Back to Journals » Journal of Inflammation Research » Volume 15
SPP1/AnxA1/TIMP1 as Essential Genes Regulate the Inflammatory Response in the Acute Phase of Cerebral Ischemia-Reperfusion in Rats
Authors Nie QQ, Zheng ZQ, Liao J, Li YC, Chen YT, Wang TY, Yuan GQ, Wang Z , Xue Q
Received 11 April 2022
Accepted for publication 16 August 2022
Published 24 August 2022 Volume 2022:15 Pages 4873—4890
DOI https://doi.org/10.2147/JIR.S369690
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Qian-Qian Nie,1,* Zong-Qing Zheng,2,* Juan Liao,1 Yu-Chao Li,3 Yan-Ting Chen,1 Tian-Ye Wang,1 Gui-Qiang Yuan,4 Zhong Wang,2 Qun Xue1
1Department of Neurology & Brain and Nerve Research Laboratory, The First Affiliated Hospital of Soochow University, Suzhou, People’s Republic of China; 2Department of Neurosurgery & Brain and Nerve Research Laboratory, The First Affiliated Hospital of Soochow University, Suzhou, People’s Republic of China; 3Department of Nuclear Medicine, Changhai Hospital, Naval Medical University (Second Military Medical University), Shanghai, People’s Republic of China; 4Department of Neurosurgery & Brain and Nerve Research Laboratory, The First Affiliated Hospital of Soochow University, Changshu Second People’s Hospital, Suzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qun Xue; Zhong Wang, Department of Neurology & Brain and Nerve Research Laboratory, The First Affiliated Hospital of Soochow University, Ganjiang Road, Suzhou, 215021, People’s Republic of China, Email [email protected]; [email protected]
Background: Ischemic injury in stroke is followed by extensive neurovascular inflammation and changes in ischemic penumbra gene expression patterns. However, the key molecules involved in the inflammatory response during the acute phase of ischemic stroke remain unclear.
Methods: Gene expression profiles of two rat ischemic stroke-related data sets, GSE61616 and GSE97537, were downloaded from the GEO database for Gene Set Enrichment Analysis (GSEA). Then, GEO2R was used to screen differentially expressed genes (DEGs). Furthermore, 170 differentially expressed intersection genes were screened and analyzed for Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. Candidate genes and miRNAs were obtained by DAVID, Metascape, Cytoscape, STRING, and TargetScan. Finally, the rat middle cerebral artery occlusion-reperfusion (MCAO/R) model was constructed, and qRT-PCR was used to verify the predicted potential miRNA molecule and its target genes.
Results: GO and KEGG analyses showed that 170 genes were highly associated with inflammatory cell activation and cytokine production. After cluster analysis, seven hub genes highly correlated with post-stroke neuroinflammation were obtained: Cxcl1, Kng1, Il6, AnxA1, TIMP1, SPP1, and Ccl6. The results of TargetScan further suggested that miR-340-5p may negatively regulate SPP1, AnxA1, and TIMP1 simultaneously. In the ischemic penumbra of rats 24 h after MCAO/R, the level of miR-340-5p significantly decreased compared with the control group, while the concentration of SPP1, AnxA1, and TIMP1 increased. Time-course studies demonstrated that the mRNA expression levels of SPP1, AnxA1, and TIMP1 fluctuated dramatically throughout the acute phase of cerebral ischemia-reperfusion (I/R).
Conclusion: Our study suggests that differentially expressed genes SPP1, TIMP1, and ANXA1 may play a vital role in the inflammatory response during the acute phase of cerebral ischemia-reperfusion injury. These genes may be negatively regulated by miR-340-5p. Our results may provide new insights into the complex pathophysiological mechanisms of secondary inflammation after stroke.
Keywords: SPP1, AnxA1, TIMP1, miR-340-5p, ischemic stroke, acute phase
Introduction
Stroke is one of the leading causes of death and disability worldwide, which causes substantial economic and social burdens.1 Ischemic stroke is caused by insufficient blood and oxygen supply to the brain,2 accounting for about 85% of the casualties of stroke patients.3 The concept of treatment for ischemic injury and acute stroke has changed considerably over time.4 Although the latest American Heart Association/American Stroke Association Guidelines have recommended extending the endovascular treatment window to 6–16 hours or even 24 hours,5 recent data have shown that less than 10% of stroke patients benefit from this treatment. Ineffective treatment seems to be associated with a short time window, multiple exclusion criteria, hemorrhagic complications, and the highly specialized resources required for thrombectomy.6 Therefore, more and more attention has been focused on the acute phase of neuroprotective treatment within 7 d after ischemia due to its extended treatment window and low hemorrhagic complications.
Recently, brain-protective agents related to inflammatory immune factors have shown potential application prospects in basic and Phase II–III clinical studies.7 Since the concept of ischemic penumbra was introduced,8 researchers have found that changes in gene expression drive inflammation and programmed cell death in the brain after the interruption of blood supply. Changes in gene expression patterns triggered by ischemic injury cause an inflammatory cascade that develops gradually over hours to days, promoting the growth of the ischemic core toward the penumbra and extending brain tissue damage to the entire ischemic region.4,9,10 Although molecular neuroinflammation secondary to cerebral ischemia causes further damage to ischemic brain tissue, increasing evidence has shown that these events have a beneficial role in post-stroke recovery.11,12 The initial release of inflammatory signals is intended to clear infarcted tissue fragments, rebuild the extracellular matrix (ECM), and restore normal brain structure.13 Given that inflammation has a positive and negative role in the occurrence and development of cerebral nerve injury and recovery after ischemia,14 scientists have not been able to well explain the detailed inflammation process after stroke. Therefore, it is of critical importance to profoundly investigate the pattern of genetic alterations during the acute phase of stroke, identify more critical inflammation-related molecules as early as possible, and construct a precise network of these molecules to obtain new targets for intervention.
