Back to Journals » OncoTargets and Therapy » Volume 13
SNHG3 Knockdown Suppresses Proliferation, Migration and Invasion, and Promotes Apoptosis in Non-Small Cell Lung Cancer Through Regulating miR-216a/ZEB1 Axis
Authors Zhao S, Gao X, Zhong C, Li Y , Wang M, Zang S
Received 1 June 2020
Accepted for publication 24 August 2020
Published 4 November 2020 Volume 2020:13 Pages 11327—11336
DOI https://doi.org/10.2147/OTT.S263637
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr William C. Cho
Shasha Zhao, Xinyuan Gao, Chunlei Zhong, Yunxia Li, Ming Wang, Shuzhi Zang
Department of Respiratory Medicine, The First Hospital Affiliated to the Xinxiang Medical College, Weihui, Henan 453100, People’s Republic of China
Correspondence: Shasha Zhao Department of Respiratory Medicine
The First Hospital Affiliated to the Xinxiang Medical College, No. 88 Jiankang Road, Weihui, Henan 453100, People’s Republic of China
Tel +86-0373-4403114
Email [email protected]
Background: Tumour growth and development are dependent on many factors including long noncoding RNAs (lncRNAs). However, limited information is available on the involvement of lncRNAs in non-small cell lung cancer (NSCLC) and the molecular mechanisms have not been defined. Here, we examined the expression of small nucleolar RNA host gene 3 (SNHG3) and its contribution to the development of NSCLC.
Methods: We detected SNHG3, miR-216a, and ZEB1 expression in tissues from NSCLC patients and lung adenocarcinoma cell lines using quantitative real-time polymerase chain reaction. Proliferation, migrations, invasion, and apoptosis of tumour cells were assessed using cell counting kit-8, transwell experiments, and flow cytometry after SNHG3 knockdown by small interfering RNAs. Bioinformatics and luciferase reporter assays were employed for analysing the interactions between SNHG3, miR-216a, and ZEB1.
Results: We found highly upregulated SNHG3 in tissues and cells from NSCLC patients, which was linked to poor prognosis. SNHG3 silencing diminished the ability of NSCLC cells to proliferate, migrate, and invade and promoted apoptosis. Furthermore, SNHG3 competed with endogenous RNA and enhanced the expression of ZEB1 by interfering with miR-216a. ZEB1 overexpression or miR-216a blockade reversed SNHG3-induced tumour inhibition. Similar effects were observed in vivo where SNHG3 knockdown inhibited NSCLC tumour growth by reducing expression of miR-216a while increasing that of ZEB1.
Conclusion: Knockdown of SNHG3 inhibits NSCLC tumour development and progression by upregulation of ZEB1 and interference with miR-216a, revealing an attractive alternative target for patients with NSCLC.
Keywords: lncRNA SNHG3, competing endogenous RNA, NSCLC, tumour proliferation, tumour migration, tumour invasion, tumour apoptosis
Introduction
Cancer mortality across the world can be mainly attributed to non-small cell lung cancer (NSCLC),1 which has a 5-year survival of only 16.6%.2 Despite the discovery of several oncogenes and tumour suppressive genes related to NSCLC,3,4 the precise pathogenesis and the mechanisms of action are undefined. Therefore, the identification of novel molecules and pathways that contribute to tumour development is important for further advancements in diagnostics and treatment strategies for patients with NSCLC to improve outcome.
