Back to Journals » Journal of Inflammation Research » Volume 17
Shared Immune Associations Between COVID-19 and Inflammatory Bowel Disease: A Cross-Sectional Observational Study in Shanghai, China
Authors Li S , Zhang F, Lin R, Sun Q, Qu L, Zhong L
Received 14 November 2023
Accepted for publication 20 March 2024
Published 27 March 2024 Volume 2024:17 Pages 1929—1940
DOI https://doi.org/10.2147/JIR.S449746
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Adam D Bachstetter
Shan Li,1,* Fengdi Zhang,2,* Ritian Lin,1 Qinjuan Sun,1 Lihong Qu,2 Lan Zhong1
1Department of Gastroenterology, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, People’s Republic of China; 2Department of Infectious Diseases, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lan Zhong, Department of Gastroenterology, Shanghai East Hospital, Tongji University School of Medicine, 150, Jimo Road, Pudong New Area, Shanghai, 200120, People’s Republic of China, Email [email protected] Lihong Qu, Department of Infectious Diseases, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, People’s Republic of China, Email [email protected]
Purpose: The rapid global spread of the SARS-CoV-2 Omicron variant introduces a novel complication: the emergence of IBD (inflammatory bowel disease)-like ulcers in certain patients. This research delves into this new challenge by juxtaposing the clinical manifestations and genetic expression patterns of individuals affected by the Omicron variant of COVID-19 with those diagnosed with IBD. It aims to decode the link between these conditions, potentially shedding light on previously unexplored facets of COVID-19 pathophysiology. This investigation emphasizes gene expression analysis as a key tool to identify wider disease correlations and innovative therapeutic avenues.
Patients and Methods: From March to December 2022, patients with SARS-CoV-2 Omicron infection and inflammatory bowel disease and healthy controls were recruited in Shanghai East Hospital, Shanghai, China. The epidemiological and clinical characteristics of the patients were compared. Four RNA sequencing datasets (GSE205244, GSE201530, GSE174159, and GSE186507) were extracted from the Gene Expression Omnibus database to detect mutually differentially expressed genes and common pathways in patients with SARS-CoV-2 infection and inflammatory bowel disease.
Results: Compared to patients with active inflammatory bowel disease, patients with SARS-CoV-2 infection were more likely to have elevated interferon-α levels and an increased lymphocyte count and less likely to have high interleukin-6, tumor necrosis factor-α, and C-reactive protein levels and an elevated neutrophil count. A total of 51 common differentially expressed genes were identified in the four RNA-sequencing datasets. Enrichment analysis suggested that these genes were related to inflammation and the immune response, especially the innate immune response and nucleotide oligomerization domain-like receptor signaling pathway.
Conclusion: The inflammation and immune-response pathways in COVID-19 and inflammatory bowel disease have several similarities and some differences. The study identifies the NLR signaling pathway’s key role in both COVID-19 and IBD, suggesting its potential as a target for therapeutic intervention and vaccine development.
Keywords: viral-induced inflammation, omicron infection, RNA sequencing, gene expression in infection, inflammatory bowel disease
Introduction
Coronavirus disease 2019 (COVID-19) is a highly contagious respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has spread worldwide rapidly over the past 4 years.1 After infection with SARS-CoV-2, some patients have been reported to develop long-term symptoms or health problems, a phenomenon known as Long-COVID.2 Long COVID, impacting at least 10% of individuals infected by SARS-CoV-2, presents a compelling case for further investigation due to its extensive symptomatology affecting multiple organ systems. Gastrointestinal symptoms of Long COVID are associated with persistent SARS-CoV-2 replication in the intestinal mucosa, including nausea, abdominal pain, loss of appetite, and constipation, with significant changes in the composition of the gut microbiota.3,4 Interestingly, during the COVID-19 epidemic in Shanghai, some patients who underwent colonoscopy for gastrointestinal symptoms following COVID-19 were found to have inflammatory bowel disease (IBD)-like ulcers. Similar cases have also been reported by other researchers.5
IBD is a group of heterogenous, chronic, non-specific intestinal inflammatory diseases with elusive etiology and pathogenesis, clinically classified as Crohn’s disease (CD) and ulcerative colitis (UC). UC is confined to the colon and can lead to ulceration, severe bleeding, toxic megacolon, and fulminant colitis.6 CD can involve any part of the digestive tract and is characterized by transmural inflammation that can lead to complications such as fibrotic strictures, fistulas, and abscesses.7 Although potential differences between UC and CD have been observed, such as differential enrichment of immune cell subpopulations and genetic variants (eg, NOD and PTPN22), the pathogenesis of both diseases has been linked to disruption of the epithelial barrier, innate immune response, and dysregulation of adaptive immunity.8 To date, certain viruses have been demonstrated to be linked to IBD. For example, phages and specific factors derived from the virome can damage intestinal barrier; Enterovirus B and Picornaviridae are implicated in intestinal inflammation; and Pseudomonas aeruginosa affects innate immunity.9 Nonetheless, limited research has explored the connection between COVID-19 and IBD. Given that IBD-like ulcers have been observed in the colon of patients with COVID-19, it’s imperative to investigate the association between these two diseases. SARS-CoV-2 infection leads to elevated levels of various pro-inflammatory markers, such as tumor necrosis factor-α (TNF-α) and various interleukins (IL-6, IL-1, and IL-17), that can trigger a cytokine storm. Similar phenomena are also observed in IBD, suggesting similarities in the patterns of immune activation between these two diseases. Therefore, the specific relationship between the immune response induced by SARS-CoV-2 infection and IBD needs further investigation.
