Back to Journals » OncoTargets and Therapy » Volume 13
SNHG15 Contributes To Cisplatin Resistance In Breast Cancer Through Sponging miR-381
Authors Mi H, Wang X, Wang F, Li L, Zhu M, Wang N, Xiong Y, Gu Y
Received 15 July 2019
Accepted for publication 22 October 2019
Published 21 January 2020 Volume 2020:13 Pages 657—666
DOI https://doi.org/10.2147/OTT.S223321
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Hailong Mi,* Xiaochun Wang,* Fang Wang, Lin Li, Mingzhi Zhu, Nan Wang, Youyi Xiong, Yuanting Gu
Department of Breast Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 475000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yuanting Gu
Department of Breast Surgery, The First Affiliated Hospital of Zhengzhou University, 1 Jianshe East Road, Zhengzhou 450000, Henan Province, People’s Republic of China
Tel +86-0371-67967172
Email [email protected]
Background: Increasing evidence implies the participation of long non-coding RNAs (lncRNAs) in chemoresistance to cancer treatment. Their role and molecular mechanisms in breast cancer chemoresistance, nevertheless, are yet not considerably elucidated. In this work, we research the function of small nucleolar RNA host gene 15 (SNHG15) in cisplatin (DDP) resistance of breast cancer and uncover the underlying molecular mechanism.
Methods: SNHG15 and miR-381 expression levels were detected using Quantitative real-time PCR (qRT-PCR) analysis. The functional roles of SNHG15 and miR-381 in breast cancer were determined using MTT assay and flow cytometry analysis. The effect of SNHG15 on miR-381 expression was determined using Luciferase reporter assay, RNA immunoprecipitation (RIP) assay and qRT-PCR analysis.
Results: SNHG15 was found to be up-regulated in cisplatin resistant breast cancer tissues and cell lines. Breast cancer patients with high SNHG15 expression had a poor prognosis. SNHG15 silencing enhanced cisplatin sensitivity of MCF-7/DDP and MDA-MB-231/DDP cells. Additionally, SNHG15 could function as a miR-381 sponge. miR-381 overexpression could overcome cisplatin resistance. miR-381 knockdown countered SNHG15 knockdown-mediated enhancement of cisplatin sensitivity in MCF-7/DDP and MDA-MB-231/DDP cells. Besides, SNHG15 knockdown facilitated cisplatin sensitivity of cisplatin resistant breast cancer cells in vivo.
Conclusion: In summary, SNHG15 knockdown overcame cisplatin resistance of breast cancer by sponging miR-381, providing a novel therapeutic target for breast cancer.
Keywords: breast cancer, cisplatin, small nucleolar RNA host gene 15, miR-381
Introduction
Breast cancer is a most common malignancy in women worldwide and a major serious health threat to women.1 Even though impressive improvements have been achieved in diagnosis and therapy of breast cancer in the past decade, prognosis for advanced breast cancer remains relatively poor.2 Platinum-based chemotherapy is a significant therapeutic strategy for breast cancer patients.3 Nevertheless, chemoresistance remains a key barrier to the efficacy of chemotherapy drugs for cancers, including breast cancer.4,5 Hence, to sequentially elucidate the underlying mechanism and discover new therapeutic targets are essential for developing effective therapies for breast cancer patients.
Long non-coding RNAs (lncRNAs) are a group of endogenous non-protein-coding RNAs with more than 200 nucleotides in length.6 Recently, lncRNAs were discovered to play crucial regulatory roles in various biological processes, including tumorigenesis process.7 Increasing evidence indicates that lncRNAs are usually deregulated in malignant tumors’ progression.8–10 Moreover, dysregulated lncRNAs have been reported to be implicated with cisplatin resistance in various cancers.11 LncRNA small nucleolar RNA host gene 15 (SNHG15), an intergenic lncRNA located on chromosome 7p13, belongs to a family of non-coding RNAs that hosting snoRNAs.12 Previous studies suggest that SNHG15 could promote cancer cell proliferation and invasiveness and is able to play as a crucial determinant role in the chemoresistance.13 However, the functional role of SNHG15 in cisplatin (DDP) resistance in breast cancer has not been evaluated.