Long non-coding RNAs (lncRNAs) contain over 200 nucleotides without protein-coding functions and serve as a principal regulator of various cellular processes by regulating gene expression.5 lncRNAs not only directly regulate the expression of genes but also interact with microRNAs (miRNAs), which are 21–24 nucleotides-long sequences of non-coding RNA that trigger mRNA degradation and translational inhibition by interacting with the 3ʹ-untranslated region (UTR).7 These interactions counter the action of miRNAs on the target mRNAs.6 In recent years, multiple studies have demonstrated the competitive endogenous RNA (ceRNA) function of lncRNAs, which participate in regulating chemoresistance and tumour progression.8 For example, LINC00665 has been demonstrated to interfere with miR-138-5p and increase E2F3 expression, which decreases NSCLC cell proliferation and invasion.9 The lncRNA small nucleolar RNA host gene 3 (SNHG3) plays a role in various cancers and studies have demonstrated the function of SNHG3 as a ceRNA and its association with tumour progression and poor outcome. SNHG3 expression is significantly increased in patients with colorectal cancer and SNHG3 silencing results in upregulation of runt-related transcription factor 2 (RUNX2) expression, which subsequently inhibits tumour cell growth and metastasis.10 Similar effects have been shown in papillary thyroid carcinoma where SNHG3 silencing dampened cell migration, invasion, proliferation, and colony formation by interfering with miR-214-3p, leading to upregulation of PSMD10.11 Furthermore, SNHG3 has recently been discovered to promote proliferation of NSCLC tumour cells and acts as an oncogene,12 although its precise role and mechanisms of action remain unclear.
Here, we observed the remarkable upregulation of SNHG3 expression in tissues from NSCLC patients and lung adenocarcinoma cell lines. Moreover, knockdown of SNHG3 prevented tumour cells from proliferating, migrating, and invading and induced apoptosis. Furthermore, SNHG3 competed with miR-216a, leading to enhanced ZEB1 expression and downstream oncogenic effects. Our data further clarified the role of lncRNAs and highlighted a potential new treatment strategy for NSCLC patients.
Methods
NSCLC Tissue Collection and Cell Culture
NSCLC tissue specimens were acquired from 42 patients in The First Hospital Affiliated to The Xinxiang Medical College, China. Liquid nitrogen was used to snap-freeze the tissues, which were stored at −80°C. Informed consent was given by all study participants before surgery and all procedures were reviewed and authorised by the institute’s ethics committee (2017–37). The lung cancer cell lines A549, H322, H1299, GLC-82, and SPC-A1 (ATCC, USA) and normal bronchial epithelial cell line NBE (NBE-3, CRL-6254; ATCC, USA) were expanded in RPMI-1640 medium supplemented with 10% foetal bovine serum and 1% penicillin/streptomycin.
Quantitative Real-Time Polymerase Chain Reaction
We isolated total RNA using TRIzol reagent (Invitrogen, USA) and generated cDNA by M-MLV Reverse Transcriptase (Invitrogen, USA). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed with an ABI Prism 7900 using VeriQuest Fast SYBR Green qPCR Master Mix (Thermo Fisher Scientific, USA). 2−∆∆Ct analysis was performed to obtain gene expression and normalised to GAPDH or U6. Normalised lung adenocarcinoma and lung squamous cell carcinoma RNA-seq data were obtained from the database of The Cancer Genome Atlas (TCGA).
Cell Transfection
Small interfering RNA targeting SNHG3 (si-SNHG3 #1, 5ʹ-GGGAGAGUAGGUAAACUGA-3ʹ; si-SNHG3 #2, 5ʹ-UGGUACUGUU GAAGGAAAC-3ʹ; SNHG3 #3, 5ʹ-GCUAAGAGGAGUGAGGCAG-3ʹ), si-control (5ʹ-TTCTCCGAACGTGTCACGT-3ʹ), miR-216a (5ʹ-UAAUCUCAGCUGGCAAC UGUGA-3ʹ), miR-control (5ʹ-CCGACUGCAGUCGC-3ʹ), and anti-miR-216a (5ʹ-GCAGCGCCTGTGAGAGGGAT GAAAA-3ʹ) were acquired from GenePharma (China). The sequences of SNHG3 (NR_036473.1) were amplified with primers (Forward 5ʹ-TGACGGAGTCGGTTTGTCACTC-3ʹ; Reverse 5ʹ-ACGAATGGGGCTGA CTCATCTG-3ʹ) and incorporated into the pcDNA-3.1 vector (Invitrogen, USA) to create pcDNA-SNHG3 (SNHG3). Cells were transfected using Lipofectamine 2000 (Invitrogen, USA).