With the completion of human genome sequencing, the development of biological sciences has entered the post-genomic era. The focus of genomics research has shifted from the structure of the genome to the function of genes.15 Bioinformatics has been widely used to identify DEGs and functional pathways associated with stroke onset and progression due to its high-throughput and large sample size.16,17 However, these independent datasets are highly heterogeneous, and differences in animal species, experimental models, and duration of ischemia can lead to different results.18 Moreover, the inflammatory response in the acute phase of ischemic stroke involves a series of time-dependent events such as microglia activation, astrocyte proliferation,19,20 clearance of dead tissue by macrophages, and the involvement of various matrix molecules in scar formation.13 This sequence of inflammatory events has been shown to change over time with different molecular expression patterns. In addition, microRNAs (miRNAs) have an essential role in the inflammatory cascade of cerebral ischemia-reperfusion injury. The construction of miRNA-mRNA networks is of theoretical importance for discovering potential molecular therapeutic targets.21
The human middle cerebral artery (MCA) and its branches are commonly involved arteries in ischemic stroke (accounting for 70% of cerebral infarction), which are prone to occlusion or ischemic injury.22 The transient or permanent middle cerebral artery occlusion (MCAO) model has been verified by more than 2000 experiments and is widely used to investigate ischemic stroke in rats.22,23 It is suitable for simulating focal cerebral ischemia and is characterized by brain cell death, inflammation, and blood-brain barrier damage.
Therefore, we screened two rat stroke mRNA datasets in the GEO database, respectively, in the early and late acute stages of the stroke so as to identify essential genes involved in the progression of inflammatory response and miRNAs that may regulate the inflammatory response. In addition, we performed qRT-PCR validation of critical genes and miRNAs in combination with stable rat MCAO/R models and complete description of the temporal profile of three hub genes during the acute phase of cerebral ischemia-reperfusion, aiming to further elucidate the key molecular mechanisms of inflammation in the acute phase of stroke as a basis for designing effective therapies in the acute phase of stroke.
Materials and Methods
Acquisition of RNA Information
The microarray expression datasets GSE6161624 and GSE9753725 were acquired from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). A total of 10 samples were selected from GSE61616, including 5 Middle cerebral artery occlusion-reperfusion (MCAO/R) models and 5 Sham models (SHAM). A total of 12 samples were chosen from GSE97537, including 7 Middle cerebral artery occlusion reperfusion (MCAO/R) models and 5 Sham models (SHAM). The RNA data from the selected samples were downloaded for further analysis.
Differentially Expression Analysis
The GEO2R online analysis tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to explore the DEGs between MCAO/R and SHAM samples,26 and the adjusted P-value and |logFC| were calculated. Differential expressed genes (DEGs) were defined as adjusted P-value<0.05, and |logFC| >1. Statistical analysis was carried out for each dataset, and the intersecting part was identified using the Venn diagram webtool (bioinformatics.psb.ugent.be/webtools/Venn/).
Gene Set Enrichment Analysis (GSEA)
Gene Set Enrichment Analysis (GSEA)27 sequenced the genes according to the differential expression degree of the two groups and then detected whether the preset gene set was enriched at the top or bottom of the sequencing table All genetic information of MCAO/R and SHAM samples was uploaded to GSEA software (version 4.1.0) for further analysis.
DAVID Analysis
To reveal the functions and pathways of 170 overlapping genes, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analyses by the DAVID 6.8 online tools (https://david.ncifcrf.gov/). P<0.05 was regarded as the criterion for statistical significance.
Metascape Functional Annotation and Pathway Analysis
One hundred seventy overlapping genes were uploaded to Metascape online analysis tools (https://metascape.org/) to perform Gene Ontology (GO) analysis, including molecular function (MF), biological process (BP), and cell composition (CC) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. Min Overlap=3, P-value Cutoff=0.01, Min Enrichment=1.5.
Module Analysis and Protein–protein Interaction (PPI) Network
DEGs were inputted into the Metascape (https://metascape.org/) and STRING (https://www.string-db.org/) online database to the protein network interaction diagram. Cytoscape software (version 3.8.2) was used to perform visualization of the PPI network. One hundred seventy overlapping genes were uploaded to Cytoscape software to visualize the interaction network of the biological process by using the Cytoscape ClueGo plug‐in. Only pathways with a P value ≤0.05 were shown.
Prediction of Vital miRNAs and Construction of mRNA–miRNA Correlation Network
Target genes from DEGs were inputted into the TargetScan7.1 (http://www.targetscan.org/) online program to predict the critical miRNAs. Finally, those results were uploaded to Cytoscape software to construct of mRNA–miRNA correlation network. The selection conditions were set as the seed match > 7 mer-1A, the target gene binding region was 3′UTR, and the context++ score percentile > 90.
Middle Cerebral Artery Occlusion-Reperfusion (MCAO/R) Model
A total of 55 adult male Sprague-Dawley rats, 6–8 weeks old, weighing 280–330 g, were obtained from Zhaoyang Animal Company. All the animals were housed in an environment with a temperature of 22 ± 1 °C, relative humidity of 50 ± 1%, and a light/dark cycle of 12/12 hr. All animal studies (including the rat’s euthanasia procedure) were done in compliance with the regulations and guidelines of Soochow University institutional animal care and conducted according to the AAALAC and the IACUC guidelines.
Rats were randomly divided into the Sham group and MCAO/R group, with six rats in each group. After anesthesia, an incision was made along the midline of the neck, the common carotid artery was exposed, the external carotid artery was ligated, and the internal carotid artery was isolated. A silicon-rubber tip-coated nylon monofilament was inserted through the external carotid artery into the internal carotid artery until slight resistance was felt. The blood flow was then blocked for 2 hours, after which the monofilament was removed to restore the blood supply to the brain.28,29 The penumbra brain tissue was removed 24h/3d/5d/7d/9d after reperfusion for further experiments.