miRNAs are a class of small, non-coding RNA with about 22 nucleotides in length, which contribute to their target genes’ inhibition, through mRNA degradation or translation inhibition.14,15 MiRNAs is recently implicated by more and more reports as critical determinants that are involved in various cellular processes, including chemoresistance and tumorigenesis.16 MiRNAs’ aberrant expression has been regarded as a powerful regulator of CDDP resistance. For instance, miR-34a has been discovered to be down-regulated and contributed to DDP resistance in DDP-resistant prostate cancer cells.17 miR-381 could overcome cisplatin resistance in non-small cell lung cancer (NSCLC) cells through inactivating nuclear factor-κB signaling.18 Moreover, miR-381 overexpression could improve DDP sensitivity of breast cancer cells through targeting MDR1.19 However, how miR-381 was regulated in breast cancer cisplatin resistance remains largely unknown.
In this study, we aimed to investigate the expression pattern and functional role and underlying molecular mechanism of SNHG15 in breast cancer cisplatin resistance. Our study revealed that SNHG15 expression was up-regulated in breast cancer tissues and cell lines, especially in DDP-resistant breast cancer tissues and cells. Functionally, SNHG15 knockdown re-sensitized DDP-resistant breast cancer cells to DDP. Mechanically, knockdown of SNHG15 facilitated the sensitivity of breast cancer cells towards cisplatin through increasing miR-381 expression. Our work demonstrated a novel SNHG15/miR-381 regulatory axis, overcoming cisplatin resistance in breast cancer.
Materials And Methods
Tumor Tissue Samples And Cells
The paired tumor tissues (n=42) and adjacent normal tissues (n=42) were obtained from breast cancer patients who underwent surgery at the First Affiliated Hospital of Zhengzhou University. Written informed consent was obtained from all participants. This study had acquired the approval of the ethics committee of the First Affiliated Hospital of Zhengzhou University. Normalized RNA-seq data of Breast adenocarcinoma (BRCA) were downloaded from the TCGA data portal website (https://cancergenome.nih.gov/).
Human breast cancer cell lines (MCF-7 and MDA-MB-231) and human normal breast epithelial cell line MCF-10A were obtained from ATCC (Manassas, VA, USA). DDP-resistant cell lines (MCF-7/DDP and MDA-MB-231/DDP) were selected from MCF-7 and MDA-MB-231 cells after continuous exposure to stepwise increasing concentrations of DDP for 12 months. All cells were grown in RPMI-1640 medium (Gibco BRL, Grand Island, NY, USA) supplement with 10% FBS at 37°C with 5% CO2.
Cell Transfection
Mimic control (miR-con), miR-381 mimic (miR-381) and miR-381 inhibitor (anti-miR-381) were purchased from Genepharma (Shanghai, China). SNHG15 overexpressing vector pcDNA3.1-SNHG15 (SNHG15) or the empty vector pcDNA3.1 (Vector) or and small interfering RNAs against SNHG15 (si-SNHG15 #1, si-SNHG15 #2, and si-SNHG15 #3) and their negative control (si-con) were designed and synthesized by Genepharma (China). Lipofectamine 2000 (Invitrogen) was used for cell transfections.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA extraction was carried out through using Trizol reagent (TaKaRa, Tokyo, Japan). Then, the RNA was reverse transcripted to cDNA using PrimeScript RT Reagent Kit (TaKaRa). The qRT-PCR was carried out using SYBR green qRT-PCR assay. GAPDH and U6 were used for internal controls for SNHG15 and miR-381, respectively. The data was analyzed using the 2−ΔΔCt method.
DDP Sensitivity Assay
The sensitivity of breast cancer cells was evaluated by MTT assay. DDP sensitivity was determined using the IC50 value (half maximal inhibitory concentration). GraphPad Prism 7.0 Software was used to calculate the IC50 value.
Flow Cytometric Analysis
Cell apoptosis was identified using Annexin V-FITC/PI apoptosis detection kit (MultiScience Biotech, Hangzhou, China) as described previously.20
Subcellular Fraction Assays
Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA) was used to separate the nuclear and cytoplasmic RNA of MCF-7/DDP and MDA-MB-231/DDP cells.
