Back to Journals » Journal of Inflammation Research » Volume 15
Screening Key lncRNAs of Ankylosing Spondylitis Using Bioinformatics Analysis
Authors Wang JX , Zhao X, Xu SQ
Received 31 August 2022
Accepted for publication 26 October 2022
Published 4 November 2022 Volume 2022:15 Pages 6087—6096
DOI https://doi.org/10.2147/JIR.S387258
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Jian-Xiong Wang,* Xu Zhao,* Sheng-Qian Xu
Department of Rheumatology & Immunology, The First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Sheng-Qian Xu, Email [email protected]
Background: Long non-coding RNAs (lncRNAs) are important regulators in ankylosing spondylitis (AS). Few studies have examined the lncRNA-RNA binding protein (RBP) interaction in AS. This study performed bioinformatics analysis and clinical verification to identify key lncRNAs and propose their RBP interaction.
Methods: Three GEO datasets of AS were analyzed by differential expression analysis. The differentially expressed lncRNAs between the AS and control groups were screened out, and the intersecting lncRNAs were regarded as target lncRNAs. Functional was performed to identify target lncRNAs by enrichment analysis, co-expressed RNA analysis, and lncRNA-RBP interaction analysis. Finally, this study analyzed the differential expression level and clinical value of lncRNAs between the AS and control groups.
Results: Linc00304, linc00926, and MIAT were differentially expressed and upregulated. Enrichment analysis indicated that the key KEGG terms were the T-cell receptor signaling pathway and B-cell receptor signaling pathway. The key molecular function term was protein binding, and the key biological process term was adaptive immune response. In qRT-PCR results, 44 samples were validated. linc00304 expression was positively correlated with bath ankylosing spondylitis disease activity index (BASDAI), bath ankylosing spondylitis functional index (BASFI), erythrocyte sedimentation rate (ESR), and c-reactive protein (CRP). linc00926 expression was only positively correlated with ESR, whereas MIAT expression was positively correlated with BASFI, ESR, and CRP. Logistic regression revealed that linc00304, ESR, and CRP were the independent risk factors for BASDAI activation. The area under the curve (AUC) of serum linc00304 level in the diagnosis of AS was 0.687 (cutoff value: 0.413, specificity: 0.423, sensitivity: 0.900). AUC of linc00926 was 0.664 (cutoff value: 0.299, sensitivity: 0.882, specificity: 0.417). AUC of MIAT was 0.623 (cutoff value: 0.432, specificity: 0.443, sensitivity: 0.890) (all P < 0.05).
Conclusion: Overall, this study uncovered three novel lncRNAs, which were upregulated in AS, and proposed a new lncRNA-RBP-mRNA interaction that might regulate adaptive immune response.
Keywords: ankylosing spondylitis, long non-coding RNA, RNA binding protein, RNA sequencing
Introduction
Ankylosing spondylitis (AS) is a long-term inflammatory arthritis caused by autoimmune imbalance. It is often accompanied by inflammatory back pain, three peripheral joint manifestations (arthritis, enthesitis, and dactylitis), three extrarticular manifestations (uveitis, psoriasis, and crohn’s disease), two genetic signatures (family history of AS and HLA-B27), and two inflammatory signatures (good response to NSAIDs and elevated CRP level). When the trigger point of structural damage is activated (magnetic resonance imaging showed post-inflammatory fat deposition or bone marrow edema),1 the process of structural damage will not cease, even if the activity is controlled. The mechanism underlying the cessation of the process remains unclear. In recent years, several studies have shown that long noncoding RNAs (lncRNAs) are pervasively dysregulated in different cell types or tissues. lncRNAs in whole blood cells, peripheral blood mononuclear cells (PBMCs), T cells, hip joint cells, and osteogenically differentiated bone marrow mesenchymal stem cells participate in the regulation of key proinflammatory cytokines, such as IL-1β, TNF-α, IL-6, and IL-23/IL-17.2–5 More mechanistic studies are needed to identify more important lncRNAs and their downstream pathways that are critical to the pathogenesis of AS. lncRNAs are longer than other RNAs; they contain multiple functional domains6 or several RNA binding protein (RBP) binding sites. Most lncRNAs have been identified to have RBPs that bind to their binding sites.7 On the one hand, lncRNAs can regulate the bioactivity and stability of RBPs in ribonucleoprotein complexes, thereby affecting the interaction between RBPs and mRNAs,8 and directly bind with RBPs to regulate other target mRNAs.9 On the other hand, RBP can regulate the expression, stability, and function of lncRNAs.10 Therefore, the interaction between RBPs and lncRNAs plays an important role in the regulation of other target genes, thereby participating in various disease processes. However, the interaction mechanism in AS has been rarely studied.
