Back to Journals » Journal of Hepatocellular Carcinoma » Volume 10
Six-Transmembrane Epithelial Antigen of Prostate 4: An Indicator of Prognosis and Tumor Immunity in Hepatocellular Carcinoma
Authors Ju MH, Jang EJ, Kang SH , Roh YH , Jeong JS, Han SH
Received 4 November 2022
Accepted for publication 14 March 2023
Published 20 April 2023 Volume 2023:10 Pages 643—658
DOI https://doi.org/10.2147/JHC.S394973
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr David Gerber
Mi Ha Ju,1 Eun Jeong Jang,2 Sung Hwa Kang,2 Young Hoon Roh,2 Jin Sook Jeong,1,* Song-Hee Han1,*
1Department of Pathology, Dong-A University College of Medicine, Busan, Republic of Korea; 2Department of Surgery, Dong-A University College of Medicine, Busan, Republic of Korea
*These authors contributed equally to this work
Correspondence: Song-Hee Han, Department of Pathology, Dong-A University College of Medicine, 26, Daesingongwon-ro, Seo-gu, Busan, 49201, Republic of Korea, Tel +82-51-240-2863, Fax +82-51-240-7396, Email [email protected]
Purpose: The six-transmembrane epithelial antigen of prostate 4 (STEAP4) has been linked to tumor progression via its involvement in inflammatory responses, oxidative stress, and metabolism. However, STEAP4 has rarely been studied in hepatocellular carcinoma (HCC). We explored STEAP4 expression associated with tumor prognosis to understand its role in tumor biology in HCC.
Patients and Methods: STEAP4 mRNA and protein expressions were primarily analyzed using bioinformatics tools based on The Cancer Genome Atlas database to understand the expression pattern, molecular mechanism, prognostic impact, and association with immune cell infiltration. We further investigated the association between STEAP4 protein expression and clinicopathological parameters and their predictive value in HCC patients using immunohistochemical staining of tissue microarrays.
Results: The expression of STEAP4 mRNA and protein in HCC tissues was significantly lower than in normal liver tissues. Reduced expression of STEAP4 was linked to advanced HCC stages, poor recurrence-free survival (RFS), and overall survival. Furthermore, reduced STEAP4 expression was a significant predictor of worse RFS in univariate and multivariate analyses in the immunohistochemical cohort. GO, KEGG, and GSEA analyses revealed that STEAP4 is related to numerous biological processes and pathways, including drug metabolism, DNA replication, RNA metabolism, and immune response. In terms of the immune system, the decreased level of STEAP4 was correlated with the immunosuppressive microenvironment.
Conclusion: Our data indicated that reduced STEAP4 expression was significantly associated with tumor aggressiveness and poor prognosis, possibly because of its link to various biological processes and induction of HCC immune evasion. Therefore, STEAP4 expression may serve as a potential prognostic biomarker for cancer progression and immunity, as well as a therapeutic target in HCC.
Keywords: immunohistochemistry, bioinformatics, Hepatocellular carcinoma, prognostic factor
Introduction
Hepatocellular carcinoma (HCC) is the most prevalent primary liver carcinoma, accounting for > 80% of all cancers and the third leading cause of cancer-related deaths globally.1,2 According to global cancer statistics in 2020, there have been 905,677 new cases of liver and intrahepatic bile duct cancers and 830,180 liver cancer-related deaths worldwide. Patients with chronic liver disease, such as hepatitis B (HBV) or C virus (HCV), are primarily affected.3 Currently, the decreasing prevalence of HBV and HCV and the rising prevalence of metabolic conditions, such as obesity, diabetes, and nonalcoholic fatty liver disease, appear to have altered the primary risk factors for HCC in many countries.4 Notably, the fundamental essence of the onset and progression of HCC, which requires a multistep process of genetic alterations in hepatocytes through persistent inflammatory stimulation, remains unchanged.5
As tumor immunology research has advanced, a relationship between chronic inflammation and carcinogenesis has been confirmed.6 During chronic liver injury induced by various pathogens and toxins, the liver experiences immunological dysfunction caused by excessive activation of pro-inflammatory cytokines, leading to hepatocarcinogenesis.7,8 The complex relationships between immune cell subsets and effector molecules in the microenvironment are responsible for establishing an immunosuppressive or immune-active milieu that suppresses or promotes HCC progression, respectively. The six-transmembrane epithelial antigen of prostate 4 (STEAP4), also known as the six-transmembrane protein of prostate 2 or tumor necrosis factor-alpha (TNF-α)–induced protein 9, is a member of the STEAP family and has multiple biological functions, including cell proliferation and apoptosis, oxidative stress, and molecular signaling pathways.9 When compared to the other STEAP proteins, STEAP4 has the most extensive enzymatic activity and is involved in these processes by forming homo- or heterodimers. In addition, STEAP4 is involved in inflammatory regulation by modulating iron and copper homeostasis.10 STEAP4 expression has been correlated with inflammatory signaling and the proliferation and progression of prostate cancer, highlighting its critical role in carcinogenesis.11,12 Colitis-associated colon cancer has also been shown to facilitate inflammatory tissue injury resulting from mitochondrial iron dysfunction via STEAP4.13 Furthermore, STEAP4 has been implicated in the mitochondrial iron pathway and has been pharmacologically targeted in human epidermal growth factor receptor 2 (HER2)+ breast cancer.14 The role of STEAP4 in HCC is yet to be determined.