Luciferase Reporter Assay
The wild or mutant SNHG15 sequence containing predicted miR-381 binding site were subcloned respectively into the luciferase reporter vectors to generate SNHG15-WT and SNHG15-MUT vectors. Then, MCF-7/DDP and MDA-MB-231/DDP cells were co-transfected with these constructs and miR-381 or miR-con. Finally, luciferase activity in MCF-7/DDP and MDA-MB-231/DDP cells was determined using Luciferase Reporter assay system (Promega, Madison, WI, USA).
RNA Immunoprecipitation (RIP) Assays
RIP experiments were performed using the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) and antibodies against Ago2 and IgG (Cell Signaling Technology, Danvers, MA, USA). The co-precipitated RNAs were purified and subjected to qRT-PCR analysis.
Animal Experiments
The animal experiments were conducted in strict accordance with the guiding principles of the institutional animal ethics committee and approved by the institutional research committee of the First Affiliated Hospital of Zhengzhou University. MCF-7/DDP cells stably expressing sh-SNHG15 were transplanted into BALB/c nude mice (six-week-old) from Slac Laboratory (Shanghai, China), and then mice were injected intraperitoneally with 5 mg/kg cisplatin or same volume of PBS every seven days. Every seven days, tumor sizes were monitored, and tumor volumes were calculated based on the formula: volume = 0.5 × length × width2. Mice were euthanized at 35th day post injection and tumors were excised and weighed.
Statistical Analysis
All data were evaluated as means±standard deviation (SD). The statistic difference was calculated using Student’s t-test and one-way ANOVA. P value <0.05 was considered statistically significant.
Results
SNHG15 Was Up-Regulated In DDP-Resistant Breast Cancer Tissues And Cell Lines
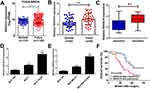
To investigate the function of SNHG15 in breast cancer, we firstly examined the expression of SNHG15 in breast cancer tissues from TCGA databases. Compared with normal tissues, SNHG15 expression was dramatically increased in breast cancer tumor tissues (Figure 1A). To further prove the result from TCGA databases, SNHG15 expression in breast cancer tumor tissues (n=42) and adjacent normal tissues (n=42) was further determined by qRT-PCR analysis. Consistently, SNHG15 was higher in breast cancer tissues than that in adjacent normal tissues (Figure 1B). Additionally, SNHG15 expression was extremely increased in DDP-resistant breast cancer tissues when compared with DDP-sensitive breast cancer tissues (Figure 1C). Furthermore, the expression of SNHG15 was significantly improved in MCF-7 and MDA-MB-231 cells compared with normal MCF-10A cells (Figure 1D and E). Notably, compared with their parental cells, MCF-7/DDP and MDA-MB-231/DDP cells displayed high SNHG15 expression level (Figure 1D and E). Moreover, the breast cancer patients with high SNHG15 level had a poor prognosis (P = 0.0162) (Figure 1F). Collectively, these data suggested that up-regulated SNHG15 may be implicated with cisplatin resistance in breast cancer.
SNHG15 Knockdown Overcame Cisplatin Resistance Of Breast Cancer Cells
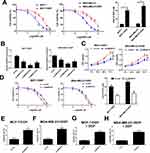
To evaluate the resistance of MCF-7/DDP and MDA-MB-231/DDP cells to DDP, IC50 of DDP was measured by MTT assay in DDP-resistant MCF-7/DDP and MDA-MB-231/DDP cells and parental MCF-7 and MDA-MB-231 cells. Compared with the parental cells, MCF-7/DDP and MDA-MB-231/DDP cells displayed poor response to DDP (Figure 2A). To further confirm the role of SNHG15 in DDP-resistant breast cancer cells, MCF-7/DDP and MDA-MB-231/DDP cells were transfected with SNHG15 siRNAs (si-SNHG15 #1, si-SNHG15 #2 or si-SNHG15 #3) or si-con. qRT-PCR analysis indicated that introduction of SNHG15 siRNAs evidently declined SNHG15 expression in MCF-7/DDP and MDA-MB-231/DDP cells (Figure 2B), especially in si-SNHG15 #2 treated group. Therefore, si-SNHG15 #2 (si-SNHG15) was used for further experiments. Remarkably, SNHG15 silencing suppressed the cell viability and enhanced cisplatin sensitivity in MCF-7/DDP and MDA-MB-231/DDP cells (Figure 2C and D). To further determine the role of SNHG15 in DDP-induced apoptosis, flow cytometry analysis was conducted in MCF-7/DDP and MDA-MB-231/DDP cells with or without 10 μM DDP treatment. SNHG15 knockdown could increase cell apoptosis in MCF-7/DDP and MDA-MB-231/DDP cells (Figure 2E and F). Prominently, inhibition of SNHG15 in combination with DDP exposure could exert their synergistic effect contributing to significant enhancement in cell apoptosis in MCF-7/DDP and MDA-MB-231/DDP cells (Figure 2G and H). Collectively, SNHG15 knockdown facilitated cisplatin sensitivity in breast cancer cells.