This cross-sectional, observational study aimed to compare the immune response induced by SARS-CoV-2 Omicron variant infection and IBD at different stages of the disease by analyzing clinical data and transcriptome sequencing. Patients with COVID-19 or IBD were also compared with healthy controls. Further, we investigated the potential immune association between these two diseases. Our findings may help identify potential therapeutic targets for the treatment of these diseases.
Materials and Methods
Study Design and Patient Enrollment
This study was conducted at Shanghai East Hospital in Shanghai, China, from March 2022 to December 2022. A total of 199 patients with different stages of SARS-CoV-2 Omicron variant infection (120 with primary infection, 14 with reinfection, and 65 convalescent patients), 98 patients with IBD, and 60 healthy controls were enrolled. The inclusion criteria were as follows: (1) Sufficient clinical information (medical history, clinical manifestations, physical signs, and laboratory examinations) available from medical records; (2) Diagnosis of COVID-19 according to the diagnostic criteria in the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 9);10 or assessment of disease activity of IBD using the Crohn’s Disease Activity Index (CDAI) for CD11 and the modified Mayo score for patients with UC.6 Clinical remission was defined as CDAI score <150 or modified Mayo score <2 for UC and CD, respectively. Volunteers who underwent health examinations at the Health Examination Center during the same period served as healthy controls. Patients with incomplete clinical data were excluded. In accordance with our inclusion and exclusion criteria, we excluded 22 participants who did not meet the requirements. Demographic information, epidemiological data, and laboratory results of all patients were collected from the hospital’s electronic medical record system.
Clinical and Laboratory Assessments
Cytokines were detected by ”cytokine combined detection kit (Jiangxi Saiji Biotechnology Co., LTD) and Fluorescence Flow Cytometry (BriCyte E6, Mindray Medical Devices Co., LTD). Blood count and C-reactive protein (CRP) were measured using a haematology analyzer (Sysmex XN-3000, Sysmex Co., Japan). Colonoscopy was performed using a CF-HQ2901 electronic colonoscope (Olympus Co., Japan).
Analyses of Public Gene Expression Profile Data
The gene expression profiles (raw count) processed from RNA-seq of patients with COVID-19 (GSE205244, GSE201530) and patients with IBD (GSE174159, GSE186507) were collected from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Among them, the GSE201530 dataset was derived from peripheral blood mononuclear cell (PBMC) samples of 8 healthy individuals and 47 patients with primary SARS-CoV-2 Omicron infection. The GSE205244 dataset was derived from PBMC samples of 17 patients with primary SARS-CoV-2 Omicron infection and 39 patients with reinfection. The GSE174159 dataset was derived from intestinal biopsy samples of three patients with active CD, three patients with CD in remission, and five healthy individuals. The GSE186507 dataset was derived from blood samples of 68 patients with active CD, 288 patients with CD in remission, 44 patients with active UC, 334 patients with UC in remission, and 209 healthy controls.
Differentially expressed gene (DEG, |log2[fold change (FC)]| > log2 [1.5] [FC > 1.5 or FC < 2/3] and P < 0.05) analysis was performed using DESeq2 version 1.28.112 in R version 4.0.2 (https://www.R-project.org/). The intersection of DEGs under different conditions was demonstrated using VennDiagram version 1.7.313 and UpSetR version 1.4.014 in R. The enrichment analysis of these genes was performed using Metascape version 3.5 (https://metascape.org/gp/index.html).15 A schematic representation of the entire workflow in this study is shown in Figure 1.