2,3,5-Triphenyltetrazolium Chloride (TTC) Staining
Twenty-four hours after ischemia-reperfusion, the rats were deeply anesthetized and perfused with PBS via the heart. The brains were quickly taken out, snap frozen at −20°C for 20 min, and cut into 2 mm thick brain slices. Tissues were then mixed with 2% TTC (Jiancheng Biotech) solution and incubated at 37°C for 15–30min in the dark.
Hematoxylin-Eosin (HE) Staining
Paraffin sections were dewaxed sequentially with xylene, anhydrous ethanol, and alcohol, stained with hematoxylin (Shanghai yuanye Bio-Technology Co., Ltd; S19007) and eosin (Aladdin, E110818), and dehydrated. Samples were then incubated with xylene, sealed with Neutral Balsam Neutral Balsam (Solarbio, G8590), and observed under the microscope.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from rat cerebral ischemic penumbra tissue or sham-operated tissue using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The total RNA concentration was then determined using a One Drop OD-1000+ spectrophotometer. Synthesis of cDNA was achieved with the RevertAid First Strand cDNA Synthesis Kit (#K1621, Thermo Scientific, USA). PCR amplification was performed using fluorescent dye SYBR™ Green PCR Master Mix (#43,091,055, ThermoFisher, USA), and primers were provided by RIBOBIO, China (Supplementary Material). Gene expression was quantified by calculating 2-ΔΔCt by normalizing the amount of RNA to tubulin content.
Statistical Analysis
Quantitative data are presented as means ± SEM, and comparisons between two groups were performed by Student’s t-test. Statistical comparisons between multiple groups were evaluated using one-way analysis of variance (ANOVA) followed by the least significant difference (LSD) test. A p-value < 0.05 was considered statistically significant.
Results
DEGs Identification
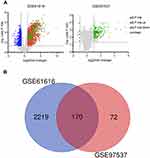
We screened two target microarrays in the GEO database to search for essential inflammatory genes in the acute phase of ischemic stroke. The GSE97537 chip includes 7 MCAO/R samples and 5 SHAM samples, and the GSE61616 chip includes 5 MCAO/R samples and 5 SHAM samples (Table 1). The sequencing information of the two data sets is from Rattus norvegicus, and both belong to the GPL1355 platform. According to the standard adjusted P-value<0.05 and |logFC| >1, 2389 DEGs were obtained from the GSE61616 data set, and 242 DEGs were obtained from the GSE97537 data set. The volcano plot was made as shown in Figure 1A. These genes were further filtered and then mapped by the Venn diagram. As shown in Figure 1B, the two sets of data share 170 overlapping genes that are differentially expressed.
 |
Table 1 Basic Information of the Selected Datasets |
The GSE97537 microarray data were obtained from rat brain tissue 24 h after ischemia-reperfusion, and the GSE61616 microarray data were obtained from rat brain tissue 7 d after ischemia-reperfusion. One hundred seventy genes were differentially expressed in the early and late acute phase, and most were upregulated (Supplementary Material), despite the 6-day time span. Unfortunately, we could not determine whether these 170 genes were consistently differentially expressed throughout the acute phase or experienced fluctuations. Still, their expression was significantly increased during the time window of 24 h-7 d after ischemia-reperfusion; thus, we speculated there might be a close association between those genes and critical events that determine injury and repair of ischemic brain tissue.
Gene Set Enrichment Analysis (GSEA)
To gain a preliminary understanding of the main roles of all genes in the two selected databases, we performed a GSEA analysis. All gene expression information in the two selected GEO data sets was uploaded to the GSEA software, and the C2, C5, and C7 gene set databases were selected to analyze gene expression profiles at the overall level. The fundamentally enriched gene sets were situated in a default cut-off as P-value < 0. 05, FDR < 0. 25. The analysis results showed that both gene sets were significantly enriched in the defense response, complement, and coagulation cascades (Figure 2A and B), which may be associated with neurovascular inflammation after stroke.
 |
Figure 2 GSEA analysis of the two GEO datasets. (A) GSEA analysis results for GSE61616. (B) GSEA analysis results for GSE97537. |
GO and KEGG Enrichment Analysis of DEGs
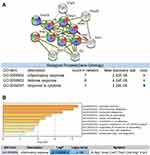
Next, we focused on these 170 intersecting differential genes from the two datasets. GO and KEGG enrichment analyses were performed for 170 overlapping genes using DAVID and Metascape online sites. The analysis results of the DAVID database are presented in a bubble plot, as shown in Figure 3A. The biological processes of the 170 overlapping genes were significantly enriched in the inflammatory response, response to lipopolysaccharide, and neutrophil chemotaxis. Cellular components were significantly enriched in the extracellular exosome, extracellular space, and cell surface. As for molecular function, changes were primarily enriched in integrin binding and chemokine activity. KEGG analyses revealed that MCMs were involved in TNF signaling pathway.
The results of the analysis using Metascape are shown in Figure 3B. These overlapping genes were mainly enriched in the inflammatory response, wound response, chemokine activity, and apoptosis-related pathways. The top 20 GO and KEGG enrichment items were classified into four functional groups: biological process (17 items), cellular component (1 item), molecular function (1 item), and KEGG pathway (1 item). The 170 overlapping genes were mainly enriched in response to wounding, inflammatory response, immune effector process, regulation of cell adhesion, regulation of cytokine production, response to lipopolysaccharide, angiogenesis, positive regulation of cell death, regulation of hemopoiesis, apoptotic signaling pathway, positive regulation of reactive oxygen species metabolic process, regeneration, acute inflammatory response, phagocytosis, negative regulation of response to external stimulus, response to interleukin-1, and response to extracellular stimulus. The cellular component that these genes were involved in was the side of the membrane. The molecular function for these genes was integrin binding.
