Back to Journals » Infection and Drug Resistance » Volume 16
Risk Factors and Outcomes for Isolation with Polymyxin B-Resistant Enterobacterales from 2018–2022: A Case-Control Study
Authors Yan W , Wu J, Wang S, Zhang Q , Yuan Y, Jing N, Zhang J, He H, Li Y
Received 7 September 2023
Accepted for publication 14 December 2023
Published 22 December 2023 Volume 2023:16 Pages 7809—7817
DOI https://doi.org/10.2147/IDR.S435697
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Wenjuan Yan,1,* Jiaojiao Wu,2,* Shanmei Wang,1 Qi Zhang,1 Youhua Yuan,1 Nan Jing,1 Jiangfeng Zhang,1 Hangchan He,3 Yi Li1
1Department of Clinical Microbiology, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of Henan University, Zhengzhou, Henan, People’s Republic of China; 2Department of Clinical Microbiology, Xiayi People’s Hospital, Shangqiu, Henan, People’s Republic of China; 3Department of Clinical Laboratory, Baofeng Traditional Chinese Medicine Hospital, Pingdingshan, Henan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yi Li, Department of Clinical Microbiology, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of Henan University, Zhengzhou, Henan, People’s Republic of China, Tel +8615939039006, Email [email protected]
Purpose: To analyze the risk factors and clinical outcomes of patients isolated with polymyxin B-resistant (PR) Enterobacterales from various clinical specimens to prevent and control the spread of these strains.
Methods: This retrospective case-control study included 72 PR Enterobacterales-positive cases and 144 polymyxin B-susceptible (PS) Enterobacterales controls from 2018 to 2022. Patients with PR Enterobacterales isolated in various clinical cultures were defined as cases. Patients with PS Enterobacterales cultures at similar anatomic sites during the same period were randomly selected as controls. Data were collected from clinical and laboratory test records. Bivariable logistic regression and Pearson’s chi-square tests were used to assess risk factors.
Results: PR strains were predominantly Klebsiella pneumoniae (72.2%) and Salmonella enteritidis (8.3%). Of the patients, 66.04% were admitted to an intensive care unit (ICU). Risk factors for isolation with PR strains included chronic heart disease (P = 0.012; odds ratio [OR] 1.15; 95% confidence interval [CI] 1.03– 1.28), immunosuppressant use (P = 0.016; OR 1.04 [1.0– 1.07), drainage tube [head] (P = 0.006; OR 1.1 [1.0– 1.1]), and polymyxin B exposure (P = 0.007; OR 1.03 [1.0– 1.06]. With respect to outcomes, admission to an ICU (P = 0.003; OR 7.1 [1.9– 25.4]), hypertension (P = 0.035; OR 1.4 [1.02– 1.83]), and drainage tube [head] (P = 0.044; OR 1.1 [1.0– 1.15]) were associated with treatment failure. Additionally, treatment failure was more frequent in patients (45.83%) than in controls (14.58%).
Conclusion: The major risk factors for isolation with PR strains were chronic heart disease, exposure to immunosuppressants, use of drainage tubes, and polymyxin B exposure. The isolation of PR strains in patients was a predictor of unfavorable outcomes. These findings provide a basis for monitoring the spread of PR Enterobacterales.