Cell Proliferation
We performed cell counting kit-8 assay (CCK-8) on the A549 and H1299 cell lines. Briefly, the transfected cells were inoculated into 96-well culture plates. Subsequently, 10 μL of CCK-8 solution was added into each well of the plate. Finally, the absorbance at 450 nm at 0, 24, 48, 72 and 96 h was measured to assess the proliferation and viability of the cells.
Transwell Migration and Invasion
Migration and invasion of tumour cells and cell lines were assessed by transwell chambers (Millipore, USA) as described previously.13 Migration and invasion cell numbers were counted from five randomly selected microscopic fields.
Flow Cytometry
Annexin V-FITC/PI kit (BD PharMingen, USA) was used to stain transfected cells and flow cytometry was employed to assess apoptosis.
Luciferase Reporter Assay
Wildtype and mutant reporter plasmids of SNHG3 (SNHG3-WT and SNHG3-MUT, respectively) and ZEB1 3ʹ UTR were individually synthesised by GenePharma (China) and were transfected into A549 and H1299 cells in conjunction with miR-control or miR-216a. We assessed the luciferase activity after 48 hours by Dual-Luciferase Reporter Assay (Promega, USA).
In vivo Tumour Xenograft Model
BALB/c nude mice were supplied by Vital River Laboratories (China) and all procedures and protocols were approved by the institutional animal ethics committee of the First Hospital Affiliated to The Xinxiang Medical College (2019–21) and performed in strict accordance with the guiding principles of the institutional animal management committee of the First Hospital Affiliated to The Xinxiang Medical College. A549 cells were infected with recombinant lentivirus carrying SNHG3 (Lenti-sh-SNHG3, Hanbio, China) for SNHG3 overexpression or empty vector (Lenti-sh-control, Hanbio, China) as control, which were intraperitoneally injected in the mice. Tumour growth was measured every 7 days and calculated as described previously.2 Mice were sacrificed 35 days after injection and the tumours were extracted for further analysis.
Statistical Analysis
Statistical comparisons between groups were executed using Student’s t-test or one-way ANOVA with Turkey’s post hoc test in SPSS 20.0 (IBM, USA). Chi-square test was used to analyze the association between SNHG3 and NSCLC patient characteristics. For comparison of survival curves, the Log rank (Mantel-Cox) test was used. Pearson’s correlation analysis was used to identify the correlation between genes. Statistical significance was set at P < 0.05.
Results
SNHG3 Upregulation in Tissues and Cell Lines from NSCLC Patients
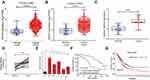
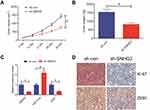
We analysed SNHG3 expression in lung adenocarcinoma tissues from the databases lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) of TCGA to explore the role of SNHG3 and found significant upregulation of SNHG3 in tissues from LUAD and LUSC patients (Figure 1A and B). We confirmed these findings by qRT-PCR in our cohort of 42 patients with NSCLC and detected a remarkable increase in SNHG3 expression in tumour samples (Tumor) compared with adjacent normal tissues (Normal) (Figure 1C and D). Analysis of SNHG3 expression in NSCLC cell lines showed a similar dramatic increase, which was particularly high in A549 (a human alveolar basal epithelial cell adenocarcinoma cell line) and H1299 (a lymph node metastatic lung cancer cell line) cells (Figure 1E). The expression of SNHG3 had a relationship with lymph node metastasis (P=0.013) and clinical stage (P=0.030) of NSCLC patients (Table 1). The Kaplan–Meier curves showed a poor prognosis for patients with elevated SNHG3 expression (P = 0.0237, Figure 1F), which was in agreement with the dataset from TCGA (Figure 1G). Taken together, these results suggested that aberrantly increased SNHG3 expression is a contributor for developing NSCLC.