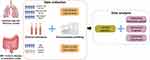 |
Figure 1 Schematic diagram of the workflow (produced using FigDraw). |
Statistical Analysis
Continuous data were reported as the mean ± standard deviation (SD) or median and interquartile range (IQR). The Student’s t-test or Wilcoxon rank-sum test was used to compare groups, as appropriate. Categorical data were reported as frequencies and percentages. The chi-squared test or Fisher’s exact test was used to compare groups, as appropriate. R Software version 4.1.3 (R Foundation for Statistical Computing, Vienna, Austria; https://www.R-project.org/) was used to conduct all statistical analyses. All analyses were two-sided, and p-values < 0.05 were considered statistically significant.
Results
Demographic and Clinical Characteristics of Participants
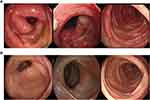
The 199 patients with SARS-CoV-2 Omicron variant infection included in the study had a median age of 50 (interquartile range [IQR], 12 to 96) years, of whom 37% (n = 73) were female and 63% (n = 126) were male. Baseline characteristics of all patients, including age, sex, vaccination history, disease duration, clinical manifestations, comorbidities, laboratory test results, and medical treatment, are shown in Table 1. The main comorbidities were hypertension and diabetes mellitus. The most common symptoms in patients with primary Omicron infection were cough (29%, 35/120), expectoration (22%, 26/120), sore throat (27%, 32/120), myalgia (24%, 20/120), and headache (15%, 12/120). As shown in Table 1, reinfected patients were significantly older, with a median age of 61.5 years and male predominance (71%, 10/14). One of the patients, a 30-year-old man, developed abdominal pain and diarrhea after the SARS-CoV-2 infection, and colonoscopy revealed diffuse edema of the entire colon with multiple UC-like ulcers (Figure 2A). One month later, the patient’s SARS-CoV-2 DNA load was below the limit of detection, and a repeat colonoscopy showed mucosal healing (Figure 2B).
 |
Table 1 Characteristics of Patients Infected with the Omicron Variant |
Sixty healthy controls were enrolled, with a median age of 47.5 (IQR, 33.75 to 59.25) years, of whom 62% were men (Table 2), and the majority had inflammatory cytokine levels within the normal ranges.
 |
Table 2 Univariate Analysis Comparing Healthy Individuals with Patients with Primary SARS-CoV-2 Omicron Infection |
A total of 98 patients with IBD (47 with CD, 51 with UC) were enrolled, with a median age of 44 (IQR, 31 to 58.75) years, of whom 43% (n = 42) were women and 57% (n = 56) were men. Sixty patients (61%) with IBD were in remission, of whom 29 had CD and 31 had UC, and 38 patients were in the active stage, of whom 18 had CD and 20 had UC. Anemia was observed in 37% of patients (n = 14) with active IBD, which was related to chronic gastrointestinal bleeding and malnutrition (Additional file 1). Compared with patients in remission, patients with active disease were significantly (all p<0.001) more likely to have elevated TNF-α, IL-6, and CRP levels.
Blood Levels of Inflammatory Markers in Patients with SARS-CoV-2 Infection, IBD, and Healthy Controls
Compared with the healthy controls, the proportion of patients with elevated IL-6, IL-10, interferon (IFN)-α, neutrophil, and CRP levels were significantly higher among those with primary SARS-CoV-2 infection, which were 63% (p < 0.001), 23% (p < 0.001), 37% (p < 0.001), 18% (p = 0.01), and 24% (p = 0.001), respectively, and 44% had a lower lymphocyte count (Table 2). In our study, we have thoughtfully categorized IBD into two principal stages for analysis: active and remission. Patients with SARS-CoV-2 infection showed significantly higher levels of IFN-α (>8.50 pg/mL; 34%, p < 0.001) and lower levels of lymphocytes (<1.1 x 109/L; 43%, p = 0.03), whereas the proportions of patients with increased IL-6 (>5.30 pg/mL; 61%, p = 0.014), TNF-α (>4.60 pg/mL; 5%, p < 0.001), neutrophil (≥ 6.3 × 109/L; 18%, p = 0.001), and CRP (>10 mg/L; 21%, p < 0.001) levels was less than in patients with active IBD (Table 3). Compared with patients with IBD remission, elevated levels of IL-10, IL-6, and IFN-α were found in 22% (p = 0.003), 61% (p = 0.001), and 34% (p < 0.001) patients with SARS-CoV-2 infection respectively, and a lower percentage of patients with elevated neutrophil counts (18%, p < 0.001) was found in the same population (Additional file 2).