Next, the ClueGo plug-in of Cytoscape software was further used to visualize the interaction network of biological processes enriched by 170 genes. As shown in Figure 4A and B, the pie chart shows the top 20 enriched items, which are the regulation of cytokine production (18.18%), leukocyte migration (14.77%), inflammatory response (7.73%), leukocyte chemotaxis (5.91%), response to wounding (4.55%), positive regulation of angiogenesis (3.86%), leukocyte differentiation (3.64%), macrophage migration (3.41%), tube morphogenesis (3.18%), granulocyte migration (2.95%), myeloid leukocyte migration (2.73%), response to lipopolysaccharide (2.73%), regulation of wound healing (2.50%), positive regulation of tumor necrosis factor superfamily cytokine production (2.05%), blood coagulation (2.05%), leukocyte activation involved in immune response (1.82%), regulation of leukocyte activation (1.82%), positive regulation of tumor necrosis factor production (1.82%), extrinsic apoptotic signaling pathway (1.82%), and cellular response to interleukin-1 (1.82%). Most of these modules are highly associated with inflammatory cell activation and inflammatory processes.
Construction of Protein–Protein Interaction (PPI) Network to Mine Gene Clusters Related to the Inflammatory Response
To dig out core genes from DEGs, we uploaded the 170 intersected genes to Metascape for further analysis, obtaining seven gene clusters (Figure 5A and B). Among them, the red cluster contained the most significant number of genes, with a total of 13 genes involved in the biological processes of cytokine activity (log10(p) = −8.7), inflammatory response (log10(p) = −8.1), and receptor-ligand activity (log10(p) = −8), which are highly correlated with inflammation. The 13 genes were TIMP1, IL6, KNG1, ANXA1, SPP1, FBN1, CCL6, S1PR3, CP, PRSS23, CXCL16, CXCL1, and P2RY12. The second-largest cluster is the blue cluster, with a total of ten genes that are highly correlated with immune response. The remaining small clusters are mainly related to immunity and calcium ion activity. In this study, we selected the largest cluster associated with inflammation for subsequent analysis.
Screening of Genes Associated with the Inflammatory Response
Thirteen genes highly associated with inflammation in the red cluster were uploaded to STRING (Figure 6A) and Metascape (Figure 6B) for further analysis. In the visible protein–protein interaction network, the genes involved in the inflammatory response contain red patches, and there are a total of seven (FDR=2.42E-06): AnxA1, Ccl6, Cxcl1, Il6, Kng1, SPP1, and TIMP1. In GO and KEGG enrichment analysis, eight genes were found to be enriched in the inflammatory response pathway (LogP=−8.115286814): AnxA1, Ccl6, Cxcl1, Il6, Kng1, SPP1, TIMP1, and S1pr3. Except for S1pr3, the remaining seven genes overlapped in the results obtained by the two analysis methods.
Further miRNA Mining and Interaction Network Analysis
The aforementioned analysis software yielded seven intersection genes highly related to inflammatory response: AnxA1, Ccl6, Cxcl1, Il6, Kng1, SPP1, and TIMP1. To obtain the miRNAs that may regulate these genes and further understand the pathophysiological roles, gene-miRNAs analysis was performed using TargetScan7.1 software. The selection conditions were set as the seed match > 7 mer-1A, the target gene binding region was 3′UTR, and the context++ score percentile > 90. Cytoscape was used to draw the interaction network, as shown in Figure 7. Our results suggested that miR-340-5p simultaneously regulates the three candidate target genes of AnxA1, SPP1, and TIMP1, which may jointly participate in inflammatory response after cerebral ischemia.
 |
Figure 7 miRNA Prediction with Target Scan. Interaction network between genes involved in the inflammatory response and its targeted miRNAs. Genes are colored in red, and miRNAs are colored in pink. |
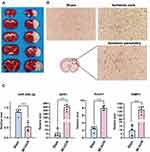
Validation of Critical Genes and miRNAs Highly Associated with Inflammation in a Rat MCAO/R Model
To verify the association of miR-340-5p with AnxA1/SPP/TIMP1, we constructed a rat MCAO/R model. The brain was collected 24 h after ischemia-reperfusion, and TTC staining and HE staining confirmed the operation’s success (Figure 8A and B). We used qRT-PCR to detect the expression levels of candidate genes and miRNAs in the rat ischemic penumbra and the corresponding control group. The experimental results showed that the expression of miR-340-5p was significantly decreased at 24h after ischemia-reperfusion (p-value<0.0001, Figure 8C), while the expression of AnxA1, SPP1, and TIMP1 increased (p-value<0.0001, Figure 8C). These results suggest that miR-340-5p may negatively regulate three post-ischemic upregulated genes: SPP1, AnxA1, and TIMP1, and have an essential role in the inflammatory response in the early post-stroke acute phase.
Temporal Expression of SPP1/AnxA1/TIMP1 mRNA in Rat Ischemic Penumbra After MCAO/R
To provide insights for future anti-inflammatory strategies targeting the acute phase of ischemic brain injury, we further elucidated the temporal dynamics of three hub genes. The expression levels of SPP1/TIMP1/AnxA1 mRNA in the ischemic penumbra of rats were continuously measured from 24 hours after I/R to 9 days. Our results showed that SPP1/TIMP1/AnxA1 exhibited significantly different time courses (Figure 9). SPP1 reached a small peak at 24 h and then gradually decreased, followed by a gradual increase and a peak at 7 d (Figure 9A). Timp1 reached a peak at 24 h; it then continued to fluctuate until a second small peak at 7 d (Figure 9B). Anxa1 increased gradually over 9 d, except for a slight drop back on day 7 (Figure 9C). Our data show that in the complex network of inflammation and brain injury after ischemic stroke, the expression of genes highly associated with inflammation fluctuates wildly, indicating that the inflammatory response has changed dramatically over time and may involve critical pathologic mechanisms.