Unfortunately, most HCC cases are diagnosed at unresectable advanced stages, and the prognosis for these patients is poor.15 Sorafenib and regorafenib were the only systemic drugs for advanced HCC prior to the worldwide approval of atezolizumab plus bevacizumab.16,17 Currently, atezolizumab and bevacizumab are the first-line treatments for advanced HCC.18 Despite the efficacy of immune checkpoint inhibitors (ICIs), a large cohort of patients remain unresponsive to ICI.18 Identifying predictive biomarkers is one of the main challenges for proper patient selection. Thus, this study aimed to identify STEAP4-related biological characteristics and their interactions with the tumor microenvironment that influence HCC prognosis to evaluate its utility as a biomarker.
Materials and Methods
Analysis of the Expression of STEAP4 mRNA and Protein Using Bioinformatics Tools
Based on The Cancer Genome Atlas (TCGA) initiatives, the Tumor Immune Estimation Resource (TIMER) database was used to evaluate the differential expression of STEAP4 mRNA across various cancers and associated normal tissues.19 The University of Alabama at Birmingham Cancer data analysis Portal (UALCAN) platform was used to compare mRNA expression and protein levels of STEAP4 between TCGA and The Genotype-Tissue Expression databases and to explore their relationship with clinicopathological features of HCC.20 To determine the genetic alterations and their relationship with mRNA expression, we interactively analyzed multidimensional genomics data for cancer using the cBio Cancer Genomics Portal platform, which included data from over 5000 tumor samples from 20 studies.21 We next analyzed the relationship between STEAP4 mRNA and survival rates, such as recurrence-free survival (RFS) and overall survival (OS), in patients with HCC using the Kaplan–Meier plotter.22 Moreover, we explored the correlated diverse biological functions involving STEAP4 based on enrichment analysis for biological process (BP) and molecular function (MF) of gene ontology (GO) using the ClueGO plug-in in the Cytoscape software. Pathway enrichment analysis was carried out according to the Kyoto Encyclopedia of Genes and Genomes (KEGG),23 . In HCC, we explored the type of genetic mutations that affected STEAP4 expression. To illustrate the immune microenvironment of HCC with STEAP4, we used the TIMER 2.0 database,19 Gene Expression Profiling Interactive Analysis (GEPIA) database,24,25 and Tumor Immune System Interaction Database (TISIDB),26 which provide the immune infiltration levels of multiple tumors in TCGA. Immune infiltration was estimated using CIBERSORT and Estimate methods.
Patients and Sample Collection
The study population comprised 173 HCC patients treated at the Dong-A University Hospital (Busan, Korea) between 2010 and 2013. None of the patients received pre-operative neoadjuvant therapy. Surgical pathology reports and clinical information, including sex, age, and stage, were reviewed. HCC stage was categorized based on the eighth edition of the Cancer Staging Manual of the American Joint Committee on Cancer. The clinicopathological characteristics of the HCC patients are shown in Table 1. RFS was defined as the time interval between the first surgery and the last follow-up visit when the patient did not show any detectable recurrence. The clinical study protocol was approved by the Institutional Review Board of Dong-A University Hospital, Busan, Republic of Korea (DAUHIRB-16-023). The requirement for informed consent from patients was waived because of the use of archived formalin-fixed and paraffin-embedded tumor tissues. The study complies with the Declaration of Helsinki.
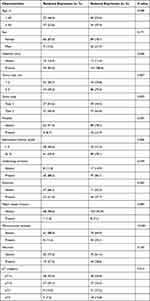 |
Table 1 Clinicopathological Characteristics According STEAP4 Expression in Hepatocellular Carcinoma immunohistochemical Cohort |
Tissue Microarray Construction and Immunohistochemistry Protocol
Tissue microarray (TMAs) were used for immunohistochemical staining (IHC). We reviewed the hematoxylin and eosin-stained slides and selected the most representative areas of the tumors. Three cores of 3 mm diameter from different representative areas of the carcinoma and one from an adjacent non-neoplastic area were reconstructed into a tissue array and arranged into new recipient blocks using a tissue arrayer (Unitma Co., Ltd., Seoul, Korea). IHC staining for STEAP4 was performed using a BenchMark XT automated immunostainer (Ventana Medical Systems Inc., Tucson, AZ, USA) and Ultraview Universal DAB Detection Kit (Ventana Medical Systems Inc.). After deparaffinization, rehydration, and antigen retrieval, diluted primary rabbit polyclonal anti-human STAMP2 antibody (Rosemont, IL, USA, 1:20) was added and incubated.