 |
Table 3 Univariate Analysis Comparing Patients with SARS-CoV-2 Omicron Infection with Patients with Active Inflammatory Bowel Disease |
Patients with SARS-CoV-2 reinfection were mostly asymptomatic, with 7% (1/14, p = 0.034) being positive for IFN-α, and none had elevated CRP levels compared with those with primary infection (Additional file 3). Furthermore, there was no significant difference in other inflammatory markers between the patients with SARS-CoV-2 reinfection and healthy controls except for increased IL-6 levels (50%, 7/14, p = 0.03) (Additional file 4).
Identification and Enrichment Analysis of Differentially Expressed Genes in Patients with SARS-CoV-2 Omicron Infection and IBD
Given the similarities in the inflammatory immune responses between IBD and SARS-CoV-2 Omicron infection, we investigated whether SARS-CoV-2 Omicron infection and IBD had common molecular mechanisms. For this purpose, we used several published gene expression profiles of patients with SARS-CoV-2 Omicron infection and patients with IBD. Initially, we screened 397 DEGs (131 up-regulated and 261 down-regulated) in patients with primary SARS-CoV-2 Omicron infection (control, n = 17) vs patients with SARS-CoV-2 Omicron reinfection (n = 39) (GSE205244 – PBMC). We then screened 433 DEGs (383 up-regulated and 45 down-regulated) in healthy controls (control, n = 8) and patients with primary SARS-CoV-2 Omicron infection (n = 47) (GSE201530 – PBMC). Similarly, 57 DEGs (26 up-regulated and 31 down-regulated) were identified in the patients with active CD (n = 68) compared with healthy controls (control, n = 209) and 399 DEGs (388 up-regulated and 11 down-regulated) in the patients with active UC (n = 44) and healthy subjects (control, n = 209) (GSE186507 – PBMC). In addition, considering the tissue specificity of inflammatory bowel mucosa in IBD, we further collected expression profiles from intestinal biopsy specimens and screened 1003 DEGs (668 up-regulated and 335 down-regulated) in the active CD (n = 3) and healthy controls (control, n = 5) (GSE174159 – intestine). We described the common DEGs between these datasets using UpSet and Venn plots (Figure 3A). Our analyses identified 172 common DEGs between patients with primary SARS-CoV-2 Omicron infection (control) and patients with SARS-CoV-2 Omicron reinfection. However, only 24 common DEGs were identified from the two sets among patients with active UC (PBMC), active CD (PBMC), and active CD (intestine), suggesting tissue-specificity and different mechanisms between UC and CD. A total of 51 shared genes (present in at least one DEG set of patients with SARS-CoV-2 Omicron and patients with IBD) were identified as DEGs common to the two diseases.
In addition, patients with CD in remission had eight DEGs (two up-regulated and six down-regulated) in PBMCs that differed from those of healthy controls, whereas patients with UC in remission had only four DEGs (one up-regulated and three down-regulated) in PBMCs that differed from those of healthy controls (GSE186507) (data not shown). This suggests that the gene expression profile in the circulation of patients with IBD in remission is similar to that of healthy individuals.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed to investigate the potential biological functions of the shared DEGs.16,17 Figure 3B shows the enrichment of common DEGs between patients with primary infection and the reinfection with SARS-CoV-2 Omicron was consistent with the clinical results, including a stronger immune response to the virus in the primary infection, especially the IFN response. Between the SARS-CoV-2 Omicron variant infection and IBD, GO analysis showed that shared DEGs were significantly involved in the intrinsic immune response, positive regulation of programmed cell death, and positive and negative regulation of the immune response (Figure 3C). Further, KEGG pathway enrichment analysis showed that shared DEGs were involved in the NOD-like receptor (NLR) signaling pathway, viral protein-cytokine, and cytokine receptor interactions. (Figure 3D). The heat map shows the names of shared DEGs (Figure 3E). These DEGs are implicated in functional enrichment associated with inflammation and immune response.