Keywords: polymyxin B resistance, treatment failure, Klebsiella pneumoniae, Salmonella enteritidis
Introduction
Multidrug-resistant Gram-negative bacilli have increased worldwide and pose a significant and growing threat that limits the effectiveness of therapeutic regimens.1 Conventional drugs, such as polymyxin E (colistin) and polymyxin B, are being reconsidered as the last line of defense against multidrug-resistant Gram-negative bacilli infections.2 Owing to the increased use of colistin in clinical settings, resistance to this antibiotic has increased, particularly among Klebsiella pneumoniae isolates, which should be monitored.3–5 Polymyxin B-resistant (PR) bacterial infections have been reported in both nosocomial and community settings.6
The mechanisms underlying polymyxin B resistance in Gram-negative bacilli mainly involve intrinsic mechanisms, mutations, or adaptation. The selective pressure applied by the polymyxin dose regulates the dynamics of genetic variants within the population. To this end, the super minimum inhibitory concentration (super-MIC) levels (> 4 x MIC) of polymyxin B were reported to have caused the development of irreversible resistance in Acinetobacter baumannii.7 It has been suggested that the principal resistance to this antibiotic in Enterobacterales is associated with lipopolysaccharide modification, which is mediated by charged 4-amino-4-deoxy-L-arabinose (L-Ara4N; mediated by arnBCADTEF) and/or phosphoethanolamine (pEtN; mediated by pmrC, or mobile colistin resistance gene).8–10 The mobile colistin resistance gene (mcr-1) and its variants, mcr-2 to mcr-10, mediate polymyxin B resistance.6,11–13 A recent study showed that New Delhi metallo-β-lactamase (NDM)-producing MDR K. pneumoniae can rapidly attain high-level polymyxin resistance through cellular metabolism plasticity, lipid A modification, and outer membrane remodeling.14
In addition to monitoring the rate of resistance to polymyxins and underlying mechanisms, the identification of relevant risk factors in patients with colistin-resistant bacteria is important to prevent the spread of nosocomial infection outbreaks. Several studies have identified risk factors related to patients with colistin-resistant and/or carbapenem-resistant Enterobacterales infection or colonization.4,15–17 However, studies focused on risk factors in Henan Province, China, are limited.
The relevant risk factors and outcomes of patients carrying PR Enterobacterales were retrospectively analyzed to provide a reference for the prevention and control of PR Enterobacterales infection outbreaks in hospital.
Materials and Methods
Study Participants
This case-control study was performed at a tertiary care teaching hospital with 5000 beds in Henan Province, China, between January 2018 and December 2022. The risk factors related to the isolation of PR Enterobacterales were investigated. The case group (n = 72) included patients with PR Enterobacterales isolated from various clinical specimens during the study period. Patients with polymyxin B-susceptible (PS) Enterobacterales isolated from clinical cultures were controls. The ratio of control to case samples was 2:1. Controls were matched for hospital ward, date of hospitalization, and pathogen with the case in a random manner. The exclusion criteria (n = 23) were as follows: patients with PR Enterobacterales isolation from intestinal screening samples during the study period and inadequate clinical or laboratory data, such as transfer or discharge without relevant clinical diagnoses and treatment processes, resulting in unclear patient outcomes. Patients with clinical cultures of Enterobacterales (eg, Providencia spp., Proteus spp., Morganella morganii, and Serratia spp.) intrinsically resistant to polymyxins were also excluded.
Definition of Outcomes
The outcomes of patients in this study were defined as follows: Treatment failure included patients who died for any reason or were discharged without improvement after the isolation of polymyxin B-resistant strains.
Clinical Data
Data were obtained from electronic medical records. Potential risk factors included demographic properties (age and sex), medical history (hospitalization, transfers between hospitals and departments, intensive care unit [ICU] admission, and surgery), comorbidities, inpatient time, admission diagnosis, antibiotic use, use of invasive medical devices (urinary catheter, mechanical ventilation, drainage tube, and nasogastric tube), and use of immunosuppressive drugs. Diagnosis was classified using the International Classification of Diseases-11 (ICD-11) based on the patient’s admission diagnosis.18
Microbiological Methods
Bacterial species identification and antimicrobial susceptibility testing were performed using the Phoenix100 automated system according to the manufacturer’s recommendations (Becton Dickinson Co., Sparks, MD, USA). The identification of Salmonella spp. was based on a serum agglutination test. The polymyxin B MICs of the isolates were determined by broth microdilution. The results were interpreted using the clinical breakpoints of the European Committee on Antimicrobial Susceptibility Testing.19 Isolates with an MIC higher than 2 mg/L for polymyxin B were considered PR. Escherichia coli ATCC 25922 and Escherichia coli NCTC 13846 (mcr-1-positive) were used as quality control strains.