SNHG15 Acted As A miR-381 Sponge In Breast Cancer Cells
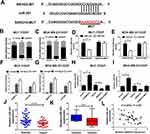
Starbase2.0 predicts that the sequence of SNHG15 harbors a binding site of miR-381 (Figure 3A). Moreover, subcellular fraction assay revealed that SNHG15 was mainly distributed in the cytoplasm of MCF-7/DDP and MDA-MB-231/DDP cells (Figure 3B and C). Hence, we further investigated whether SNHG15 functioned as a miR-381 sponge in DDP-resistant breast cancer cells. Luciferase reporter assay indicated that miR-381 overexpression obviously reduced the relative luciferase activity of SNHG15-WT but not SNHG15-MUT, which suggests SNHG15 could interact with miR-381 (Figure 3D and E). RNA RIP assay confirmed that SNHG15 and miR-381 were greatly enriched in the Ago2 antibody-treated group compared with the IgG antibody-treated group (Figure 3F and G). To further explore the regulatory role of SNHG15 in miR-381 expression, MCF-7/DDP and MDA-MB-231/DDP cells were transfected with si-SNHG15 or si-con, Vector, SNHG15 or SNHG15-MUT. In both MCF-7/DDP and MDA-MB-231/DDP cells, knockdown of SNHG15 markedly elevated miR-381 expression. Oppositely, the expression level of miR-381 was remarkably decreased by SNHG15 up-regulation. However, transfection with SNHG15-MUT has no effect on miR-381 expression (Figure 3H and I). Especially, miR-381 expression was dramatically reduced in breast cancer tissues (Figure 3J). miR-381 expression was lower in DDP-resistant breast cancer tissues than that in DDP-sensitive ones (Figure 3K). Moreover, SNHG15 is negatively correlated with miR-381 expression in breast cancer tissues (Figure 3L). All these data evidenced that SNHG15 sponges miR-381 in breast cancer cells.
miR-381 Overexpression Enhanced Cisplatin Sensitivity Of Breast Cancer Cells
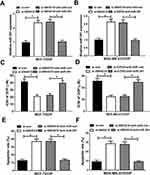
To further study the effect of miR-381 on DDP-resistant breast cancer cells, MCF-7/DDP and MDA-MB-231/DDP cells were transfected with miR-381 mimics or miR-con. qRT-PCR analysis revealed that miR-381 expression was strangely increased in miR-381 transfecting MCF-7/DDP and MDA-MB-231/DDP cells (Figure 4A and B). Moreover, overexpression of miR-381 enhanced DDP sensitivity in MCF-7/DDP and MDA-MB-231/DDP cells (Figure 4C and D). Additionally, flow cytometry analysis showed that miR-381 overexpression strangely enhanced DDP-induced apoptosis in MCF-7/DDP and MDA-MB-231/DDP cells (Figure 4E and F). Together, miR-381 overexpression facilitated cisplatin sensitivity in breast cancer cells.