Scoring System of Immunohistochemical Staining
TMA tissue sections were imaged using a virtual slide scanner (Pannoramic MIDI, 3d Histech, Hungary) at the Neuroscience Translational Research Solution Center (Busan, South Korea) and scored by two pathologists who were blinded to the clinicopathological features. Expression patterns and staining proportions were explored to evaluate STEAP4 expression. The expression pattern of STEAP4 in non-neoplastic hepatocytes was used as a reference point to determine abnormal expression changes in HCC. In molecular biology, protein aggregation is a phenomenon in which intrinsically disordered or misfolded proteins aggregate.27,28 It has negative effects on cellular functions. Thus, reduced expression was determined when ≥ 30% of the tumor cells showed clumped cytoplasmic expression or when < 30% of the tumor cells retained expression.
Statistical Analysis
Statistical analyses were performed using the SPSS software (version 22.0; SPSS, Inc.; Chicago, IL, USA). The relationship between STEAP4 expression and clinicopathological characteristics was assessed using the chi-squared test. RFS based on STEAP4 expression was evaluated using the Kaplan–Meier method with the Log-rank test. Univariate survival analysis with individual covariates and multivariate survival analysis was performed using the Cox proportional hazards regression model to assess whether the expression level of STEAP4 was an independent predictor of disease relapse or survival in patients with HCC. In all tests, statistical significance was defined as p < 0.05.
Results
The mRNA and Protein Expression of STEAP4 in HCC
To determine the expression of STEAP4, the online databases TIMER 2.0 and UALCAN were utilized. PanCancer analysis revealed that STEAP4 expression was significantly lower in HCC tissues than in normal liver tissues (p < 0.001) (Figure 1A). Analysis of expression patterns between HCC and normal liver tissues in UALCAN also showed that the mRNA expression level of STEAP4 was significantly lower in HCC tissues than in normal tissues (p < 0.001) (Figure 1B). We then analyzed the protein expression levels of STEAP4 in patients with HCC using the UALCAN database. The STEAP4 proteomic expression level in HCC tissues was lower than that in normal tissues (p < 0.001) (Figure 1C).
Additionally, we used the cBioPortal database, composed of data from four studies encompassing 1225 cases, to explore the genetic variation of STEAP4 (243 cases in INSERM Nat Genet 2015; 231 cases in AMC, Hepatology 2014; 379 cases in TCGA, PanCancer Atlas; 372 cases in TCGA, Firehose Legacy). As shown in Figure 1D, amplification and mutations were detected in STEAP4 in 1225 samples. The frequency of genetic alterations was 1.6%. The type and rate of STEAP4 alterations varied according to data type (Figure 1D). Amplification is a genetic variation with the highest incidence rate in the TCGA database.
To identify the type of alteration that affects STEAP4 expression in HCC, we explored the relationship of STEAP4 expression with copy number variation, somatic mutations, and methylation. STEAP4 expression was not significantly correlated with copy number variation or mutation type (Figure 1E). However, we found that the methylation level of STEAP4 in HCC was significantly negatively correlated with STEAP4 mRNA expression (Pearson’s r = −0.28, p < 0.001) (Figure 1F).
The Prognostic Significance of STEAP4 Associated with Various Clinicopathologic Characteristics of HCC
We investigated the STEAP4 mRNA expression patterns in a histopathological variant of HCC using the UALCAN platform. STEAP4 expression tended to be lower in conventional HCC than in combined hepatocellular-cholangiocarcinoma cases (Figure 2A). Histological grading of HCC based on a modified Edmondson-Steiner system reflects its biological behavior, which means that higher grades correlate with a worse prognosis. The mRNA expression of STEAP4 was significantly decreased when the grade was higher than that of the normal tissue (Figure 2B). We also explored the correlation between pathologic stage and mRNA expression of STEAP4. STEAP4 mRNA expression level was significantly lower in pathologic tumor stage and clinical stages 3 and 4, compared to stages 1 and 2 (p = 0.05 and p=0.03, respectively) (Figure 2C and 2). Notably, the relatively low mRNA expression level of STEAP4 in HCC was closely related to the advanced status of the tumor. We then used the Kaplan–Meier plotter to determine the survival state of STEAP4 in HCC patients (Figure 2E and 2). Our results revealed that lower mRNA expression was remarkably correlated with poor RFS and OS (p < 0.001) in patients with HCC.