Discussion
Stroke is now the second leading cause of death globally, responsible for a significant proportion of disability-adjusted life years.30 Neuroinflammation that develops constantly has a vital role in many pathologic processes in the acute phase of ischemic stroke.31–33 However, the specific mechanisms of inflammation and the changes in gene patterns are still not fully uderstood.19 This study obtained seven critical genes involved in neuroimmune activation/inflammation after cerebral ischemia by screening and enrichment analysis of gene expression profiles of rat cerebral ischemia-related datasets. We combined bioinformatics and animal experiments to further confirm three genes, SPP1, AnxA1, and TIMP1, which are upregulated after I/R and continue to fluctuate throughout the acute phase, and an essential downregulated microRNA miR-340-5p, which can theoretically regulate them simultaneously. Our results may provide new directions for elucidating the molecular mechanisms of inflammation after stroke.
MicroRNAs (miRNAs) are small single-stranded non-coding RNA molecules that can silence the expression of target genes by binding to the 3’-untranslated regions (3ʹUTRs) of target mRNA.34 In vitro and in vivo studies have shown that miRNAs can inhibit inflammation and oxidative stress and prevent excitotoxic injury after stroke.35–37 Our results confirmed that miR-340-5p expression was decreased in periinfarct brain tissues of rats 24 h after ischemia-reperfusion, which is similar to previous findings. For instance, Yoo et al reported that the expression level of miR-340-5p was decreased in peripheral blood circulation in patients with acute ischemic stroke.38 Moreover, a study of rat hippocampal neurons exposed to oxygen and sugar deprivation/reoxygenation (OGD/R) in vitro found that miR-340-5p is down-regulated in injured primary hippocampal neurons; overexpression of miR-340-5p reduced OGD/ R-induced cellular inflammation, and TNF-α, IL-1β, and other levels were lower than those of the control group.39 Unlike previous studies, we proposed three new candidate target genes for miR-340-5p. Our study further confirmed that miR-340-5p is a promising target in the acute phase of cerebral ischemia-reperfusion injury. We also associated this target with more genes and focused on their mutual regulation in the inflammatory process. These results may provide new insights into the complex mechanisms of post-ischemic inflammation.
SPP1 (encoding Osteopontin, OPN) is a possible target gene of miR-340-5p. We found that SPP1 is significantly upregulated in the ischemic penumbra after temporarily blocking the middle cerebral artery in rats, which was consistent with the report of Wang et al.40 SPP1 is a secreted multifunctional acidic phosphate protein,41 involved in various biological and disease processes, including inflammation, immune response, vascular remodeling, wound healing, bone turnover, etc.42–44 But the pathophysiological significance of its upregulation after cerebral ischemia is still controversial. For example, Carbone et al showed that serum OPN levels peaked on the 7th day after stroke and were positively correlated with cerebral infarction volume and poor neurological score.45 Notably, some recent work strongly supports OPN as an effective neuroprotective agent against ischemic injury. The ischemic challenge of cortical cells (120-minute OGD) followed by incubation with OPN for 24 h reduces cell death due to hypoxia and glucose deprivation. Our time-course data showed two peak expressions of SPP1 at 24 h and 7 d after I/R, which may explain the previous contradictory results suggesting that the role of OPN changes significantly with the disease course. OPN may exert a cerebral protective impact early in the acute phase of the ischemic stroke while mediating damage in the late phase. Of course, the different effects may also be caused by varying levels of upregulation, where low levels of upregulation may mediate repair, whereas a gradual increase with a peak after 7 d may lead to infarct core expansion and poor prognosis. In addition, according to the bioinformatics data obtained in this study, OPN was significantly upregulated in both the early and late acute phase of stroke, with the levels at 7d (|logFC|=9.74404, P-value=1.03E-05) appearing to be higher than at 24h (|logFC|=2.66, P-value=2.28E-03), which is consistent with our experimental results, and provides further evidence that it may be a key molecule in the inflammatory process. However, due to the multiple roles and subtypes of OPN in the nervous system, more data are needed to elucidate the complex role in ischemic stroke.
TIMP1 (Tissue inhibitor metalloproteinase-1) is another target candidate gene of miR-340-5p. Experimental evidence from histological sections of human specimens shows that infarcted brain tissue has a higher concentration of TIMP1 compared with healthy brain tissues,46 which is consistent with our findings in rats. TIMP1 belongs to the TIMP family, which codes for proteins that are endogenous inhibitors of matrix metalloproteinases (MMPs) and can cause irreversible inactivation of most known MMPs. MMPs are a group of peptidases involved in extracellular matrix degradation.47 The latest large prospective study of patients with acute ischemic stroke demonstrates that elevated TIMP1 levels are strongly and independently associated with an increased risk of severe disability and death after stroke.47 Furthermore, our study confirmed that TIMP1 is consistently highly expressed throughout the acute period of I/R and shows large fluctuations at 24h and 7d, suggesting that TIMP1 may be involved in the occurrence and development of inflammation in the acute stage of post-stroke as a critical gene.
According to our data, the third gene identified by our team that may interact with miR-340-5p is annexin A1 (AnxA1), which is upregulated in the ischemic penumbra of rats. Interestingly, Senchenkova et al showed that the plasma levels of AnxA1 decrease after stroke in humans and mice.48 However, there are more data in support of our conclusion. For example, Datta et al compared infarcts in three different brain regions and position-matched control samples, finding that anti-inflamma-related AnxA1 levels showed an upward trend.49 In recent years, research on AnxA1 has focused on its subcellular location. Many reports indicated that the extracellular AnxA1 is involved in regulating inflammation, while its nuclear translocation induces neuronal apoptosis after stroke.50,51 Our study confirmed that AnxA1 is significantly upregulated in the acute phase of ischemic stroke and may be regulated by miR-340-5p. Overall, AnxA1 is a highly related protein to the inflammatory process and has potential application value.
It is well known that one miRNA can bind to multiple target genes.38 Although miR-340-5p has been confirmed to negatively regulate the expression of several mRNAs, its main target genes remain to be discovered.39,52 In this study, we identified three miR-340-5p candidate target genes, ie, SPP1, AnxA1, and TIMP1, which were differentially expressed in the rat cerebral ischemic penumbra using bioinformatics methods and made preliminary verifications, providing a new promising target for studying the inflammatory mechanism of neuronal damage after stroke.