Consequently, this study attempted to find reliable lncRNAs that were highly expressed in multiple RNA-sequencing (RNA-seq) database. Additionally, the study explored the lncRNA–RBP interaction and evaluated and clinically validated the value of the target lncRNAs.
Materials and Methods
AS Data Collection
The flow chart and overall design of this case–control study are shown in Figure 1. Data were obtained from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo). The GSE181364 dataset included lncRNA expression profiles of whole blood from three healthy individuals and five patients with AS and associated bone bridge formation. The GSE18781 dataset included array-based lncRNA expression profiles of peripheral blood from 18 patients with AS and 24 control human neonates. The GSE39340 dataset included lncRNA expression profiles of peripheral blood from five patients with AS, 10 patients with rheumatoid arthritis (RA), and 10 patients with osteoarthritis (OA).
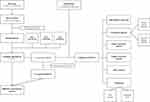 |
Figure 1 The overall design and flowchart. |
Differential Analysis
lncRNA profiles were established for each sample of patients with AS, OA, or healthy individuals for control groups. Differential expression analyses of lncRNA expression profiles of patients with AS and control groups in the GSE181364, GSE18781, or GSE39340 dataset were performed using the limma package of the R software and significance criterion of P <0.05. Thereafter, the differentially expressed lncRNAs (DElncRNAs) between patients with AS and control groups were identified. DElncRNAs were screened for intersection analysis with three GEO databases to obtain the intersecting lncRNAs.
Enrichment Analysis and Coexpression Analysis and RBP Network Analysis
The lncRNAs and predicted target genes of mRNAs were annotated, in terms of their GO categories and KEGG pathways, using the online DAVID tool11 (https://david.ncifcrf.gov). COXPRESdb (https://coxpresdb.jp) could provide co-expressed genes of lncRNAs, and enrichment analysis of co-expressed genes was helpful in estimating lncRNA function.12 ENCORI (https://starbase.sysu.edu.cn) is a database of RBP expression and lncRNA information.13 The lncRNA–RBP interaction network was constructed using the Cytoscape software (version 3.6.1).
qRT-PCR
During the clinical validation stage, RNA was extracted from PBMCs of 24 patients with AS and 20 healthy controls. Real-time quantitative polymerase chain reaction (qRT-PCR) was performed using the Trizol method. cDNA was transcribed using PrimeScript™RT Master Mix. PCR was performed on the ABI7500 PCR platform. The RT-qPCR thermocycling protocol was performed as follows: 50°C for 2 min, 95°C for 10 min, 94°C for 30s, and 60°C for 60s in 40 cycles. qPCR data were relatively quantified using 2−ΔΔCt(ΔCt = Cttarget − Ctreference, −ΔΔCt = sample ΔCT − β-actin ΔCT).14 The primer information are shown in Supplementary Material 1.