Clinical Significance of STEAP4 Expression in IHC Analysis of HCC Cohort
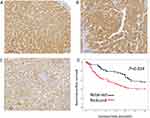
We conducted an IHC analysis of the HCC cohort and compared it with adjacent non-neoplastic tissues. Normal tissue showed diffuse and even cytoplasmic expression (Figure 3A). The IHC analysis of the HCC cohort revealed cytoplasmic clumping (Figure 3B and 3). Reduced expression was found in 114 (62.3%) cases (Figure 3C). Normally, protein folding into a functional shape is an essential process in cell biology.27,28 Failure of protein folding results in abnormal aggregates, leading to loss of function. Thus, the cytoplasmic clumping expression of STEAP4 observed in HCC was interpreted as a reduced expression related to the loss of function. Reduced STEAP4 expression was associated with HCC size ≥ 2 cm (p = 0.007), gross tumor type 2 (p = 0.029), tumor multifocality (p = 0.025), high-grade Edmondson-Steiner grade (p = 0.006), presence of microvascular invasion (p <0.001), and higher pathologic T stage (p = 0.014) (Table 1).
Kaplan–Meier analysis demonstrated that reduced STEAP4 expression was significantly associated with poor RFS (p = 0.034, Figure 3D). In the multivariate analysis, reduced STEAP4 expression (hazard ratio [HR] = 1.739, p = 0.023), higher Edmondson-Steiner grade (HR = 1.975, p = 0.003), higher pathologic T stage (HR = 2.273, p = 0.007), and presence of microvascular invasion (HR = 2.379, p = 0.023) were independent prognostic factors for shorter RFS in HCC (Table 2). These findings suggested that reduced STEAP4 expression influences HCC progression.
 |
Table 2 Univariate and Multivariate Analyses of Recurrence-Free Survival in the Hepatocellular Carcinoma |
STEAP4-Related Biological Pathway
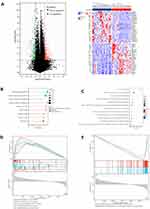
To further illustrate the potential role of STEAP4 in the tumor biology of HCC, we divided the TCGA-LIHC samples into low and high groups based on the median STEAP4 mRNA expression and the differential expression profile was conducted to identify the differentially expressed genes (DEG) according to the criteria of P-value <0.01 and a fold-change (FC) >1. The volcano plot was presented in Figure 4A. The heatmap depicted the top 50 genes that were positively and negatively related to STEAP4 expression in HCC (Figure 4A). We then systematically evaluated the potential biological functions of the DEGs utilizing the GO term and KEGG pathway enrichment analyses. KEGG analysis revealed relationships between different signaling pathways and metabolic processes in cells that control different physiological functions (Figure 4B). According to the GO enrichment analysis, STEAP4 participated in inflammation and cell growth and differentiation, both of which are often associated with cancer progression (Figure 4C). An analysis of GSEA was conducted to determine which biological processes and signaling pathways are enriched for STEAP4. The data showed that STEAP4 with low expression levels was primarily enriched in DNA and RNA replication and repair (Figure 4D). A negative enrichment was found for the genes involved in cytochrome P450 drug metabolism (Figure 4E).
Relation of STEAP4 Expression with the Somatic Mutation of HCC
Genomic analysis was conducted to identify the molecular characteristics associated with various STEAP4 expression levels. As shown in Figure 5A, the top 20 somatic mutations in HCC are summarized by group. There are several hub genes that influence HCC carcinogenesis, such as TP53, CTNNB1, and AXIN1.29,30 In both groups, the tumor suppressor TP53, protein-encoding gene TTN, and proto-oncogene CTNNB1 were significantly mutated. However, TP53 and CTNNB1 were more altered in the low STEAP4 expression group compared to the high STEAP4 expression group. Mutations in TP53 are associated with hepatocarcinogenesis and clinical outcomes.41,42 CTNNB1 and AXIN1 are also linked to the development of HCC through the Wnt signaling pathway. Thus, mutations related to specific molecules were associated with clinical outcomes.
Analysis of the Role of STEAP4 in HCC Tumor Microenvironment
The tumor microenvironment (TME) has been implicated in genetic damage responses.43 The STEAP4 expression in HCC was analyzed to comprehend its relationship with immune cell infiltration. According to the ESTIMATE algorithm, we found a positive correlation between STEAP4 expression and stromal (p = 3.2e-6) and estimate (p = 1.4e-3) scores with an increasing tendency (Figure 5B). The results suggested that STEAP4 expression may be involved in regulating the tumor microenvironment.