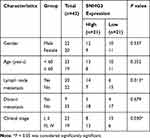 |
Table 1 Correlation of SNHG3 Expression with Clinicopathological Features of NSCLC Patients |
Inhibition of Tumour Growth and Migration by SNHG3 Knockdown
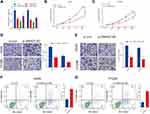
We transfected A549 and H1299 cells with siRNAs against SNHG3 to examine the functionality of SNHG3 in NSCLC. SNHG3 siRNAs significantly suppressed SNHG3 expression in these cell lines, of which si-SNHG3 #2 was especially effective (Figure 2A). We observed that knocking SNHG3 by si-SNHG3 #2 or si-SNHG3 #3 led to an inhibition of A549 and H1299 proliferation (Figure 2B and C, Supplement Figure 1A and B). In particular, our transwell experiments indicated that si-SNHG3 #2 and si-SNHG3 #3 also effectively blocked the capacity of these cells to migrate and invade (Figure 2D and E, Supplement Figure 1C and D) and flow cytometric analyses confirmed that SNHG3 knockdown promoted apoptosis (Figure 2F and G, Supplement Figure 1E and 1F). In summary, SNHG3 silencing reduced proliferation, migration, and invasion of lung adenocarcinoma cell lines and promoted apoptosis.
SNHG3 Interference with miR-216a Function
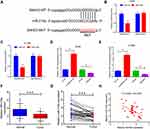
The molecular pathway employed by SNGH3 in NSCLC was investigated by predicting target miRNAs using starBase v3.0. We located a binding domain in SNHG3 for miR-216a (miR-216a-5p) (Figure 3A) and our luciferase reporter assays in A549 and H1299 cell lines supported this, showing that overexpression of miR-216a diminished luciferase activity in SNHG3-WT, but not SNHG3-MUT (Figure 3B and C). Conversely, knockdown of SNHG3 increased expression of miR-216a while SNHG3 overexpression resulted in the opposite effect (Figure 3D and E). We observed significantly lowered expression of miR-216a tumour tissue from NSCLC patients (Figure 3F and G), which negatively correlated with SNGH3 expression (Figure 3H). These resulted suggested that SNHG3 may interfere with miR-216a in NSCLC.
Blocking miR-216a Countered the Anti-Tumour Effects of SNHG3 Knockdown and Promoted Tumorigenesis
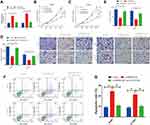
To explore the functional role of miR-216a in NSCLC cells, we first transfected A549 and H1299 cell s with anti-miR-con or anti-miR-216a. Our results indicated that miR-216a expression was remarkably reduced after anti-miR-216a transfection (Supplement Figure 2A). Moreover, miR-216a inhibition led to increased capacity to proliferate, migrate, and invade paired with a reduction in apoptosis (Supplement Figure 2B-G). Then, we transfected A549 and H1299 cell lines with si-SNHG3 #2 alone or si-SNHG3 #2 combined with anti-miR-216a to further investigate the relationship between SNHG3 and miR-216a. We observed that blocking miR-216a diminished si-SNHG3 #2-mediated increase in miR-216a expression (Figure 4A) and consequently negated the effects of SNHG3 knockdown, leading to increased capacity to proliferate, migrate, and invade paired with a reduction in apoptosis (Figure 4B-G). These results suggested the contribution of SNHG3 to tumour progression was propagated by miR-261a.