Statistical Analyses
Categorical variables are reported as n (percentages); continuous variables with a normal distribution are expressed as means ± standard deviations (SD); and continuous variables with a non-normal distribution are described as medians (interquartile ranges: IQRs). Categorical data were analyzed using chi-squared or Fisher’s exact tests; the Student’s t-test or the Mann–Whitney U-test was used to analyze continuous variables. Odds ratios (ORs) with 95% confidence intervals (CIs) and P-values were used for univariate analyses. Variables with a P–value < 0.05 in the univariate analyses were entered into multivariate logistic regression. Survival curves (150 day) were created using the Kaplan–Meier method. Log rank tests were performed using Prism 8.0 (GraphPad). Other statistical analyses were performed using SPSS (version 25.0; IBM Corporation, Armonk, NY, USA). P < 0.05 was considered statistically significant.
Results
Characteristics of PR Isolates
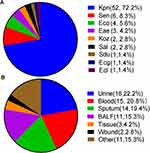
As shown in Figure 1, the most commonly identified PR Enterobacterales was K. pneumoniae (72.2%; 52/72), followed by Salmonella enteritidis (8.3%; 6/72), and Escherichia coli (5.6%; 4/72). Among the PR isolates, 34.7% (25/72) were isolated from sputum and BALF, 22.2% (16/72) from urine, 20.8% (15/72) from blood, and the remainder from other samples.
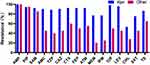
Antimicrobial susceptibility testing revealed that the resistance rate for 52 K. pneumoniae isolates was significantly higher than that of the other 20 Enterobacterales isolates. In particular, 76.9% (40/52) of K. pneumoniae isolates were carbapenem-resistant (Figure 2).
Patient Characteristics
A total of 72 patients carrying PR Enterobacterales were included during the 5-year study period, and 144 patients carrying PS Enterobacterales were included as controls. The median ages of cases and controls were 63 (51–73) and 62 (48–73) years, respectively (P = 0.391), and 66.67% of patients were male. Of the study patients, 63.89% were admitted to the ICU. Patient characteristics are shown in Table 1.
 |
Table 1 Univariate and Multivariate Analyses of Risk Factors in Patients Infected with Polymyxin-Resistant Enterobacterales Strains |
Risk Factors for Isolation with PR Strains
Table 1 compares the clinical characteristics of patients with PR and PS controls. Univariate analysis revealed that chronic heart disease, chronic kidney disease, organ transplantation, immunosuppressant use, invasive procedures (central venous catheter, chest drainage tube [chest], and head drainage tube [head]), and polymyxin B exposure were associated with the isolation of PR Enterobacterales.
Chronic heart disease (P = 0.012; odds ratio [OR] 1.15; 95% confidence interval [CI] 1.03–1.28), immunosuppressant use (P = 0.016; OR 1.04 [1.0–1.07]), drainage tube [head] (P = 0.006; OR 1.1 [1.0–1.1]), and polymyxin B exposure (P = 0.007; OR 1.03 [1.0–1.06]) were independently related to the isolation of PR Enterobacterales in a multivariate analysis (Table 1).
Treatment Failure Among Cases and Controls
Among the 72 patient cases, 33 (45.83%) experienced treatment failure; Conversely, within the cohort of 144 controls, 21 (14.58%) experienced treatment failure. The survival rate was significantly lower in patients in the case group than in the control group in the survival curve analysis (Figure 3).
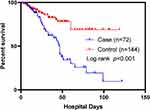 |
Figure 3 Survival curves between patients with (case) and without (control) polymyxin B-resistant Enterobacterales isolation. |
Univariate analyses revealed that a history of ICU admission, hypertension, nervous system disease, mechanical ventilation, urinary catheter, central venous catheter, and head drainage were associated with treatment failure (Table 2). In multivariate analysis, independent risk factors for treatment failure included admission to an ICU (P = 0.003; OR 7.1 [1.9–25.4]), hypertension (P = 0.035; OR 1.4 [1.02–1.83]), and drainage tube [head] (P = 0.044; OR 1.1 [1.0–1.15]) (Table 2).
 |
Table 2 Univariate and Multivariate Analyses of Risk Factors for Treatment Failure in Patients with Polymyxin B-Resistant Enterobacterales Infection |
Discussion
Increased polymyxin consumption for multidrug-resistant Gram-negative bacilli treatment has resulted in the development of polymyxin B resistance by these species in several countries.20 This is an important public health issue. Monitoring the resistance mechanism and elucidating risk factors for polymyxin B resistance may be helpful for preventing the spread of resistant strains.