SNHG15 Knockdown Improved Cisplatin Sensitivity Of Breast Cancer Cells Through Sponging miR-381
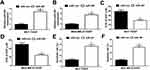
To further investigate whether SNHG15 contributed to cisplatin resistance in breast cancer through sponging miR-381 expression, MCF-7/DDP and MDA-MB-231/DDP cells were transfected with si-con, si-SNHG15, si-SNHG15+anti-miR-con or si-SNHG15+anti-miR-381. Transfection of si-SNHG15 elevated miR-381 expression, which was extremely reversed by miR-381 inhibition (Figure 5A and B). MTT assay revealed that down-regulation of SNHG15 facilitated cisplatin sensitivity of MCF-7/DDP and MDA-MB-231/DDP cells, however, SNHG15 inhibition-mediated enhancement of cisplatin sensitivity in MCF-7/DDP and MDA-MB-231/DDP cells was patently abolished by miR-381 down-regulation (Figure 5C and D). Furthermore, introduction of anti-miR-381 extremely demolished the inductive effect of silenced SNHG15 on apoptosis in MCF-7/DDP and MDA-MB-231/DDP cells (Figure 5E and F). Taken together, these results demonstrated that SNHG15 led to cisplatin resistance of breast cancer cells through sponging miR-381.
SNHG15 Knockdown Enhanced Cisplatin Sensitivity In Tumors In Vivo
To validate whether SNHG15 contributed to cisplatin resistance in vivo, a mouse breast cancer xenograft model was created. We made an sh-SNHG15 stably expressing MCF-7/DDP cell line and inoculated them subcutaneously into nude mice. Thenceforth, mice were injected intraperitoneally with PBS or CDDP, and tumor size was evaluated every 7 days. SNHG15 silencing evidently suppressed tumor growth. cisplatin treatment also inhibited the growth of tumors. Moreover, SNHG15 silencing in combination with cisplatin administration displayed slower tumor growth (Figure 6A). At the 35th day post treatment, mice were euthanized, and xenograft tumors were excised and weighed. The tumor weight was reduced after SNHG15 knockdown or cisplatin exposure, especially when SNHG15 knockdown and cisplatin treatment simultaneously (Figure 6B). In addition, SNHG15 silencing or cisplatin treatment inhibited the expression of SNHG15 while up-regulated the expression of miR-381, and this effect could be strengthened by combination of SNHG15 silencing and cisplatin exposure (Figure 6C). All these data demonstrated that knockdown of SNHG15 improved cisplatin sensitivity in breast cancer cells in vivo.
Discussion
Therapeutic outcome has been restricted by chemoresistance severely for breast cancer patients. Consequently, to reveal the underlying mechanism and uncover novel therapeutic strategies for chemoresistance is not dispensable. In this study, we found that SNHG15 expression was significantly increased in DDP-resistant breast cancer tissues and cells. Moreover, knockdown of SNHG15 re-sensitized DDP-resistant breast cancer cells to DDP. Prominently, SNHG15 silencing improved the response of breast cancer cells to cisplatin through elevating miR-381 expression. Therefore, SNHG15 is a positive regulator in breast cancer cisplatin resistance and targeting SNHG15 may be an effective scheme for cisplatin chemoresistance in breast cancer.
Elucidating the molecular mechanism underlying DDP resistance was helpful to discover reasonable and effective targeted therapeutic strategies to overwhelmed DDP resistance. Our study found that SNHG15 expression was increased in DDP-resistant breast cancer tissues and cells, and SNHG15 silencing overcame DDP resistance in DDP-resistant breast cancer cells. In line with our findings, increasing evidence demonstrated that dysregulated SNHG15 was implicated with chemoresistance in various malignancies. For instance, knockdown of SNHG15 could suppress cell proliferation and invasion, and sensitize colorectal cancer cells to 5-FU.13 Moreover, SNHG15 was overexpressed in epithelial ovarian cancer, and contributed to the proliferation, migration, invasion, and chemoresistance of epithelial ovarian cancer cells towards cisplatin.21 All these findings demonstrated that targeted inhibiting SNHG15 maybe a hopeful therapeutic strategy for CDDP chemoresistance.