SNHG3 Elevated ZEB1 Expression by Interfering with miR-216a
The current literature describes the sponging action of lncRNAs, which disrupts miRNA function and releases their downstream targets14,15 We further explored the effects of SNHG3 on ZEB1, a target gene of miR-216a, to examine whether SNHG3 interference with miR-216a affected its expression. Multiple studies have demonstrated that ZEB1 heavily involved in the metastasis of tumour cells.16,17 Moreover, miR-216a has been shown to target ZEB1, leading to a suppression of cervical cancer cells’ ability to proliferate and migrate.18 Overexpression of miR-216a diminished luciferase activity of ZEB1-WT and this effect was rescued overexpression of SNHG3 (Figure 5A and B). Similar findings were found by qRT-PCR and Western blot analyses, which showed a reduction in ZEB1 expression by miR-216a overexpression or SNHG3 knockdown. However, anti-miR-216 elevated ZEB1 expression (Supplement Figures 3A and B). Interestingly, blockade of miR-216a negated the inhibitory effects of si-SNHG3 on ZEB1 expression (Figure 5C and D). We detected elevated ZEB1 expression in tumour tissues from patients with NSCLC (Figure 5E and F), which positively correlated with SNHG3 expression (Figure 5G). Taken together, these results suggested that SNHG3 enhanced ZEB1 expression by interfering with miR-216a.
Suppression of Tumour Growth in vivo by Knockdown of SNHG3
We extrapolated our findings on the role of SNHG3 on tumorigenesis in vitro and examined the effects of SNHG3 knockdown in vivo. We inoculated A549 cells that stably expressed Lenti-sh-SNHG3 or Lenti-sh-control into nude mice and found that SNHG3 knockdown caused a marked inhibition of tumour growth and reduced tumour volume and weight (Figure 6A and B). In addition, we observed a decline in the expression of SNHG3 and ZEB1 paired with a striking elevation in miR-216a expression (Figure 6C) and reduction in Ki-67 expression (Figure 6D). Taken together, our in vivo experiments indicated that knockdown of SNHG3 inhibited tumour growth.
Discussion
NSCLC is one of the primary causes of cancer deaths across the world and despite continued development of NSCLC therapies, advanced disease remains poorly treatable and novel treatment strategies are urgently needed for these patients. lncRNAs have been demonstrated to contribute to tumorigenesis in various cancer types,19 although their exact function and molecular mechanisms in NSCLC are still undefined. Here, we documented heightened SNHG3 expression in tissues from NSCLC patients and lung adenocarcinoma cell lines. Our data suggested that SNHG3 increased proliferation, migration, and invasion of NSCLC cells, which was paired with a suppression of apoptosis. These effects were caused by SNGH3’s interference with miR-216a function that led to an increase in ZEB1 expression and other downstream effects.
lncRNA SNHG3 is aberrantly expressed in various cancers and exhibits oncogenic properties. Li et al have described that SNHG3 suppression in prostate cancer cells blocked pro-tumour activities while promoting apoptosis by interfering with miR-577 and subsequently causing an upregulation of Smad ubiquitination regulatory factor 1 (SMURF1).20 Similar observations have been made in hepatocellular carcinoma cells by Wu et al where SNHG3 disrupts miR-139-5p and elevates BMI1 expression.21 Other studies have demonstrated that SNHG3 overexpression promotes metastasic capabilities of tumour cells in gastric cancer through the suppression of MED18 expression.22 We measured an increased SNHG3 expression in NSCLC samples from the database of TCGA as well as samples from our patients with NSCLC and lung adenocarcinoma cell lines. We correlated the excessive expression of SNHG3 with a poor prognosis in NSCLC patients and observed that it promoted processes involved in tumour formation and metastasis, which is consistent with the studies outlined above and recent literature.12,23 Taken together, these studies and our data demonstrate the scope of SNHG3’s ability to promote tumour formation and show its potentially new avenue for cancer therapy.
lncRNAs are known to act as a ceRNAs and interfere with miRNAs, resulting in the upregulation of their target proteins.14,15 For instance, upregulation of SNHG7 accelerates the progression of lung cancer by competing with miR-193b and increasing Fas apoptosis inhibitory molecule 2 expression.24 Similarly, UCA1 promoted ERBB4 expression by interfering with miR-193a-3p and lead to the progression of NSCLC tumours.25 We used bioinformatics combined with luciferase reporter assays to predict the target of SNHG3 and found that it interfered with miR-216a, leading to elevated ZEB1 expression in NSCLC cells. Moreover, the tumour suppressing effects of SNHG3 knockdown were negated by inhibiting miR-216a or overexpressing ZEB1. In summary, SNHG3 contributed to the development of NSCLC tumours by modulating the miR-216a/ZEB1 axis.