Interestingly, we noticed that there might be some connection between the three candidate genes selected. The characteristics of the inflammatory response after ischemic stroke have been widely described,14,19,53 but the initial purpose of its occurrence is to play a protective role,13 which has become a new focus of scholars. Matrix proteases, extracellular matrix molecules, and integrins are the three key molecules that initiate the classic wound healing response. Both OPN and TIMP1 are involved in the degradation and remodeling of ECM after stroke. For example, as an integrin ligand, OPN can interact with various specific integrin receptors through its arginine, glycine, asylate domain (RGD) and mediate the repair of brain tissue damage. The matrix protease inhibitor TIMP1 can regulate excessive proteolytic activity and maintain ECM homeostasis in the acute stage of focal brain injury.47 According to our data, SPP1 and TIMP1 fluctuate to peak at the same time points, which may reflect their synergistic effects. AnxA1 can regulate cell adhesion and migration and interact with cytoskeletal proteins.54 In addition, all three genes are involved in the occurrence and development of atherosclerosis.47,55,56 Studies have also found that the neuroprotective effect mediated by OPN is the same as miR-340-5p, which can be activated through the PI3K/Akt signaling pathway, while PI3K inhibitors can block the protective effect of OPN.57
The dual roles of the above genes have plagued scholars for obvious reasons as they are interconnected and constantly changing in the inflammatory network of ischemia. Herein, we constructed the interaction network of Spp1, Anxa1, and Timp1 and comprehensively analyzed their fluctuations during the acute phase of I/R. Our study confirmed that all three screened genes were significantly upregulated from 24h to 7d after ischemia-reperfusion, and it is reasonable to speculate that the course stage they are in or the changes in their levels may influence their effects. MiR-340-5p was deduced from our screened genes. As bioinformatics analysis revealed it regulates three of our seven critical genes, we believe that it has a pivotal role in regulating inflammation during the acute phase of stroke. Further animal experiments confirmed that miR-340-5p was significantly down-regulated, and Spp1, Anxa1, and Timp1 were upregulated 24 hours after cerebral ischemia-reperfusion.
Our results point toward an apparent new direction. Future studies should focus on how changes in Spp1, Anxa1, and Timp1 levels during acute ischemic stroke affect their function. For example, how the different environmental and temporal processes of the inflammatory cascade determine whether they lead to injury or mediate recovery. The network of interactions among them and with miRNAs should also receive more attention in the future.
This study has some limitations. All the data we analyzed came from online databases and from rodents. Further research requires more samples and human tissue data to support our results. Secondly, we did not directly verify the relationship between SPP1, TIMP1, AnxA1, and miR-340-5p. Their potential connection mechanism in ischemic brain injury also requires more in-depth exploration.
Conclusion
Our study suggests that differentially expressed genes SPP1, TIMP1, and ANXA1 may have a vital role in the inflammatory response during the acute phase of ischemic stroke. These genes may be negatively regulated by miR-340-5p. Our results may provide new insights into the complex pathophysiological mechanisms of secondary inflammation after stroke.
Data Sharing Statement
- The datasets analyzed during the current study are available in the GEO repository, [https://www.ncbi.nlm.nih.gov/geo/].24,25
- The GEO2R online analysis tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/).
- Venn diagram webtool (bioinformatics.psb.ugent.be/webtools/Venn/).
- GSEA software (version 4.1.0).
- DAVID 6.8 online tools(https://david.ncifcrf.gov/).
- Metascape online analysis tools (https://metascape.org/).
- STRING (https://www.string-db.org/).
- Cytoscape software (version 3.8.2).
- TargetScan7.1 (http://www.targetscan.org/).
Ethics Approval and Consent to Participate
The experimental protocols involving animals were approved by the Animal Care and Use Committee of Soochow University. All animal use, care, and operative procedures complied with the Guide for the Care and Use of Laboratory Animals by the National Institution of Health.
Acknowledgments
The authors thank the Department of Neurosurgery and Brain and Nerve Research Laboratory for providing the experimental platform.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work is supported by the Jiangsu Province Key Research and Development Program(Social Development)(BE2019666).
Disclosure
The authors declare that they have no competing interests.
References
1. Kim J, Thayabaranathan T, Donnan GA, et al. Global stroke statistics 2019. Int J Stroke. 2020;15(8):819–838. doi:10.1177/1747493020909545