Discussion
SARS-CoV-2 infects humans by binding its spike protein to the angiotensin-converting enzyme 2 (ACE2) receptor in the mucosa. ACE2 is expressed in intestinal epithelial cells, and its expression in the small intestine is higher than that in the colon.18 Consequently, approximately 5–20% patients with SARS-CoV-2 infection develop gastrointestinal symptoms, intestinal injuries, and even intestinal IBD-like ulcers.3,5 Although the etiologies of COVID-19 and IBD are different, there are some similarities in their response to treatments. Recent studies have shown that patients with IBD have a decreased risk of developing COVID-19. This may be due to previous anti-TNF-α treatment in patients with IBD; anti-TNF-α antibody is associated with lower hospitalization rates and less severe COVID-19 symptoms.19,20 Patients with IBD or COVID-19 develop inflammation associated with a dysregulated immune response, and classic treatments for IBD, such as corticosteroids, tofacitinib, and anti-TNF-α therapy, are also effective in severe cases of COVID-19; however, the underlying mechanisms remain unclear.19,21–23
Although SARS-CoV-2 replicates in patients with reinfection, it generally causes only a mild inflammatory response that does not differ much from that in healthy people. The proportion of patients with elevated IL-6 levels exceeded 50% in the patients with SARS-CoV-2 infection and those with active IBD. The increase in TNF-α level was more pronounced in patients with IBD, whereas the increase in IFN-α level was the main factor involved in the patients with SARS-CoV-2 infection. Furthermore, we compared the transcriptomic data of patients with IBD and patients with COVID-19, demonstrating that the two diseases share a similar innate immune response and NLR signaling pathways from the perspective of gene expression regulation.
Accumulating evidence suggests that tissue damage in these two disorders is due to a dynamic interplay between immune cells and non-immune cells, which is mediated by cytokines produced within the inflammatory microenvironment. Moreover, cytokines link infections with an antiviral immune response and also bridge the innate and adaptive arms of the immune response.24 For SARS-CoV-2 infection, as with other viral infections, the type I IFN response is the most important component of innate antiviral immunity, resulting in the rapid induction of antiviral proteins in response to viral infection.25 This study showed a significant increase in IFN-α levels in the patients with SARS-CoV-2 Omicron variant infection, which was further confirmed with transcriptomic analysis. However, in cases of severe COVID-19, a type I IFN response coexists with TNF/IL-1β-driven inflammation, and more severe inflammation-driven progressive injury may occur without intervention.25 The type I IFN canonical and noncanonical signaling partially relies on a high level of signal transducer and activator of transcription 1 (STAT1) or phosphorylated STAT1 (pSTAT1).26 These findings are consistent with our transcriptomics data. Levels of other cytokines, such as IL-6, are generally elevated in critically ill patients with COVID-19. IL-6 levels are also elevated in serum and intestinal tissues of patients with CD and correlate with clinical disease activity and inflammation severity, signaling through the Janus kinase (JAK)/STAT pathways.27 IFN-γ/TNF-α can cooperate to dose-dependently kill IECs via the CASP8-JAK1/2-STAT1 module.28 In human monocytes, a synergistic interaction between TNF-α and IFN triggers the activation of the JAK/STAT1/IFR1 axis. Lung cell death is proportional to TNF-α and IFN concentrations.29 Our findings show that STAT1 was elevated in the patients with COVID-19 and those with IBD. This could explain why corticosteroids and tofacitinib could be used to treat these two distinct diseases. In patients with COVID-19, corticosteroids inhibit the pathologic interferon response of monocytes by down-regulating STAT1, and tofacitinib prevents IL-mediated hyperinflammation by blocking the JAK/STAT pathway.26,30 Our findings and those of other studies suggest that currently available medications for IBD do not aggravate or increase the rate of SARS-CoV-2 infection, and patients with IBD can continue to receive IBD-related medications during the pandemic.31
While the peak of COVID-19 in China and other countries may have subsided, the ramifications of the pandemic, such as Long COVID, continue to persist, proving to be enduring and widespread in their impact. Some viruses have been shown to have a detrimental effect on intestinal barrier functions, which contribute to the development of IBD.32 However, it remains unclear whether COVID-19 patients will develop IBD. Activation of the NLR signaling pathway in both COVID-19 and IBD is another novel finding of our study. The NLR pathway is responsible for detecting various pathogens and generating innate immune responses, driving the activation of pathways such as nuclear factor-kappa B and mitogen-activated protein kinase, and playing a role in inflammatory responses.33 NLRs can interact with receptor-interacting protein 2 (Rip2) kinase. This interaction through CARD-CARD binding leads to the activation of pathways such as NF-κB or MAPK, resulting in the secretion of pro-inflammatory cytokines like IL-6, IL-1β, and TNF-α. Additionally, NLRs can mediate autophagy by interacting with ATG16L and participate in recognizing damage-associated molecular patterns (DAMPs) produced by cell damage.