Clinical Verification
This study enrolled 24 patients with AS and 20 age-sex matched healthy controls from the Department of Rheumatology and the Physical Examination Center of the First Affiliated Hospital of Anhui Medical University from May 2021 to May 2022 (Clinical details available in Supplementary Material 2). All patients with AS met the modified New York 1984 criteria.15 All clinical data and participant information were obtained by questionnaire administration. The indicators included general patient information, disease course, laboratory index [erythrocyte sedimentation rate (ESR, mm/h) and C-reactive protein (CRP, mg/l)], disease activity score [Ankylosing Spondylitis Disease Activity Score with C-reactive protein (ASDAScrp),16 Bath ankylosing spondylitis disease activity index (BASDAI),17 Bath ankylosing spondylitis functional index (BASFI),18 and modified stoke ankylosing spondylitis spine score (mSASSS)].19 BASDAI score >4 was defined as AS disease activity.16 All recruiters were first examined. The exclusion criteria included incomplete clinical data, history of any acute or other chronic diseases, and chronic medication use. This study was approved by the Ethics Committee (No. 20200740) and was performed in accordance with the Declaration of Helsinki. All participants provided informed consent.
All statistical analyses were performed using SPSS (version 24.0). The Chi-square test was used for qualitative data. Differences in normally distributed data and skewed data were assessed using t-test and nonparametric test, respectively. Pearson and Spearman correlation analyses were used to assess the relationship between different parameters. Multivariate logistic regression analysis was performed to identify independent predictors of disease activity (BASDAI) by analyzing relevant clinical indicators and target lncRNA levels. Findings from this analysis were used to establish a forest map. P <0.05 indicated statistical significance in all analyses. To further indicate the predictive performance of target lncRNA levels, we plotted receiver operating characteristic (ROC) curves for the diagnosis of AS dependence and calculated the area under the curve (AUC) for each dataset value.
Results
Identification and Value of Target lncRNAs
Comparing the profiles of DElncRNA in three AS GEO databases, we found that linc00304, linc00926, and MIAT in the GSE181364 database were differentially expressed in both GSE18781 and GSE39340 databases (Figure 2A–C). The Heatmap showed the association between the target lncRNA expression levels and these AS-related biomarkers (Figure 2D). Target lncRNAs expressed significantly differently in patients with AS than in healthy controls, and they were significantly correlated with such AS biomarkers.
Functional Analysis of Target lncRNAs
Target prediction of co-expressed mRNAs was performed using COXPRESdb programs, and the top 50 co-expressed mRNAs were predicted for three lncRNAs. Thereafter, GO and KEGG analyses were performed on the co-expressed mRNAs. The results showed that 532 terms were annotated for co-expression of linc00304, 564 terms were annotated for co-expression of linc00926, and 670 terms were annotated for co-expression of MIAT. The top 10 items of statistical significance in the GO and KEGG analyses are shown in Figure 3. According to our findings, binding proteins, immune system, T-cell receptor signaling pathway (TCR signaling pathway), B-cell receptor signaling pathway (BCR signaling pathway), integral component of membrane, and adaptive immune system were frequently enriched, which might be the core factors involved in the systemic inflammatory process of AS.
Exploration of the Potential lncRNA-RBP Interaction
Using the ENCORI website, this study predicted 13 RBPs that were associated with target lncRNAs and 16 key co-expressed mRNAs (Figure 4). Therefore, we considered linc00304, linc00926, and MIAT to be related with mRNAs through those RBPs and involved in the pathogenesis of AS by regulating cellular immunity.
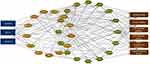 |
Figure 4 lncRNA-RBP-mRNA interaction in AS. Blue represents lncRNAs, brown represents miRNAs, green represents mRNAs, and red represents the key functional enrichment terms. |
Validation by qRT-PCR
Three target lncRNA expression levels were verified by qRT-PCR in 20 healthy individuals and 24 patients with AS. The results indicated that linc00304, linc00926, and MIAT were upregulated and showed differentially expressed levels between patients with AS and healthy controls (Figure 5A). The results were consistent with those of the GEO data, confirming reliability.