2. Hui C, Tadi P, Patti L. Ischemic stroke. In: StatPearls. Treasure Island (FL); 2021.
3. Musuka TD, Wilton SB, Traboulsi M, Hill MD. Diagnosis and management of acute ischemic stroke: speed is critical. Can Med Assoc J. 2015;187(12):887–893. doi:10.1503/cmaj.140355
4. Iadecola C, Buckwalter MS, Anrather J. Immune responses to stroke: mechanisms, modulation, and therapeutic potential. J Clin Invest. 2020;130(6):2777–2788. doi:10.1172/JCI135530
5. Powers WJ, Rabinstein AA, Ackerson T, et al. Guidelines for the early management of patients with acute ischemic stroke: 2019 update to the 2018 guidelines for the early management of acute ischemic stroke: a guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2019;50(12):e344–e418. doi:10.1161/STR.0000000000000211
6. Rinaldo L, Rabinstein AA, Cloft H, Knudsen JM, Castilla LR, Brinjikji W. Racial and Ethnic Disparities in the Utilization of Thrombectomy for acute stroke. Stroke. 2019;50(9):2428–2432. doi:10.1161/STROKEAHA.118.024651
7. Woodruff TM, Thundyil J, Tang SC, Sobey CG, Taylor SM, Arumugam TV. Pathophysiology, treatment, and animal and cellular models of human ischemic stroke. Mol Neurodegener. 2011;6(1):11. doi:10.1186/1750-1326-6-11
8. Astrup J, Siesjö BK, Symon L. Thresholds in cerebral ischemia - The ischemic penumbra. Stroke. 1981;12(6):723–725. doi:10.1161/01.STR.12.6.723
9. Moskowitz MA, Lo EH, Iadecola C. The science of stroke: mechanisms in search of treatments. Neuron. 2010;67(2):181–198. doi:10.1016/j.neuron.2010.07.002
10. Weinstein PR, Hong S, Sharp FR. Molecular identification of the ischemic penumbra. Stroke. 2004;35(11 Suppl 1):2666–2670. doi:10.1161/01.STR.0000144052.10644.ed
11. Hu XM, Li PY, Guo YL, Wang HY, Gao YQ, Chen J. Microglia/macrophage polarization dynamics reveal novel mechanism of injury expansion after focal cerebral Ischemia. Stroke. 2013;44:2. doi:10.1161/str.44.suppl_1.AWP98
12. Liu ZW, Li Y, Cui YS, et al. Beneficial effects of gfap/vimentin reactive astrocytes for axonal remodeling and motor behavioral recovery in mice after stroke. Glia. 2014;62(12):2022–2033. doi:10.1002/glia.22723
13. Ellison JA, Barone FC, Feuerstein GZ. Matrix remodeling after stroke - De Novo expression of matrix proteins and integrin receptors. Ann Ny Acad Sci. 1999;890:204–222. doi:10.1111/j.1749-6632.1999.tb07996.x
14. Maida CD, Norrito RL, Daidone M, Tuttolomondo A, Pinto A. Neuroinflammatory mechanisms in ischemic stroke: focus on cardioembolic stroke, background, and therapeutic approaches. Int J Mol Sci. 2020;21:18. doi:10.3390/ijms21186454
15. Del Giacco L, Cattaneo C. Introduction to genomics. Methods in Molecular Biology. 2012;823:79–88.
16. Chen G, Li L, Tao H. Bioinformatics identification of ferroptosis-related biomarkers and therapeutic compounds in ischemic Stroke. Front Neurol. 2021;12:745240. doi:10.3389/fneur.2021.745240
17. Deng GX, Xu N, Huang Q, et al. Association between promoter DNA methylation and gene expression in the pathogenesis of ischemic stroke. Aging (Albany NY). 2019;11(18):7663–7677. doi:10.18632/aging.102278
18. Xiao X, Bai P, Cao S, et al. Integrated bioinformatics analysis for the identification of key molecules and pathways in the hippocampus of rats after traumatic brain injury. Neurochem Res. 2020;45(4):928–939. doi:10.1007/s11064-020-02973-9
19. Pluta R, Januszewski S, Czuczwar SJ. Neuroinflammation in post-ischemic neurodegeneration of the brain: friend, foe, or both? Int J Mol Sci. 2021;22:9. doi:10.3390/ijms22094405
20. Radenovic L, Nenadic M, Ulamek-Koziol M, et al. Heterogeneity in brain distribution of activated microglia and astrocytes in a rat ischemic model of Alzheimer’s disease after 2 years of survival. Aging (Albany NY). 2020;12(12):12251–12267. doi:10.18632/aging.103411
21. Li G, Morris-Blanco KC, Lopez MS, et al. Impact of microRNAs on ischemic stroke: from pre- to post-disease. Prog Neurobiol. 2018;163–164:59–78. doi:10.1016/j.pneurobio.2017.08.002
22. Howells DW, Porritt MJ, Rewell SSJ, et al. Different strokes for different folks: the rich diversity of animal models of focal cerebral ischemia. J Cerebr Blood F Met. 2010;30(8):1412–1431. doi:10.1038/jcbfm.2010.66
23. Fluri F, Schuhmann MK, Kleinschnitz C. Animal models of ischemic stroke and their application in clinical research. Drug Des Dev Ther. 2015;9:3445–3454.
24. Wang L, Yu Y, Yang J, Zhao X, Dissecting Xuesaitong’s LZ. mechanisms on preventing stroke based on the microarray and connectivity map. Mol Biosyst. 2015;11(11):3033–3039. doi:10.1039/C5MB00379B
25. Takuma A, Saito Y, Abe A, Asakura T, Abe K. Expression data from MCAO or Sham operated rat brain [mRNA]; 2018.
26. Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi:10.1093/bioinformatics/btm254
27. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
28. Sommer CJ. Ischemic stroke: experimental models and reality. Acta Neuropathol. 2017;133(2):245–261. doi:10.1007/s00401-017-1667-0
29. Longa EZ, Weinstein PR, Carlson S, Cummins R. Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke. 1989;20(1):84–91. doi:10.1161/01.STR.20.1.84
30. Lo JW, Crawford JD, Desmond DW, et al. profile of and risk factors for poststroke cognitive impairment in diverse ethnoregional groups. Neurology. 2019;93(24):e2257–e2271. doi:10.1212/WNL.0000000000008612
31. Jayaraj RL, Azimullah S, Beiram R, Jalal FY, Rosenberg GA. Neuroinflammation: friend and foe for ischemic stroke. J Neuroinflamm. 2019;16:1–24.