In this study, the most common PR Enterobacterales was K. pneumoniae (72.2%), consistent with previous results.16,21,22 Additionally, 76.9% (40/52) of PR K. pneumoniae isolates were carbapenem resistant. The resistance rates of carbapenem-resistant K. pneumoniae to colistin in Taiwan21 and Oman23 were 17% and 24.2%, respectively. We found that polymyxin B exposure is a risk factor for hospitalized patients with PR Enterobacterales, which is consistent with the results of previous studies.24,25 However, most of the prior literature focuses on PR combined with carbapenem resistance and/or specific populations, such as critically ill patients, or specific infection types, such as bloodstream infection (BSI). For example, previous polymyxin B exposure was a predictor of the isolation of PR carbapenem-resistant and carbapenemase-producing Enterobacterales or colistin-resistant K. pneumoniae in other studies.15,24 Colistin was reported to be related to the development of colistin-resistant K. pneumoniae BSI.26 In another study, recent exposure to colistin or polymyxin B was related to increased opportunities for colistin resistance.27 Except for polymyxin B exposure and carbapenem resistance, previous exposure to carbapenems was the risk factor for colonization or infection with PR E. coli or K. pneumoniae in a low-endemicity setting.17
Chronic heart failure, use of immunosuppressive agents, and head drainage were also risk factors for PR Enterobacterales in our study population. It was reported that coronary heart disease is an independent risk factor that predisposes patients to PR resistance.28 Chronic renal failure has been shown as a risk factor in critically ill patients with PR and carbapenemase-producing Enterobacterales in a previous study, which also included other risk factors such as surgical procedures, indwelling devices (urinary catheters), and transfer between hospital wards.16 Other risk factors associated with colistin resistance in Gram-negative bacilli include neurological disease, residence in a nursing facility prior to admission, receipt of carbapenems or an antimicrobial with anti-methicillin-resistant Staphylococcus aureus activity, and receipt of ventilatory support.4 In addition, male sex, immunosuppression, and antibiotic use, particularly carbapenems and fluoroquinolones, are associated with mcr-1-positive E. coli infection.29 Increased age (> 55 years) and antibiotic use in the preceding 90 days were associated with an increased risk in patients with colistin-resistant Enterobacterales.30
Regarding patient outcomes, our study demonstrated that the rate of treatment failure in 72 patients carrying PR strains was 45.83%, which is similar to previous results that reported an overall mortality of between 35% and 66%.16,17,22,31 In bloodstream infections caused by carbapenem-resistant K. pneumoniae, age and colistin resistance increased the 28-day mortality.32 In our study, multivariate regression analysis showed that ICU admission, hypertension, and head drainage were associated with treatment failure, different from a previous study that reported that carbapenem exposure was associated with mortality among adult ICU patients with PR Enterobacterales strains.16
This study has some limitations. First, the retrospective and single-centre research design could result in selection biases. Second, the association between possible risk factors identified in this study and the isolation of PR strains was only a statistical link and needs to be validated using more samples. Third, we did not evaluate resistance mechanisms in the PR strains. Thus, more surveillance and multicenter studies are required.
Conclusions
In conclusion, this study revealed that the common bacteria resistant to polymyxin B in Henan, China, are K. pneumoniae, S. enteritidis, and E. coli, and that more than half of K. pneumoniae are also resistant to carbapenem. Risk factor analysis showed that chronic heart failure, use of immunosuppressive agents, head drainage, and polymyxin B exposure are related to the acquisition of PR strains. ICU admission, hypertension, and head drainage are associated with treatment failure. Therefore, hospitals should take multiple measures to prevent further outbreaks of PR Enterobacterales.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethical Approval
This study was conducted with the approval of the Ethics Committee of the Henan Provincial People’s Hospital (2022-1-233). All organs for transplant recipients were given freely and with written informed consent. Additionally, the donation and transplantation processes follow the guidelines of the Declaration of Istanbul.