How elevated SNHG15 contributes to cisplatin resistance in breast cancer remains elusive. Hence, the operative mechanism of SNHG15 was further explored in this work. Emerging evidence suggested that SNHG15 inhibited the expression of miRNAs through functioning as miRNA sponges.22–24 However, whether SNHG15 could regulate cisplatin resistance in breast cancer through controlling miRNA expression was still indescribable. In our study, Starbase2.0 database prediction and luciferase reporter assays confirmed that SNHG15 could bind with miR-381 in MCF-7/DDP and MDA-MB-231/DDP cells. Mounting evidence have indicated that miR-381 played a tumor suppressor role in tumorigenesis.25–28 Moreover, down-regulated miR-381 was implicated with chemoresistance in many cancers.29–32 Prominently, miR-381 could enhance cisplatin sensitivity of breast cancer cells through targeting MDR1.19 Correspondingly, our found indicated that overexpression of miR-381 overcame cisplatin resistance in MCF-7/DDP and MDA-MB-231/DDP cells. Additionally, miR-381 inhibition reversed SNHG15 silencing-mediated enhancement of DDP sensitivity in MCF-7/DDP and MDA-MB-231/DDP cells. All these data demonstrated that SNHG15 inhibition overcame DDP resistance in breast cancer cells through sponging miR-381.
In conclusion, our study demonstrated that SNHG15 knockdown improved cisplatin sensitivity of breast cancer cells through sponging miR-381, providing a promising therapeutic target for breast cancer DDP resistance.
Ethics Approval And Consent To Participate
Written informed consent was obtained from all participants. This study had acquired the approval of the ethics committee of the First Affiliated Hospital of Zhengzhou University. The animal experiments were conducted in strict accordance with the guiding principles of the institutional animal ethics committee and approved by the institutional research committee of the First Affiliated Hospital of Zhengzhou University.
Disclosure
The authors have no conflicts of interest to declare regarding this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Ca A Cancer J Clinicians. 2016;66:1. doi:10.3322/caac.21332
2. Sutter SA, Slinker A, Balumuka DD, Mitchell KB. Surgical management of breast cancer in Africa: a continent-wide review of intervention practices, barriers to care, and adjuvant therapy. J Glob Oncol. 2017;3(2):162–168. doi:10.1200/JGO.2016.003095
3. Villarreal-Garza C, Khalaf D, Bouganim N, et al. Platinum-based chemotherapy in triple-negative advanced breast cancer. Dtsch Med Wochenschr. 2015;26(8):894–901.
4. Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discovery. 2006;5(3):219–234.
5. Coley HM. Mechanisms and strategies to overcome chemotherapy resistance in metastatic breast cancer. Cancer Treat Rev. 2008;34(4):378. doi:10.1016/j.ctrv.2008.01.007
6. Yu X, Li Z. Long non-coding RNA growth arrest-specific transcript 5 in tumor biology (Review). Oncol Lett. 2015;10(4):1953–1958. doi:10.3892/ol.2015.3553
7. Zhang F, Zhang L, Zhang C. Long noncoding RNAs and tumorigenesis: genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol J Int Soc Oncodevelopmental Biol Med. 2015;37(1):163–175.
8. Schmitt AM, Chang HY. Gene regulation: long RNAs wire up cancer growth. Nature. 2013;500(7464):536–537. doi:10.1038/nature12548
9. Yang QQ, Deng YF. Genome‐wide analysis of long non‐coding RNA in primary nasopharyngeal carcinoma by microarray. Histopathology. 2015;66(7):1022–1030. doi:10.1111/his.12616
10. Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non‐coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104(4):458–464. doi:10.1111/cas.2013.104.issue-4
11. Abu N, Hon KW, Jeyaraman S, et al. Long noncoding RNAs as biotargets in cisplatin-based drug resistance. Future Oncol. 2018;14(29):3085–3095. doi:10.2217/fon-2018-0303
12. Shen Y, Liu S, Fan J, et al. Nuclear retention of the lncRNA SNHG1 by doxorubicin attenuates hnRNPC-p53 protein interactions. EMBO Rep. 2017;18(4):536–548. doi:10.15252/embr.201643139
13. Saeinasab M, Bahrami AR, González J, et al. SNHG15 is a bifunctional MYC-regulated noncoding locus encoding a lncRNA that promotes cell proliferation, invasion and drug resistance in colorectal cancer by interacting with AIF. J Exp Clin Cancer Res. 2019;38(1):172.
14. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
15. Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35(7):368–376. doi:10.1016/j.tibs.2010.03.009
16. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi:10.1146/annurev-pathol-012513-104715
17. Liu X, Luo X, Wu Y, et al. MicroRNA-34a attenuates paclitaxel resistance in prostate cancer cells via direct suppression of JAG1/Notch1 axis. Cell Physiol Biochem. 2018;50(1):261–276. doi:10.1159/000494004
18. Huang R-S, Zheng Y-L, Zhao J, Chun X. microRNA-381 suppresses the growth and increases cisplatin sensitivity in non-small cell lung cancer cells through inhibition of nuclear factor-κB signaling. Biomed Pharmacother. 2018;98:538–544. doi:10.1016/j.biopha.2017.12.092
19. Yi D, Xu L, Wang R, Lu X, Sang J. miR‐381 overcomes cisplatin resistance in breast cancer by targeting MDR1. Cell Biol Int. 2019;43(1):12–21. doi:10.1002/cbin.v43.1
20. Wang WJ, Yao Y, Jiang LL, et al. Knockdown of lymphoid enhancer factor 1 inhibits colon cancer progression in vitro and in vivo. PLoS One. 2013;8(10):e76596.
21. Qu C, Dai C, Guo Y, Qin R, Liu J. Long noncoding RNA SNHG15 serves as an oncogene and predicts poor prognosis in epithelial ovarian cancer. Onco Targets Ther. 2019;12:101. doi:10.2147/OTT.S182657
22. Kong Q, Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Commun. 2018;495(2):1594–1600. doi:10.1016/j.bbrc.2017.12.013
23. Jin B, Jin H, Wu HB, Xu JJ, Li B. Long non‐coding RNA SNHG15 promotes CDK14 expression via miR‐486 to accelerate non‐small cell lung cancer cells progression and metastasis. J Cell Physiol. 2018;233(9):7164–7172. doi:10.1002/jcp.26543
24. Wu D-M, Wang S, Wen X, et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. 2018;9(10):947. doi:10.1038/s41419-018-0975-1
25. Tian C, Li J, Ren L, Peng R, Chen B, Lin Y. MicroRNA-381 serves as a prognostic factor and inhibits migration and invasion in non-small cell lung cancer by targeting LRH-1. Oncol Rep. 2017;38(5):3071–3077. doi:10.3892/or.2017.5956
26. Cao Q, Liu F, Ji K, et al. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J Exp Clin Cancer Res. 2017;36(1):29. doi:10.1186/s13046-017-0499-z
27. Xue Y, Xu W, Zhao W, Wang W, Zhang D, Wu P. miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4. Biomed Pharmacother. 2017;86:426–433. doi:10.1016/j.biopha.2016.12.051
28. He X, Wei Y, Wang Y, Liu L, Wang W, Li N. MiR-381 functions as a tumor suppressor in colorectal cancer by targeting Twist1. Onco Targets Ther. 2016;9:1231.
29. Wang Z, Yang J, Xu G, et al. Targeting miR-381-NEFL axis sensitizes glioblastoma cells to temozolomide by regulating stemness factors and multidrug resistance factors. Oncotarget. 2015;6(5):3147–3164. doi:10.18632/oncotarget.3061
30. Chen B, Duan L, Yin G, Jing T, Jiang X. miR-381, a novel intrinsic WEE1 inhibitor, sensitizes renal cancer cells to 5-FU by up-regulation of Cdc2 activities in 786-O. J Chemother. 2013;25(4):229–238. doi:10.1179/1973947813Y.0000000092
31. Li Y, Zhao C, Yu Z, et al. Low expression of miR-381 is a favorite prognosis factor and enhances the chemosensitivity of osteosarcoma. Oncotarget. 2016;7(42):68585–68596. doi:10.18632/oncotarget.11861
32. Xu Y, Ohms S, Li Z, et al. Changes in the expression of miR-381 and miR-495 are inversely associated with the expression of the MDR1 gene and development of multi-drug resistance. PLoS One. 2013;8(11):e82062. doi:10.1371/journal.pone.0082062
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.