Interestingly, lncRNAs appear to have a dual role in tumorigenesis and may have tumour suppressive properties in addition to oncogenic functions. Studies have reported that lncRNAs may be regulated by p53 signalling, which is a tumour suppressor that is often altered in tumour cells, and act as downstream effector molecules.26 Conversely, Schmitt et al demonstrated that lncRNAs such as damage induced noncoding (DINO) interacts with p53 to stabilise the protein while simultaneously mimicking its function. Stabilised p53 then further activates downstream targets including DINO, creating a positive feedback loop.27 In contrast to the functions reported in this study, SNHG3 appears to harbour tumour suppressive functions in papillary thyroid carcinoma where silencing SNHG3 promotes tumour formation in vivo via a pathway involving AKT, mTOR, and ERK.28 In addition, SNHG3 upregulation has been shown to have a protective role in ischemia-induced injury in neonatal brains by sponging miR-196 in hippocampal cells, thus preventing expression of XIAP and CAAP1.29 However, in a similar setting, the sponging of miR-196 by SNHG3 in osteosarcoma leads to cell growth and subsequent poor outcomes.30 Therefore, our findings in patients with NSCLC may firstly not be directly extrapolatable to other cancer types and secondly, the contribution of SNHG3 to the homeostasis of other tissues and its protective roles cannot be disregarded and need to be extensively studied before its use as a therapeutic target in cancer.
In conclusion, our study revealed that SNHG3 employs its ability to interfere with miR-216a to increase ZEB1 expression and promote tumorigenesis in NSCLC. Therefore, SNHG3 could be used as a novel biomarker and therapeutic target for patients with NSCLC.
Ethics Approval and Consent to Participate
Informed consent was given by all study participants before surgery and all procedures were reviewed and authorised by the institute’s ethics committee of the First Hospital Affiliated to The Xinxiang Medical College. All procedures and protocols of animal experiments approved by the institutional animal ethics committee of the First Hospital Affiliated to The Xinxiang Medical College and performed in strict accordance with the guiding principles of the institutional animal management committee of the First Hospital Affiliated to The Xinxiang Medical College.
Disclosure
The authors declare no potential conflicts of interest for this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global Cancer Statistic, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Ettinger DS, Akerley W, Bepler G, et al. Non-small cell lung cancer. J Natl Compreh Cancer Net. 2010;8(7):740. doi:10.6004/jnccn.2010.0056
3. Chen J, Wang R, Zhang K, Chen LB. Long non‐coding RNAs in non‐small cell lung cancer as biomarkers and therapeutic targets. J Cell Mol Med. 2014;18(12):2425–2436. doi:10.1111/jcmm.12431
4. Ricciuti B, Mencaroni C, Paglialunga L, et al. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol. 2016;33(2):18. doi:10.1007/s12032-016-0731-2
5. Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4):e944014. doi:10.4161/21541272.2014.944014
6. Fang X, Yin M, Li H, et al. Comprehensive analysis of competitive endogenous RNAs network associated with head and neck squamous cell carcinoma. Sci Rep. 2018;8(1):10544. doi:10.1038/s41598-018-28957-y
7. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235–246. doi:10.1158/2159-8290.CD-15-0893
8. Abu N, Hon KW, Jeyaraman S, Jamal R. Long noncoding RNAs as biotargets in cisplatin-based drug resistance. Future Oncol. 2018;14(29):3085–3095. doi:10.2217/fon-2018-0303
9. Wu H, Wang L, Zhang S, et al. Downregulation of LINC00665 confers decreased cell proliferation and invasion via the miR-138-5p/E2F3 signaling pathway in NSCLC. Biomed Pharmacother. 2020;127:110214. doi:10.1016/j.biopha.2020.110214
10. Dacheng W, Songhe L, Weidong J, et al. LncRNA SNHG3 promotes the growth and metastasis of colorectal cancer by regulating miR-539/RUNX2 axis. Biomed Pharmacother. 2020;125:110039. doi:10.1016/j.biopha.2020.110039
11. Sui G, Zhang B, Fei D, Wang H, Guo F, Luo Q. The lncRNA SNHG3 accelerates papillary thyroid carcinoma progression via the miR-214-3p/PSMD10 axis. J Cell Physiol. 2020.