32. Iadecola C, Anrather J. The immunology of stroke: from mechanisms to translation. Nat Med. 2011;17(7):796–808. doi:10.1038/nm.2399
33. Jin R, Liu L, Zhang S, Nanda A, Li G. Role of inflammation and its mediators in acute ischemic stroke. J Cardiovasc Transl Res. 2013;6(5):834–851. doi:10.1007/s12265-013-9508-6
34. Huang R, Ma J, Niu B, et al. MiR-34b protects against focal cerebral Ischemia-Reperfusion (I/R) injury in rat by targeting keap1. J Stroke Cerebrovasc. 2019;28(1):1–9. doi:10.1016/j.jstrokecerebrovasdis.2018.08.023
35. Yin KJ, Deng Z, Hamblin M, et al. Peroxisome proliferator-activated receptor delta regulation of miR-15a in Ischemia-induced cerebral vascular endothelial injury. J Neurosci. 2010;30(18):6398–6408. doi:10.1523/JNEUROSCI.0780-10.2010
36. Liu P, Zhao HP, Wang RL, et al. MicroRNA-424 protects against focal cerebral ischemia and reperfusion injury in mice by suppressing oxidative stress. Stroke. 2015;46(2):513–519. doi:10.1161/STROKEAHA.114.007482
37. Xu L-J, Ouyang Y-B, Xiong XX, Stary CM, Giffard RG. Post-stroke treatment with miR-181 antagomir reduces injury and improves long-term behavioral recovery in mice after focal cerebral ischemia. Exp Neurol. 2015;264:1–7. doi:10.1016/j.expneurol.2014.11.007
38. Yoo H, Kim J, Lee AR, et al. Alteration of microRNA 340-5p and Arginase-1 expression in peripheral blood cells during acute ischemic stroke. Mol Neurobiol. 2019;56(5):3211–3221. doi:10.1007/s12035-018-1295-2
39. Wang J, Liu GZ. Protective effect of microRNA-340-5p against oxygen-glucose deprivation/reperfusion in PC12 cells through targeting neuronal differentiation 4. Mol Med Rep. 2020;22(2):964–974. doi:10.3892/mmr.2020.11174
40. Wang X, Louden C, Yue TL, et al. Delayed expression of osteopontin after focal stroke in the rat. J Neurosci. 1998;18(6):2075–2083. doi:10.1523/JNEUROSCI.18-06-02075.1998
41. Doyle KP, Yang T, Lessov NS, et al. Nasal administration of osteopontin peptide mimetics confers neuroprotection in stroke. J Cereb Blood Flow Metab. 2008;28(6):1235–1248. doi:10.1038/jcbfm.2008.17
42. Jing M, Li B, Hou X, et al. OPN gene polymorphism and the serum OPN levels confer the susceptibility and prognosis of ischemic stroke in Chinese patients. Cell Physiol Biochem. 2013;32(6):1798–1807. doi:10.1159/000356613
43. Giachelli CM, Steitz S. Osteopontin: a versatile regulator of inflammation and biomineralization. Matrix Biol. 2000;19(7):615–622. doi:10.1016/S0945-053X(00)00108-6
44. Berglund LM, Lyssenko V, Ladenvall C, et al. Glucose-dependent insulinotropic polypeptide stimulates osteopontin expression in the vasculature via endothelin-1 and CREB. Diabetes. 2016;65(1):239–254. doi:10.2337/db15-0122
45. Carbone F, Vuilleumier N, Burger F, et al. Serum osteopontin levels are upregulated and predict disability after an ischaemic stroke. Eur J Clin Invest. 2015;45(6):579–586. doi:10.1111/eci.12446
46. Cuadrado E, Rosell A, Penalba A, et al. Vascular MMP-9/TIMP-2 and neuronal MMP-10 up-regulation in human brain after stroke: a combined laser microdissection and protein Array Study. J Proteome Res. 2009;8(6):3191–3197. doi:10.1021/pr801012x
47. Zhong. Tissue inhibitor metalloproteinase-1 and clinical outcomes after acute ischemic stroke (vol 93, pg e1675, 2019). Neurology. 2019;93(18):819.
48. Senchenkova EY, Ansari J, Becker F. Novel role for the AnxA1-Fpr2/ALX signaling axis as a key regulator of platelet function to Promote Resolution of Inflammation. Circulation. 2019;1404:E172–E172.
49. Datta A, Akatsu H, Heese K, Sze SK. Quantitative clinical proteomic study of autopsied human infarcted brain specimens to elucidate the deregulated pathways in ischemic stroke pathology. J Proteomics. 2013;91:556–568. doi:10.1016/j.jprot.2013.08.017
50. Li X, Zheng L, Xia Q, et al. A novel cell-penetrating peptide protects against neuron apoptosis after cerebral ischemia by inhibiting the nuclear translocation of annexin A1. Cell Death Differ. 2019;26(2):260–275. doi:10.1038/s41418-018-0116-5
51. Xia Q, Li X, Zhou HJ, Zheng L, Shi J. S100A11 protects against neuronal cell apoptosis induced by cerebral ischemia via inhibiting the nuclear translocation of annexin A1. Cell Death Dis. 2018;9:1–7.
52. Zheng YK, Zhao P, Lian YJ, Li S, Chen Y, Li LH. MiR-340-5p alleviates oxygen-glucose deprivation/reoxygenation-induced neuronal injury via PI3K/Akt activation by targeting PDCD4. Neurochem Int. 2020;134:104650.
53. Del Zoppo GJ. The neurovascular unit in the setting of stroke. J Intern Med. 2010;267(2):156–171. doi:10.1111/j.1365-2796.2009.02199.x
54. Cristante E, McArthur S, Mauro C, et al. Identification of an essential endogenous regulator of blood-brain barrier integrity, and its pathological and therapeutic implications. P Natl Acad Sci USA. 2013;110(3):832–841. doi:10.1073/pnas.1209362110
55. Shen X, Zhang S, Guo Z, Xing DM, Chen WJ. The crosstalk of ABCA1 and ANXA1: a potential mechanism for protection against atherosclerosis. Mol Med. 2020;26:1. doi:10.1186/s10020-020-00213-y
56. Icer MA, Gezmen-Karadag M. The multiple functions and mechanisms of osteopontin. Clin Biochem. 2018;59:17–24. doi:10.1016/j.clinbiochem.2018.07.003
57. Meller R, Stevens SL, Minami M, et al. Neuroprotection by osteopontin in stroke. J Cerebr Blood F Met. 2005;25(2):217–225. doi:10.1038/sj.jcbfm.9600022
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.