Tumor-infiltrating lymphocyte (TIL) is a reliable indicator of a host antitumor immune response and plays a crucial role in the development and spread of tumors. Thus, we examined how it relates to immune cell infiltration and the prognosis of HCC patients. Based on a detailed analysis of TME-infiltrated immune cells in the TIMER database, Treg (p=2.72e-02) and macrophage (p=2.35e-05) were associated with HCC prognosis (Figure 5C). The difference in TIL infiltration between the groups of HCC patients with high and low STEAP4 expression revealed that, of the 21 lymphocyte subsets, 4 TILs (naive B cells, T cells CD4 memory resting, macrophages M1, and resting mast cells) were elevated in the high STEAP4 expression cohort compared to the low expression cohort (Figure 5D). In contrast, T regs and macrophages M0 were noticeably elevated in the low STEAP4 group compared to the high cohort (Figure 5D).
In order to understand the relationship between STEAP4 expression and the abundance of TILs, TIMER2.0 was used. A significant negative correlation was seen between STEAP4 and tumor purity (r = 0.307, p = 5.76e-09), Treg cells (r = −0.244, p = 4.46e-06), macrophages M0 (r = −0.17, p = 1.56e−03), and macrophage M2 (r = −0.218, p = 4.34e-05) (Figure 6A).
We then used the GEPIA2 database to examine the expression of STEAP4 and T cell activity-related gene panels. As shown in Figure 6B, it was negatively correlated with Treg cell signature genes, such as FOXP3, CTLA4, CCR8, and TNFRSF9 (R = -0.19, p = 1e-05), and positively correlated with effective T cell signature gene sets like FCGR3A, FGFBP2, and CX3CR1 (R = 0.091, p = 0.037). As part of the tumor microenvironment, immune checkpoints are also crucial for regulating and directing the immune response. Based on the TISIDB database, we analyzed the correlation between STEAP4 expression and immune checkpoints (ICs) in HCC to estimate the association. There was a negative correlation between STEAP4 expression and PDCD1 (PD-1) (p=0.0232), CTLA4 (p=0.00251), and LAG3 (p=0.00666) (Figure 6C). The findings further support the hypothesis that STEAP4 plays an important role in regulating immunity in TME.
STEAP4 Expression and Therapy Responsiveness
GSEA analysis suggested that STEAP4 expression is correlated with drug metabolism. In consideration of the correlation between tumor progression and drug resistance, we utilized the CARE algorithm to analyze the relationship between STEAP4 and drug resistance. Based on data from the Cancer Cell Line Encyclopedia (CCLE), Genomics of Drug Sensitivity in Cancer (GDSC), and The Cancer Therapeutics Response Portal (CTRP).44 We found that STEAP4 expression is negatively correlated with CARE scores for various compounds (Figure 6D). These findings suggest that STEAP4 expression plays a role in drug resistance. STEAP4 expression and drug metabolism are related, as demonstrated in the GSEA analysis.
Discussion
Based on previous studies, we selected STEAP4 as a target to investigate its role in HCC. As a metalloreductase involved in cellular iron and copper homeostasis, STEAP4 modulates inflammation.10 Previous studies have revealed that STEAP4 is overexpressed and related to invasion and metastasis in various cancers, including prostate, lung, and colon cancers;3,31,32 STEAP4 is also associated with the immune response within the tumor microenvironment.10 However, it is unclear whether STEAP4 affects carcinogenesis and immune reactions in HCC patients. Hence, we analyzed data from multiple databases and a cohort of HCC patients to determine the effect of STEAP4 on HCC. Our analysis revealed that the expression of STEAP4 mRNA and protein levels were lower in HCC tissues compared to non-neoplastic tissues and that the expression pattern identified by IHC was distinct. Prognosis analysis based on the Kaplan–Meier plotter and HCC cohort showed that low expression of STEAP4 mRNA and protein and reduced expression of STEAP4 by the HCC cohort on IHC were associated with poor clinical outcomes. Furthermore, analysis by the histological grade and tumor stage revealed that reduced STEAP4 expression was significantly related to tumor progression. Using multiple databases, we also found that STEAP4 can promote certain immune subsets and proliferation-associated biological processes suggesting that STEAP4 has a significant prognostic value for HCC patients.
Transport of the electron from NADPH to reduce the extracellular Fe3+ to Fe2+ and Cu2+ to Cu1+ is the primary function of STEAP4.10 Thus, STEAP4 is involved in fatty acid and glucose metabolism, inflammation, and oxidative stress through cellular iron and copper homeostasis. STEAP4 appears to be located in the Golgi apparatus network and plasma membrane of cells.33 However, cellular expression patterns and functional roles of STEAP4 in the liver are not well understood. Notably, recent studies have highlighted that hepatic disease is induced by a deficiency in STEAP4, unlike diseases of the prostate and colon. Kim et al found that the downregulation of hepatic STEAP4 mRNA and protein expression was linked to disturbances in fatty acid and glucose metabolism associated with iron accumulation.34 Moreover, it increased the presence of HBV-encoded X protein, which is known to play a pivotal role in the pathogenesis of virus-induced HCC.35 In line with these in vitro results, we demonstrated that decreased STEAP4 mRNA and protein expressions were associated with tumor progression. IHC analysis of HCC tissues, compared with non-neoplastic tissues, showed that STEAP4 protein expression was significantly different showing a clumping pattern. We interpreted this as a reduced expression. This reduced expression was associated with aggressive tumor behavior, such as large tumor size, type 2 gross type, multiple presentations of HCC, microvascular invasion, and a higher pathologic T stage. Furthermore, we demonstrated that reduced STEAP4 levels were an independent prognostic factor for inferior outcomes in HCC.