12. Liu L, Ni J, He X. Upregulation of the long noncoding RNA SNHG3 promotes lung adenocarcinoma proliferation. Dis Markers. 2018;2018:5736716.
13. Niu L, Liu A, Xu W, Yang L, Zhu W, Gu Y. Downregulation of peroxiredoxin II suppresses the proliferation and metastasis of gastric cancer cells. Oncol Lett. 2018;16(4):4551–4560.
14. Wang Y, Hou J, He D, et al. The emerging function and mechanism of ceRNAs in cancer. Trends Genet. 2016;32(4):211–224. doi:10.1016/j.tig.2016.02.001
15. Sanchezmejias A, Tay Y. Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J Hematol Oncol. 2015;8(1):30. doi:10.1186/s13045-015-0129-1
16. Li Y, He Q, Wen X, et al. EZH2-DNMT1-mediated epigenetic silencing of miR-142-3p promotes metastasis through targeting ZEB2 in nasopharyngeal carcinoma. Cell Death Differ. 2018;undefined(undefined):undefined.
17. Gong P, Qiao F, Wu H, et al. LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer.%A Gong P. Cell Death Dis. 2018;9(12):1158. doi:10.1038/s41419-018-1170-0
18. Z H, Zeng Y, Zhou -C-C, et al. SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch Biochem Biophys. 2018;637:1–8. doi:10.1016/j.abb.2017.11.003
19. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006
20. Li T, Xing Y, Yang F, et al. LncRNA SNHG3 sponges miR-577 to up-regulate SMURF1 expression in prostate cancer. Cancer Med. 2020.
21. Wu J, Liu L, Jin H, et al. LncSNHG3/miR-139-5p/BMI1 axis regulates proliferation, migration, and invasion in hepatocellular carcinoma. Onco Targets Ther. 2019;12:6623–6638. doi:10.2147/OTT.S196630
22. Xuan Y, Wang Y. Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis. 2019;10(10):694. doi:10.1038/s41419-019-1940-3
23. Shi J, Li J, Yang S, et al. LncRNA SNHG3 is activated by E2F1 and promotes proliferation and migration of non-small-cell lung cancer cells through activating TGF-β pathway and IL-6/JAK2/STAT3 pathway. J Cell Physiol. 2020;235:2891–2900. doi:10.1002/jcp.29194
24. She K, Yan H, Huang J, Zhou H, He J miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer.. Cell Prolif. 2018;51(1):undefined.
25. Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi:10.1016/j.canlet.2015.11.024
26. Peng W, Koirala P, Mo Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
27. Schmitt AM, Garcia JT, Hung T, et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet. 2016;48(11):1370–1376. doi:10.1038/ng.3673
28. Duan Y, Wang Z, Xu L, Sun L, He F. lncRNA SNHG3 acts as a novel tumor suppressor and regulates tumor proliferation and metastasis via AKT/mTOR/ERK pathway in papillary thyroid carcinoma. J Cancer. 2020;11(12):3492–3501. doi:10.7150/jca.42070
29. Yang Q, Wu M, Zhu L, Qiao L, Zhao R, Xia Z. Long non-coding RNA Snhg3 protects against hypoxia/ischemia-induced neonatal brain injury. Exp Mol Pathol. 2020;112:104343. doi:10.1016/j.yexmp.2019.104343
30. Chen J, Wu Z, Zhang Y. LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma. Int J Immunopathol Pharmacol. 2019;33:2058738418820743. doi:10.1177/2058738418820743
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.