Validation of Clinical Value
This study evaluated the correlation between the expression of target lncRNAs and patient disease status. Specifically, we performed Pearson correlation analyses comparing the expression levels of target lncRNAs (linc00304, linc00926, and MIAT) and AS clinical features (ESR, CRP, ASDAScrp, BASDAI, and BASFI). Results confirmed that linc00304 expression levels were positively correlated with BASDAI, BASFI, ESR, and CRP (Figure 5B–E), linc00926 levels were positively associated with ESR (Figure 5F), and MIAT levels were positively correlated with BASFI, ESR, and CRP (Figure 5G–I) (all P < 0.05).
Additionally, multivariate logistic regression analysis was performed to test whether the prognostic performances of target lncRNAs were independently associated with disease activity (BASDAI score >4). The analysis revealed that the HR of linc00304 was 7.633 (P = 0.046, 95% confidence interval (CI) = 1.140−56.014). ESR and CRP levels were closely related to the onset of AS and were risk factors, indicating that linc00304, ESR, and CRP could independently predict the onset of AS (Figure 6A).
 |
Figure 6 (A) Forest plot of AS disease activity. (B–D) ROC curve depicting the predictive accuracy of the three lncRNAs. Abbreviation: AUC, Area under the curve. |
Lastly, this study assessed the specificities and sensitivities of lncRNAs in predicting the onset of AS by calculating the area under the curve (AUC) values and cutoff values (best Youden Index). The ROC curve for serum target lncRNAs is presented in Figure 6B–D. The AUC of the ROC for the use of serum linc00304 in the diagnosis of AS was 0.687 (95% CI = 0.532–0.842), and the cutoff value was 0.413 (specificity = 0.423, sensitivity = 0.900). The AUC of the ROC for the use of serum linc00926 in the diagnosis of AS was 0.664 (95% CI = 0.490–0.838), and the cutoff value was 0.299 (sensitivity = 0.882, specificity = 0.417). The AUC of the ROC for the use of serum MIAT in the diagnosis of AS was 0.623 (95% CI = 0.423–0.822), and the cutoff value was 0.432 (specificity = 0.443, sensitivity = 0.890) (all P <0.05).
Discussion
Currently, AS is an incurable and debilitating autoimmune disease, and controlling disease activity cannot completely inhibit structural damage.20 Over the past decade, the discovery and characterization of the function of lncRNAs have revealed multiple regulatory roles. One recent study showed that peripheral blood lncRNA NONHSAT118801.2 and NONHSAT183847.1 levels were reduced in patients with AS and negatively correlated with CRP and ESR levels in these individuals, suggesting a potential protective role of lncRNAs in this pathological context.21 RNA-sequencing data revealed that lncRNA contributed to disease progression of AS,22 as lncRNA–mRNA interaction was common. This study identified three upregulated DElncRNAs (linc00304, linc00926, and MIAT) in three AS GEO databases and validated the differential expression of those lncRNAs by qRT-PCR with 44 samples. The correlation analysis between the target lncRNA expression levels and AS activity indicators showed that linc00304 expression was positively correlated with BASDAI, BASFI, ESR, and CRP in AS (Figure 5B–E). linc00926 expression was only positively correlated with ESR (Figure 5F), whereas MIAT expression was positively correlated with BASFI, ESR, and CRP (Figure 5G–I). Linc00304, ESR, and CRP were the independent risk factors for AS activity. Although three lncRNAs could partially predict the onset of AS, further studies are needed to understand the mechanisms underlying the effect of these target lncRNAs on disease progression.
The enrichment analysis results indicated that three novel lncRNAs (linc00304, linc00926, and MIAT) were involved in specific immune cell regulations pertinent to AS pathogenesis, such as TCR signaling pathway and BCR signaling pathway. The important molecular function term was protein binding, and the important biological process term was adaptive immune response (Figure 3). Bioinformatics analysis identified those essential genes in terms, including cytotoxic T-lymphocyte-associated protein 4 (CTLA4), CD79A, heme oxygenase 1 (HMOX1), toll-like receptor 10 (TLR10), complement component receptor 2 (CR2), kinesin family member 5A (KIF5A), inducible T-cell kinase (ITK), CD22, Fc Epsilon Receptor II (FCER2), UL16-binding protein 3 (ULBP3), B lymphoid tyrosine kinase (BLK), CD180, CD79B, CD3G, lymphocyte activation gene 3 (LAG3), and CD8A.