NOD-like receptor family pyrin containing 3 (NLRP3), a member of the NLR family, operates in a canonical (inflammasome-dependent) or noncanonical (inflammasome-independent) manner. In its canonical role, NLRP3 inflammasomes convert procaspase-1 into its active form, caspase-1, while in its noncanonical role, they activate procaspase-11.34 The NLRP3 inflammasome is hyperactivated in cytokine storms induced by COVID-19,35 and NLRP3 is also up-regulated in the intestinal mucosa of patients with active IBD. The activation of the NLRP3 inflammasome plays a key role in the initiation of IBD. The Never in Mitosis A-related kinase 3, an important component of the NLRP7 inflammasome in macrophages, interacts with NLRP3 to affect IBD through pyroptosis.36,37 Based on the above results and our bioinformatics analysis, NLR signaling may be an important intersection between COVID-19 and IBD and may provide potential targets for the treatment of both diseases. Actually, some NLRP3 inhibitors have been proven the value of therapeutic interventions. It has been proven that glyburide can inhibit NLRP3 activation and IL-1β secretion induced by lipopolysaccharide in bone marrow-derived macrophages.38 Some natural product inhibitors such as oridonin and parthenolide can inhibit the NF-κB pathway,39,40 and thus attenuate lung inflammation induced by SARS-CoV infection and alleviate IBD.41,42
Furthermore, NLRs play a significant role in vaccine development, especially through their involvement in adjuvant action. Before the discovery of NLRs, vaccination strategies extensively used molecules now recognized as NLR ligands. For instance, bacterial cell wall preparations containing peptidoglycan, known to activate NOD1 and NOD2, and substances like aluminum hydroxide, shown to activate the NLRP3 inflammasome, have been used as adjuvants.43,44 This new understanding of NLRs in adjuvant action provides valuable insights for developing vaccines, highlighting their role in enhancing vaccine efficacy through specific immune pathway activation.
In discussing the limitations of our study, several points warrant attention. Firstly, the research was carried out at a solitary institution in Shanghai, China. It’s plausible that our results may not directly translate to other populations or regions characterized by distinct genetic backgrounds, healthcare frameworks, and environmental influences. Secondly, while our study identifies potential therapeutic targets that hold promise for treatment strategies, the path from research discovery to clinical application is fraught with challenges. The transition of these findings into practical treatments for patients requires a robust body of further research. This entails conducting comprehensive clinical trials aimed at validating the efficacy and safety of these targets.
Conclusion
In conclusion, this study compared patients with SARS-CoV-2 Omicron variant infection, patients with IBD, and healthy controls and described the clinical and transcriptomic features of SARS-CoV-2 infection and IBD. Our findings highlight the shared association of COVID-19 and IBD with immune-related pathways, particularly the innate immune response and NLR signaling pathway, and offer fresh insights into the use of existing drugs as well as the potential development of new therapeutic targets for both diseases.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
The study was approved by the Medical Ethics Committee of Shanghai East Hospital (Research Approval No. 277, 2022) and was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants.
Acknowledgments
The authors thank Yibin Guo for his guidance on statistical analysis, and Hongzhang Xue for his guidance on Bioinformatics analysis.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was funded by the General Program of the National Natural Science Foundation of China (Grant No. 82270639), the Scientific research project of the Shanghai Municipal Health Committee (Grant No. 202240001), the Specialty Feature Construction Project of Shanghai Pudong New Area Health Commission (Grant No. PWZzb2022-05), Technology Development Project of Pudong Science, Technology and Economic Commission of Shanghai (Grant No. PKJ2021-Y08), Key Disciplines Group Construction Project of Shanghai Pudong New Area Health Commission (Grant No.PWZxq2022-06), Medical discipline Construction Project of Pudong Health Committee of Shanghai (Grant No.PWYgf2021-02) and Peak Disciplines (Type IV) of Institutions of Higher Learning in Shanghai. The authors received research funding for statistical analysis and publishing.
Disclosure
The authors declare that they have no competing interests.