Given the aggressive behaviors associated with low STEAP4 expression, it was a compelling study to examine underlying factors correlated with poor survival. To identify potential functional changes associated with reduced STEAP4 expression, we performed bioinformatic analysis, including gene ontology (GO) or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, to better understand the molecular mechanisms underlying this relationship. GO analysis showed that the listed biological processes were related to cell development and response to stress, signaling pathways, and immune response. Based on these results, STEAP4 expression appeared to be related to immune response as well as to cell signaling and development. GSEA results showed enrichment in DNA replication, RNA processing, and drug metabolic pathways leading us to speculate that STEAP4 has potential links to drug metabolism as well as DNA and RNA maintenance and regulation. It is crucial to maintain, replicate, and regulate DNA and RNA for tumor proliferation to occur.45 According to Hanahan et al,46 maintaining proliferation signals is one of the most important signals that cell acquired as they progress from normal to malignant growth.
Furthermore, as shown in Figure 5A, TP53, CTNNB1, and AXIN were more altered in the low STEAP4 expression group. TP53, CTNNB1, and AXIN1 are dominant mutational drivers in HCC.29 Cell cycle and apoptosis are controlled by TP53, which is associated with poor clinical outcomes in HCC.36,47 It is also believed that CTNNB1 and AXIN1 promote HCC by aberrantly enhancing Wnt/β-catenin signaling.37,48 Several other studies have also shown that STEAP4 is associated with HCC progression.14,31,38 As mRNA levels increased in prostate cancer, STEAP4 overexpression increased the number and size of prostate cancer cell lines.38 STEAP4 knockdown interferes with the lipopolysaccharide-induced proliferation of prostate cancer cells.31 Orfanou et al suggested that STEAP4 silencing inhibits the proliferation of HER2+ breast cancer cells.14 Recent studies have demonstrated that STEAP4 inhibits the proliferation and metastasis of HCC cells as a tumor suppressor gene.39 Thus, our results demonstrated for the first time that low STEAP4 expression may promote the HCC through its involvement in the cell proliferating pathways, although the specific mechanism needs to be further verified.
Cancer cells are characterized by uncontrolled proliferation. Furthermore, the alteration of the tumor microenvironment is considered a significant component in tumor development.40 In the present study, we examined the changes of immune subsets based on STEAP4 expression level. According to the ESTIMATE algorithm, the stromal score and estimate score were lower in low expression group. It was revealed that tumor purity was greater in the lower expression group, regardless of the degree of immune cell infiltration. Hence, it was speculated that the composition of the immune subsets might be different depending on STEAP4 expression. In our study, we found that higher infiltration levels of Tregs and macrophages were associated with poor prognosis in HCC. We demonstrated that STEAP4 expression negatively influenced Treg and M2 macrophages and positively affected M1 macrophage infiltration. We further explored the relationship between STEAP4 expression and T-cell activity. As expected, exhausted T cell signatures were negatively correlated with STEAP4 expression levels and effective T cell signatures were positively correlated. It has been reported that patients with higher levels of Treg infiltration in HCC have a poor prognosis associated with defective cytotoxic T lymphocytes (CTLs).49,50 Macrophages also have been recognized as an important component of the tumor microenvironment that contributes to the development and progression of cancer.51 Several studies demonstrated that HCC patients with low infiltration of M1 and high infiltration of M2 macrophage levels have a poor prognosis.51,52 Furthermore, we found that immune checkpoints, such as CTLA4, LAG3, and PD-L1, were also inversely correlated with STEAP4 expression in HCC suggesting that STEAP4 may be involved in regulating the TME and immune response in HCC.
Conclusion
In summary, we examined the expression, prognosis, immune cell involvement in TME, and drug sensitivity related to STEAP in HCC patients. Although the molecular mechanisms and signaling pathways of STEAP4 in influencing the composition of the immune subset, immune escape, and tumor progression in HCC require further investigation, we first demonstrated that STEAP4 can be a potential prognostic marker for HCC patients and has considerable immunotherapeutic potential for HCC patients.