Adaptive immune response is an immunomodulatory process by T/B lymphocytes. Previous studies have indicated that T cell molecular associations, reception, and infections contribute to AS progression.23 In a 2019 RNA sequencing study, the TCR signaling pathway was a significantly enriched term of the DEmRNAs co‑expressed with DElncRNAs in patients with AS. However, CTLA-4 costimulatory signal molecules were already known participants in the pathogenesis of inflammatory arthritis.24 The CTLA-4 drug could inhibit costimulatory signals from antigen presenting cells and prevent activation of T cells,25 which could improve clinical signs and symptoms of patients with inflammatory arthritis and retard the radiological progression of structural damage of affected joints. A case–control study comprising 38 patients with AS found that these patients expressed the highest CTLA-4 levels (p < 0.001).26 Aberrant activation of TLRs can produce a wide range of immune-stimulating cytokines and chemokines, suggesting that the TLR family may play a key role in the occurrence and development of autoimmune diseases.27 New evidence suggests that lncRNAs are involved in the regulation of TLR in the pathogenesis of spondyloarthropathies, including AS.28 For example, lncrNA-PCAT1 negatively regulates miR-145-5p, promotes the expression of TLR4, and stimulates osteogenic differentiation by activating the TLR signaling pathway of human adipose-derived stem cells.29
According to the mediatory immunopathogenetic process of AS, aberrant-activated B cells were correlated with AS risk.30 In a study, phosphoproteomic analysis of PBMCs from 20 patients with AS and 20 controls revealed that TCR and BCR might be closely related to AS.31 DNA microarray analysis showed that those differential genes were involved in the BCR and TCR signaling pathways.32 Given these evidence, lncRNAs were speculated to trigger BCR signaling activation and further lead to B cell activation by binding of the ligand to BCR, which contributes to the cascade of inflammation to enhance AS risk.
Another key term is protein binding; most lncRNAs have been reported to have the ability to interact with the corresponding RBP.7 Luo et al33 showed that the mRNA expression of RBP(YTHDF2) in PBMCs was significantly decreased, and RBP was positively correlated with m6A demethylase in AS. Logistic regression analysis showed that RBP expression level was a risk factor for AS. AUC for RNA binding protein in AS and control groups was 0.692, indicating that RBP expression levels in PBMCs may be involved in the pathogenesis of AS.
In conclusion, this study found three novel lncRNAs (linc00304, linc00926, and MIAT) that were significantly expressed in patients with AS. These lncRNA-RBP-mRNA interactions may participate in the development or progression of AS by adaptive immune response. Importantly, using the combination of lncRNA expression levels with conventional clinical risk factors, such as CRP, ESR, BASDAI, and BASFI, this study found these three lncRNAs to be closely related with disease activity of AS. These data suggest that linc00304, linc00926, and MIAT may be the key regulators of AS.
Data Sharing Statement
The datasets for this study can be found in the GEO database [Number: GSE181364, GSE18781, and GSE39340].
Contribution to the Field Statement
AS is an incurable and debilitating autoimmune disease, and if not controlled, the rate of disability is high. Based on GEO public database, we propose that linc00304, linc00926, and MIAT probably regulate the adaptive immune response in AS by lncRNA-RBP-mRNA interactions. TCR and BCR signaling pathways were the most critical pathways in the development and progression of AS. According to reports, most lncRNAs interact with their corresponding RBPs. However, the RBP–lncRNA interaction in AS has not been studied. Overall, this study proposed and clinically verified that lncRNA-RBP-mRNA interaction explains a new vital regulation mechanism in AS.
