, et al. On the Regulation of the P53 tumor-suppressor, and its role in the cellular-response to DNA-damage. Philos T Roy Soc B. 1995;347:83–87.
18. Tak PP, Firestein GS. NF-kappa B: a key role in inflammatory diseases. J Clin Invest. 2001;107:7–11. doi:10.1172/JCI11830
19. Chen XF, Kandasamy K, Srivastava RK. Differential roles of RelA (p65) and c-Rel subunits of nuclear factor kappa B in tumor necrosis factor-related apoptosis-inducing ligand signaling. Cancer Res. 2003;63(5):1059–1066.
20. Yamamoto Y, Gaynor RB. Therapeutic potential of inhibition of the NF-κB pathway in the treatment of inflammation and cancer. J Clin Invest. 2001;107(2):135–142. doi:10.1172/JCI11914
21. Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2(3):175–187. doi:10.1038/nrc746
22. Gupta SC, Sundaram C, Reuter S, et al. Inhibiting NF-κB activation by small molecules as a therapeutic strategy. Bba-Gene Regul Mech. 2010;1799:775–787.
23. Pierce JW, Schoenleber R, Jesmok G, et al. Novel inhibitors of cytokine-induced IκBα phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272(34):21096–21103. doi:10.1074/jbc.272.34.21096
24. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function (Reprinted from Cell, vol 116, pg 281-297, 2004). Cell. 2007;131:11–29.
25. Novak J, Olejnickova V, Tkacova N, et al. Mechanistic Role of MicroRNAs in coupling lipid metabolism and atherosclerosis. Adv Exp Med Biol. 2015;887:79–100.
26. Santulli G. MicroRNAs and Endothelial (Dys) Function. J Cell Physiol. 2016;231(8):1638–1644. doi:10.1002/jcp.25276
27. Zeuner MT, Vallance T, Vaiyapuri S, et al. Development and Characterisation of a Novel NF-κ B reporter cell line for investigation of neuroinflammation. Mediators Inflamm. 2017;2017:1–10. doi:10.1155/2017/6209865
28. Zhao M, Howard EW, Parris AB, et al. Alcohol promotes migration and invasion of triple-negative breast cancer cells through activation of p38 MAPK and JNK. Mol Carcinogen. 2017;56(3):849–862. doi:10.1002/mc.22538
29. Eskla KL, Porosk R, Reimets R, et al. Hypothermia augments stress response in mammalian cells. Free Radical Bio Med. 2018;121:157–168. doi:10.1016/j.freeradbiomed.2018.04.571
30. Badr CE, Niers JM, Tjon-Kon-Fat LA, et al. Real-Time Monitoring of NF-κB Activity in Cultured Cells and in Animal Models. Mol Imaging. 2009;8(5):278–290. doi:10.2310/7290.2009.00026
31. Lendermon EA, Coon TA, Bednash JS, et al. Azithromycin decreases NALP3 mRNA stability in monocytes to limit inflammasome-dependent inflammation. Resp Res. 2017;18.
32. Cen XH, Zhu CZ, Yang JJ, et al. TLR1/2 specific small-molecule agonist suppresses leukemia cancer cell growth by stimulating cytotoxic T Lymphocytes. Adv Sci. 2019;6(10):1802042. doi:10.1002/advs.201802042
33. Krabbe G, Minami SS, Etchegaray JI, et al. Microglial NF-κB-TNFα hyperactivation induces obsessive-compulsive behavior in mouse models of progranulin-deficient frontotemporal dementia. P Natl Acad Sci USA. 2017;114(19):5029–5034. doi:10.1073/pnas.1700477114
34. Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12(1):86. doi:10.1186/1476-4598-12-86
35. Zhang S, Wang J, Wang J. One-Day TALEN assembly protocol and a dual-tagging system for genome editing. ACS Omega. 2020;5(31):19702–19714. doi:10.1021/acsomega.0c02396
36. Wang D, Tang H, Xu X, et al. Control the intracellular NF-κB activity by a sensor consisting of miRNA and decoy. Int J Biochem Cell Biol. 2018;95:43–52. doi:10.1016/j.biocel.2017.12.009
37. Bren GD, Solan NJ, Miyoshi H, et al. Transcription of the RelB gene is regulated by NF-κB. Oncogene. 2001;20(53):7722–7733. doi:10.1038/sj.onc.1204868
38. Hannink M, Temin HM. Structure and Autoregulation of the C-Rel promoter. Oncogene. 1990;5(12):1843–1850.
39. Capobianco AJ, Gilmore TD. Repression of the chicken C-rel promoter by vRel in chicken-embryo fibroblasts is not mediated through a consensus NF-κB Binding-Site. Oncogene. 1991;6:2203–2210.
40. Ten RM, Paya CV, Israel N, et al. The characterization of the promoter of the gene encoding the p50 subunit of NF-κB indicates that it participates in its own regulation. EMBO J. 1992;11(1):195–203. doi:10.1002/j.1460-2075.1992.tb05042.x
41. Lombardi L, Ciana P, Cappellini C, et al. Structural and functional-characterization of the promoter regions of the NFKB2 Gene. Nucleic Acids Res. 1995;23(12):2328–2336. doi:10.1093/nar/23.12.2328
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.