Abbreviations
BP, biological process; CCLE, Cancer Cell Line Encyclopedia; CTLs, cytotoxic T lymphocytes; CTRP, Cancer Therapeutics Response Portal; DEG, differentially expressed genes; FC, fold-change; GDSC, Genomics of Drug Sensitivity in Cancer; GEO, Gene Expression Omnibus; GEPIA, Gene Expression Profiling Interactive Analysis; GO, gene ontology; GSEA, Gene Set Enrichment Analysis; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HER2, human epidermal growth factor receptor 2; HLA, human leukocyte antigen; HR, hazard ratio; IC, immune checkpoint; ICI, immune checkpoint inhibitor; IHC, immunohistochemistry; KEGG, Kyoto Encyclopedia of Genes and Genomes; MDSC, myeloid-derived suppressor cells; MF, molecular function; RFS, recurrence-free survival; STEAP4, six-transmembrane epithelial antigen of prostate 4; TCGA, The Cancer Genome Atlas; TIL, tumor-infiltrating lymphocyte; TIMER, Tumor Immune Estimation Resource; TISIDB, Tumor Immune System Interaction Database; TMA, Tissue microarray; TME, tumor microenvironment; TNF-α, tumor necrosis factor-alpha; UALCAN, University of Alabama at Birmingham Cancer data analysis Portal.
Data Sharing Statement
All genetic data were obtained from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/) and Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) databases.
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D1A3B03935135). Additional this research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant No.: 2022R1C1C1003307).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi:10.3322/caac.21660
2. Marengo A, Rosso C, Bugianesi E. Liver cancer: connections with obesity, fatty liver, and cirrhosis. Annu Rev Med. 2016;67(1):103–117. doi:10.1146/annurev-med-090514-013832
3. Ahn JC, Lee YT, Agopian VG, et al. Hepatocellular carcinoma surveillance: current practice and future directions. Hepatoma Res. 2022;8:10. doi:10.20517/2394-5079.2021.131
4. Petrick JL, Florio AA, Znaor A, et al. International trends in hepatocellular carcinoma incidence, 1978–2012. Int J Cancer. 2020;147:317–330.
5. Müller M, Bird TG, Nault JC. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol. 2020;72:990–1002.
6. Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545.
7. Oura K, Morishita A, Tani J, Masaki T. Tumor immune microenvironment and immunosuppressive therapy in hepatocellular carcinoma: a review. Int J Mol Sci. 2021;22:5801.
8. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550.
9. Rocha SM, Barroca-Ferreira J, Passarinha LA, Socorro S, Maia CJ. The usefulness of STEAP proteins in prostate cancer clinical practice. In: Bott SRJ, Ng KL, editors. Prostate Cancer. Brisbane (AU): Exon Publications; 2021.
10. Scarl RT, Lawrence CM, Gordon HM, Nunemaker CS. STEAP4: its emerging role in metabolism and homeostasis of cellular iron and copper. J Endocrinol. 2017;234:R123–R134. doi:10.1530/JOE-16-0594
11. Pihlstrøm N, Jin Y, Nenseth Z, Kuzu OF, Saatcioglu F. STAMP2 expression mediated by cytokines attenuates their growth-limiting effects in prostate cancer cells. Cancers. 2021;13:1579. doi:10.3390/cancers13071579
12. Burnell SEA, Spencer-Harty S, Howarth S, et al. Utilisation of the STEAP protein family in a diagnostic setting may provide a more comprehensive prognosis of prostate cancer. PLoS One. 2019;14:e0220456. doi:10.1371/journal.pone.0220456
13. Xue X, Bredell BX, Anderson ER, et al. Quantitative proteomics identifies STEAP4 as a critical regulator of mitochondrial dysfunction linking inflammation and colon cancer. Proc Natl Acad Sci U S A. 2017;114(45):E9608–E9617. doi:10.1073/pnas.1712946114
14. Orfanou IM, Argyros O, Papapetropoulos A, Tseleni-Balafouta S, Vougas K, Tamvakopoulos C. Discovery and pharmacological evaluation of STEAP4 as a novel target for HER2 overexpressing breast cancer. Front Oncol. 2021;11:608201. doi:10.3389/fonc.2021.608201
15. Giannini EG, Farinati F, Ciccarese F, et al. Prognosis of untreated hepatocellular carcinoma. Hepatology. 2015;61(1):184–190. doi:10.1002/hep.27443
16. Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–390. doi:10.1056/NEJMoa0708857
17. Cheng AL, Kang YK, Chen Z, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a Phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi:10.1016/S1470-2045(08)70285-7
18. Gordan JD, Kennedy EB, Abou-Alfa GK, et al. Systemic therapy for advanced hepatocellular carcinoma: ASCO guideline. J Clin Oncol. 2020;38:4317–4345. doi:10.1200/JCO.20.02672
19. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi:10.1093/nar/gkaa407
20. Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi:10.1016/j.neo.2022.01.001
21. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi:10.1158/2159-8290.CD-12-0095
22. Lánczky A, Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res. 2021;23:e27633. doi:10.2196/27633
23. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi:10.1093/nar/28.1.27
24. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi:10.1093/nar/gkz430
25. Li C, Tang Z, Zhang W, Ye Z, Liu F. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021;49(W1):W242–W246. doi:10.1093/nar/gkab418
26. Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–4202. doi:10.1093/bioinformatics/btz210
27. Invernizzi G, Papaleo E, Sabate R, Ventura S. Protein aggregation: mechanisms and functional consequences. Int J Biochem Cell Biol. 2012;44:1541–1554. doi:10.1016/j.biocel.2012.05.023
28. Scheibel T, Buchner J. Protein aggregation as a cause for disease. Handb Exp Pharmacol. 2006;172:199–219.
29. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6.