Acknowledgments
This project was supported by the Wu Jieping Medical Foundation (Grant No. 320.6750) and National Natural Science Foundation of China (Grant No. 8157060087).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This project was supported by the Wu Jieping Medical Foundation (Grant No. 320.6750) and National Natural Science Foundation of China (Grant No. 8157060087).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Koo BS, Song Y, Shin JH, Lee S, Kim TH. Evaluation of disease chronicity by bone marrow fat fraction using sacroiliac joint magnetic resonance imaging in patients with spondyloarthritis: a retrospective study. Int J Rheum Dis. 2019;22(4):734–741. doi:10.1111/1756-185X.13485
2. Liu W, Huang L, Zhang C, Liu Z. lncRNA MEG3 is downregulated in ankylosing spondylitis and associated with disease activity, hospitalization time and disease duration. Exp Ther Med. 2019;17(1):291–297. doi:10.3892/etm.2018.6921
3. Ma J, Zhang X, Zhang H, Chen H. lncRNA MEG3 suppresses the progression of ankylosis spondylitis by regulating the Let-7i/SOST axis. Front Mol Biosci. 2020;7:173. doi:10.3389/fmolb.2020.00173
4. Yu HC, Huang KY, Lu MC, et al. Down-regulation of LOC645166 in T cells of ankylosing spondylitis patients promotes the NF-kappaB Signaling via decreasingly blocking recruitment of the IKK complex to K63-linked polyubiquitin chains. Front Immunol. 2021;12:591706. doi:10.3389/fimmu.2021.591706
5. Li M, Zhou X. Long noncoding RNA intersectin 1-2 gradually declines during Adalimumab treatment, and its reduction correlates with treatment efficacy in patients with ankylosing spondylitis. Inflammopharmacology. 2021;29(5):1371–1378. doi:10.1007/s10787-021-00854-3
6. Uszczyk MM, Wutz A, Rybin V, et al. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA. 2011;17(11):1973–1982. doi:10.1261/rna.2747411
7. Li A, Ge M, Zhang Y, et al. Predicting long noncoding RNA and protein interactions using heterogeneous network model. Biomed Res Int. 2015;2015:671950. doi:10.1155/2015/671950
8. Tichon A, Gil N, Lubelsky Y, et al. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun. 2016;7:12209. doi:10.1038/ncomms12209
9. Wu W, Han Q, Han Q, et al. New insights into the interplay between non-coding RNAs and RNA-binding protein HnRNPK in regulating cellular functions. Cells. 2019;8(1):62. doi:10.3390/cells8010062
10. Lubelsky Y, Ulitsky I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature. 2018;555(7694):107–111. doi:10.1038/nature25757
11. Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216–W221. doi:10.1093/nar/gkac194
12. Obayashi T, Kagaya Y, Aoki Y, Tadaka S, Kinoshita K. COXPRESdb v7: a gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2019;47:D55–D62. doi:10.1093/nar/gky1155
13. Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. doi:10.1093/nar/gkt1248
14. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi:10.1038/nprot.2008.73
15. van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984;27(4):361–368. doi:10.1002/art.1780270401
16. Machado P, Landewé R, Lie E, et al. Ankylosing Spondylitis Disease Activity Score (ASDAS): defining cut-off values for disease activity states and improvement scores. Ann Rheum Dis. 2011;70(1):47–53. doi:10.1136/ard.2010.138594
17. Garrett S, Jenkinson T, Kennedy LG, et al. A new approach to defining disease status in ankylosing spondylitis: the Bath Ankylosing Spondylitis Disease Activity Index. J Rheumatol. 1994;21(12):2286–2291.
18. Calin A, Garrett S, Whitelock H. A new approach to defining functional ability in ankylosing spondylitis: the development of the Bath AnkylosingSpondylitis Functional Index. J Rheumatol. 1994;21:2281–2285.