References
1. Koelle K, Martin MA, Antia R, et al. The changing epidemiology of SARS-CoV-2. Science. 2022;375(6585):1116–1121. doi:10.1126/science.abm4915
2. Al-Aly Z, Topol E. Solving the puzzle of long covid. Science. 2024;383(6685):830–832. doi:10.1126/science.adl0867
3. Garg M, Christensen B, Lubel JS. Gastrointestinal ACE2, COVID-19 and IBD: opportunity in the face of tragedy? Gastroenterology. 2020;159(4):1623–1624.e3. doi:10.1053/j.gastro.2020.04.051
4. Zollner A, Koch R, Jukic A, et al. Postacute COVID-19 is characterized by gut viral antigen persistence in inflammatory bowel diseases. Gastroenterology. 2022;163(2):495–506.e8. doi:10.1053/j.gastro.2022.04.037
5. Rutigliani M, Bozzo M, Barberis A, et al. Case report: a peculiar case of inflammatory colitis after SARS-CoV-2 infection. Front Immunol. 2022;13:849140. doi:10.3389/fimmu.2022.849140
6. Ordás I, Eckmann L, Talamini M, et al. Ulcerative colitis. Lancet. 2012;380(9853):1606–1619. doi:10.1016/S0140-6736(12)60150-0
7. Torres J, Mehandru S, Colombel J-F, et al. Crohn’s disease. Lancet. 2017;389(10080):1741–1755. doi:10.1016/S0140-6736(16)31711-1
8. Chang JT. Pathophysiology of inflammatory bowel diseases. N Engl J Med. 2020;383(27):2652–2664. doi:10.1056/NEJMra2002697
9. Tun HM, Peng Y, Massimino L, et al. Gut virome in inflammatory bowel disease and beyond. Gut. 2024;73(2):350–360. doi:10.1136/gutjnl-2023-330001
10. National Health Commission of the People’s Republic of China. Diagnosis and treatment plan for novel coronavirus pneumonia (trial version 9). Int J Epidemiol Infect Dis. 2022;49:73–80. Chinese, Abstract in English.
11. Harvey RF, Bradshaw JM. A simple index of Crohn’s-disease activity. Lancet Lond Engl. 1980;1(8167):514. doi:10.1016/S0140-6736(80)92767-1
12. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8
13. Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinf. 2011;12(1):35. doi:10.1186/1471-2105-12-35
14. Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. In: Hancock J, editor. Bioinformatics. Vol. 33. 2017:2938–2940.
15. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
16. Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi:10.1093/nar/28.1.27
17. Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(D1):D1049–1056. doi:10.1093/nar/gku1179
18. Verstockt B, Verstockt S, Abdu Rahiman S, et al. Intestinal receptor of SARS-CoV-2 in inflamed IBD tissue seems downregulated by HNF4A in ileum and upregulated by interferon regulating factors in colon. J Crohn's Colitis. 2021;15(3):485–498. doi:10.1093/ecco-jcc/jjaa185
19. Kokkotis G, Kitsou K, Xynogalas I, et al. Systematic review with meta-analysis: COVID-19 outcomes in patients receiving anti-TNF treatments. Aliment Pharmacol Ther. 2022;55(2):154–167. doi:10.1111/apt.16717
20. Tripathi K, Godoy Brewer G, Thu Nguyen M, et al. COVID-19 and outcomes in patients with inflammatory bowel disease: systematic review and meta-analysis. Inflamm Bowel Dis. 2022;28(8):1265–1279. doi:10.1093/ibd/izab236
21. Guimarães PO, Quirk D, Furtado RH, et al. Tofacitinib in patients hospitalized with Covid-19 pneumonia. N Engl J Med. 2021;385:406–415.
22. Ferrarini A, Vacca A, Solimando AG, et al. Early administration of tofacitinib in COVID‐19 pneumonitis: an open randomised controlled trial. Eur J Clin Invest. 2023;53(2):e13898. doi:10.1111/eci.13898
23. Rubin DT, Modesto I, Vermeire S, et al. Worldwide post‐marketing safety surveillance experience with tofacitinib in ulcerative colitis. Aliment Pharmacol Ther. 2022;55(3):302–310. doi:10.1111/apt.16619
24. Shen S, Gong M, Wang G, et al. COVID-19 and Gut Injury. Nutrients. 2022;14(20):4409. doi:10.3390/nu14204409
25. Lee JS, Park S, Jeong HW, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020;5(49):eabd1554. doi:10.1126/sciimmunol.abd1554
26. Jeong H-W, Lee JS, J-H K, et al. Corticosteroids reduce pathologic interferon responses by downregulating STAT1 in patients with high-risk COVID-19. Exp Mol Med. 2023;55:653–664.