30. Julie G, Domitile C, Jean-frederic B, et al. Hepatocellular carcinoma immune landscape and the potential of immunotherapies. Front Immunol. 2021;12:655697.
31. Li W, Yin X, Yan Y, Liu C, Li G. STEAP4 knockdown inhibits the proliferation of prostate cancer cells by activating the cGMP-PKG pathway under lipopolysaccharide-induced inflammatory microenvironment. Int Immunopharmacol. 2021;101(Pt B):108311.
32. Chen H, Xu C, Yu Q, et al. Comprehensive landscape of STEAP family functions and prognostic prediction value in glioblastoma. J Cell Physiol. 2021;236:2988–3000.
33. Yoo SK, Cheong J, Kim HY. STAMPing into mitochondria. Int J Biol Sci. 2014;10:321–326.
34. Kim HY, Kwon WY, Park JB, et al. Hepatic STAMP2 mediates recombinant FGF21-induced improvement of hepatic iron overload in nonalcoholic fatty liver disease. FASEB J. 2020;34:12354–12366.
35. Kim HY, Cho HK, Yoo SK, Cheong JH. Hepatic STAMP2 decreases hepatitis B virus X protein-associated metabolic deregulation. Exp Mol Med. 2012;44:622–632.
36. Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: i have a DREAM. Cell Death Differ. 2018;25:114–132.
37. Krutsenko Y, Singhi AD, Monga SP. β-catenin activation in hepatocellular cancer: implications in biology and therapy. Cancers. 2021;13:1830.
38. Korkmaz CG, Korkmaz KS, Kurys P, et al. Molecular cloning and characterization of STAMP2, an androgen-regulated six transmembrane protein that is overexpressed in prostate cancer. Oncogene. 2005;24:4934–4945.
39. Tang Y, Wang Y, Xu X, Sun H, Tang W. STEAP4 promoter methylation correlates with tumorigenesis of hepatocellular carcinoma. Pathol Res Pract. 2022;233:153870.
40. Xia Y, Brown ZJ, Huang H, Tsung A. Metabolic reprogramming of immune cells: shaping the tumor microenvironment in hepatocellular carcinoma. Cancer Med. 2021;10:6374–6383.
41. Chen R, Zhao M, An Y, Liu D, Tang Q, Teng G. A prognostic gene signature for hepatocellular carcinoma. Front Oncol. 2022;12:841530.
42. Gao Q, Zhu H, Dong L, et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179:561–577.
43. Giraldo NA, Sanchez-Salas R, Peske JD, et al. The clinical role of the TME in solid cancer. Br J Cancer. 2019;120(1):45–53.
44. Jiang P, Lee W, Li X, et al. Genome-scale signatures of gene interaction from compound screens predict clinical efficacy of targeted cancer therapies genome-scale signatures of gene interaction from compound screens predict clinical efficacy of targeted cancer therapies. Cell Syst. 2018;6:343–354.
45. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674.
46. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46.
47. Liu J, Ma Q, Zhang M, et al. Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: evidence from a systematic review and meta-analysis. Eur J Cancer. 2012;48(15):2328–2338.
48. Mo Z, Wang Y, Cao Z, et al. An integrative analysis reveals the underlying association between CTNNB1 mutation and immunotherapy in hepatocellular carcinoma. Front Oncol. 2020;10:853.
49. Gao Q, Qiu SJ, Fan J, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol. 2007;25:2586–2593.
50. Tu JF, Ding YH, Ying XH, et al. Regulatory T cells, especially ICOS+ FOXP3+ regulatory T cells, are increased in the hepatocellular carcinoma microenvironment and predict reduced survival. Sci Rep. 2016;6:35056.
51. Ding W, Tan Y, Qian Y, et al. Clinicopathologic and prognostic significance of tumor-associated macrophages in patients with hepatocellular carcinoma: a meta-analysis. PLoS One. 2019;14:e0223971.
52. Dong P, Ma L, Liu L, et al. CD86+/CD206+, Diametrically Polarized Tumor-Associated Macrophages, Predict Hepatocellular Carcinoma Patient Prognosis. Int J Mol Sci. 2016;17:320.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.