19. Creemers MC, Franssen MJ, Van’t Hof MA, et al. Assessment of outcome in ankylosing spondylitis: an extended radiographic scoring system. Ann Rheum Dis. 2005;64(1):127–129. doi:10.1136/ard.2004.020503
20. Ramiro S, van der Heijde D, van Tubergen A, et al. Higher disease activity leads to more structural damage in the spine in ankylosing spondylitis: 12-year longitudinal data from the OASIS cohort. Ann Rheum Dis. 2014;73(8):1455–1461. doi:10.1136/annrheumdis-2014-205178
21. Wang JX, Jing FY, Xu YC, et al. The potential regulatory mechanism of lncRNA 122K13.12 and lncRNA 326C3.7 in ankylosing spondylitis. Front Mol Biosci. 2021;8:745441. doi:10.3389/fmolb.2021.745441
22. Huang D, Liu J, Wan L, et al. Identification of lncRNAs associated with the pathogenesis of ankylosing spondylitis. BMC Musculoskelet Disord. 2021;22(1):272. doi:10.1186/s12891-021-04119-6
23. Ramos M, De Castro JAL. HLA-B27 and the pathogenesis of spondyloarthritis. Tissue Antigens. 2002;60:191–205. doi:10.1034/j.1399-0039.2002.600301.x
24. Cao J, Zou L, Luo P, Chen P, Zhang L. Increased production of circulating soluble co-stimulatory molecules CTLA-4, CD28 and CD80 in patients with rheumatoid arthritis. Int Immunopharmacol. 2012;14:585–592. doi:10.1016/j.intimp.2012.08.004
25. Song IH, Heldmann F, Rudwaleit M, et al. Treatment of active ankylosing spondylitis with Abatacept: an open-label, 24-week pilot study. Ann Rheum Dis. 2011;70(6):1108–1110. doi:10.1136/ard.2010.145946
26. Çetintepe SP, Şentürk T, Sargın G, Aydın N. Serum sCTLA-4 levels and clinical manifestations in ankylosing spondylitis patients. Eur J Rheumatol. 2018;5(2):115–117. doi:10.5152/eurjrheum.2018.17114
27. Chen JQ, Szodoray P, Zeher M. Toll-like receptor pathways in autoimmune diseases. Clin Rev Allergy Immunol. 2016;50:1–17. doi:10.1007/s12016-015-8473-z
28. Li YX, Liu T, Liang YW, et al. Integrative analysis of long non-coding RNA and messenger RNA expression in toll-like receptor 4-primed mesenchymal stem cells of ankylosing spondylitis. Ann Transl Med. 2021;9(20):1563. doi:10.21037/atm-21-5020
29. Yu L, Qu H, Yu Y, Li W, Zhao Y, Qiu G. LncRNA-PCAT1 targeting miR-145-5p promotes TLR4-associated osteogenic differentiation of adipose-derived stem cells. J Cell Mol Med. 2018;22:6134–6147. doi:10.1111/jcmm.13892
30. Ge L, Wang J, Zhu BQ, Zhang ZS. Effect of abnormal activated B cells in patients with ankylosing spondylitis and its molecular mechanism. Eur Rev Med Pharmacol Sci. 2018;22(9):2527–2533. doi:10.26355/eurrev_201805_14941
31. Luo F, Jiang N, Wang H, et al. Phosphoproteomic analysis of peripheral blood mononuclear cells of ankylosing spondylitis patients. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2020;36(5):444–450. Chinese.
32. Xu L, Sun Q, Jiang S, Li J, He C, Xu W. Changes in gene expression profiles of the Hip joint ligament of patients with ankylosing spondylitis revealed by DNA chip. Clin Rheumatol. 2012;31(10):1479–1491. doi:10.1007/s10067-012-2038-9
33. Luo Q, Guo Y, Xiao Q, et al. Expression and clinical significance of the m6A RNA-binding proteins YTHDF2 in peripheral blood mononuclear cells from new-onset ankylosing spondylitis. Front Med. 2022;9:922219. doi:10.3389/fmed.2022.922219
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