27. Lu Q, Yang MF, Liang YJ, et al. Immunology of inflammatory bowel disease: molecular mechanisms and therapeutics. J Inflamm Res. 2022;15:1825–1844. doi:10.2147/JIR.S353038
28. Woznicki JA, Saini N, Flood P, et al. TNF-α synergises with IFN-γ to induce caspase-8-JAK1/2-STAT1-dependent death of intestinal epithelial cells. Cell Death Dis. 2021;12(10):864. doi:10.1038/s41419-021-04151-3
29. Lau SKP, Lau CCY, Chan K-H, et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94(12):2679–2690. doi:10.1099/vir.0.055533-0
30. Satarker S, Tom AA, Shaji RA, et al. JAK-STAT pathway inhibition and their implications in COVID-19 therapy. Postgrad Med. 2021;133(5):489–507. doi:10.1080/00325481.2020.1855921
31. Ricciuto A, Lamb CA, Benchimol EI, et al. Inflammatory bowel disease clinical activity is associated with COVID-19 severity especially in younger patients. J Crohn's Colitis. 2022;16(4):591–600. doi:10.1093/ecco-jcc/jjab172
32. Liu S, Zhao W, Lan P, et al. The microbiome in inflammatory bowel diseases: from pathogenesis to therapy. Protein Cell. 2021;12(5):331–345. doi:10.1007/s13238-020-00745-3
33. Chen L, Cao S, Lin Z, et al. NOD-like receptors in autoimmune diseases. Acta Pharmacol Sin. 2021;42(11):1742–1756. doi:10.1038/s41401-020-00603-2
34. Jin J, Zhou T-J, Ren G-L, et al. Novel insights into NOD-like receptors in renal diseases. Acta Pharmacol Sin. 2022;43:2789–2806.
35. Rodrigues TS, de SKSG, Ishimoto AY, et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med. 2021;218(3):e20201707. doi:10.1084/jem.20201707
36. Ranson N, Veldhuis M, Mitchell B, et al. NLRP3-dependent and -independent processing of interleukin (IL)-1β in active ulcerative colitis. Int J Mol Sci. 2018;20(1):57. doi:10.3390/ijms20010057
37. Chen X, Liu G, Yuan Y, et al. NEK7 interacts with NLRP3 to modulate the pyroptosis in inflammatory bowel disease via NF-κB signaling. Cell Death Dis. 2019;10(12):906. doi:10.1038/s41419-019-2157-1
38. Hill JR, Coll RC, Sue N, et al. Sulfonylureas as concomitant insulin secretagogues and NLRP3 inflammasome inhibitors. ChemMedChem. 2017;12(17):1449–1457. doi:10.1002/cmdc.201700270
39. Wang S, Yang H, Yu L, et al. Oridonin attenuates Aβ1-42-induced neuroinflammation and inhibits NF-κB pathway. PLoS One. 2014;9(8):e104745. doi:10.1371/journal.pone.0104745
40. Saadane A, Masters S, DiDonato J, et al. Parthenolide inhibits IkappaB kinase, NF-kappaB activation, and inflammatory response in cystic fibrosis cells and mice. Am J Respir Cell Mol Biol. 2007;36(6):728–736. doi:10.1165/rcmb.2006-0323OC
41. He H, Jiang H, Chen Y, et al. Oridonin is a covalent NLRP3 inhibitor with strong anti-inflammasome activity. Nat Commun. 2018;9(1):2550. doi:10.1038/s41467-018-04947-6
42. Wang S, Zhang Y, Saas P, et al. Oridonin’s therapeutic effect: suppressing T h1/ T h17 simultaneously in a mouse model of C rohn’s disease. J Gastroenterol Hepatol. 2015;30(3):504–512. doi:10.1111/jgh.12710
43. Tsukidate T, Hespen CW, Hang HC. Small molecule modulators of immune pattern recognition receptors. RSC Chem Biol. 2023;4(12):1014–1036. doi:10.1039/D3CB00096F
44. Sun Q, Liu X, Li X. Peptidoglycan-based immunomodulation. Appl Microbiol Biotechnol. 2022;106(3):981–993. doi:10.1007/s00253-022-11795-4
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.