Additionally, the Ethics Committee of Henan Provincial People’s Hospital granted a waiver for patient consent, given that the study neither involved the collection of personal information nor subjected participants to any interventions. The study was conducted in accordance with the Declaration of Helsinki, data were anonymized, and the confidentiality of patients was guaranteed.
Author Contributions
All authors made a significant contribution to the work reported in the conception, study design, execution, acquisition of data, analysis, and interpretation. All authors took part in drafting, revising, or critically reviewing the article, gave final approval of the version to be published, and have agreed on the journal to which the article has been submitted. All authors agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. El-Sayed Ahmed MAE, Zhong LL, Shen C, Yang Y, Doi Y, Tian GB. Colistin and its role in the era of antibiotic resistance: an extended review (2000–2019). Emerg Microbes Infect. 2020;9(1):868–885. doi:10.1080/22221751.2020.1754133
2. Stefaniuk EM, Tyski S. Colistin resistance in enterobacterales strains - a current view. Pol J Microbiol. 2019;68(4):417–427. doi:10.33073/pjm-2019-055
3. Srinivas P, Rivard K. Polymyxin resistance in gram-negative pathogens. Curr Infect Dis Rep. 2017;19(11):38. doi:10.1007/s11908-017-0596-3
4. Richter SE, Miller L, Uslan DZ, et al. Risk factors for colistin resistance among gram-negative rods and Klebsiella pneumoniae isolates. J Clin Microbiol. 2018;56(9):e00149–e00218. doi:10.1128/JCM.00149-18
5. Seethalakshmi PS, Rajeev R, Prabhakaran A, Kiran GS, Selvin J. The menace of colistin resistance across globe: obstacles and opportunities in curbing its spread. Microbiol Res. 2023;270:127316. doi:10.1016/j.micres.2023.127316
6. Nang SC, Li J, Velkov T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit Rev Microbiol. 2019;45(2):131–161. doi:10.1080/1040841X.2018.1492902
7. Zhao J, Zhu Y, Lin YW, et al. Polymyxin dose tunes the evolutionary dynamics of resistance in multidrug-resistant Acinetobacter baumannii. Clin Microbiol Infect. 2022;28(7):
8. Yang TY, Wang SF, Lin JE, et al. Contributions of insertion sequences conferring colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents. 2020;55(3):105894. doi:10.1016/j.ijantimicag.2020.105894
9. Bialvaei AZ, Samadi Kafil H. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin. 2015;31(4):707–721. doi:10.1185/03007995.2015.1018989
10. Moffatt JH, Harper M, Boyce JD. Mechanisms of polymyxin resistance. Adv Exp Med Biol. 2019;1145:55–71. doi:10.1007/978-3-030-16373-0_5
11. Sun J, Zhang H, Liu YH, Feng Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018;26(9):794–808. doi:10.1016/j.tim.2018.02.006
12. Ling Z, Yin W, Shen Z, Wang Y, Shen J, Walsh TR. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother. 2020;75(11):3087–3095. doi:10.1093/jac/dkaa205
13. Rhouma M, Madec JY, Laxminarayan R. Colistin: from the shadows to a one health approach for addressing antimicrobial resistance. Int J Antimicrob Agents. 2023;61(2):106713. doi:10.1016/j.ijantimicag.2023.106713
14. Lu J, Han M, Yu HH, et al. Lipid A modification and metabolic adaptation in polymyxin-resistant, New Delhi metallo-β-lactamase-producing Klebsiella pneumoniae. Microbiol Spectr. 2023;11(4):e0085223. doi:10.1128/spectrum.00852-23
15. Teo JQ, Chang CW, Leck H, et al. Risk factors and outcomes associated with the isolation of polymyxin B and carbapenem-resistant Enterobacteriaceae spp.: a case-control study. Int J Antimicrob Agents. 2019;53(5):657–662. doi:10.1016/j.ijantimicag.2019.03.011
16. da Silva KE, Baker S, Croda J, et al. Risk factors for polymyxin-resistant carbapenemase-producing Enterobacteriaceae in critically ill patients: an epidemiological and clinical study. Int J Antimicrob Agents. 2020;55(3):105882. doi:10.1016/j.ijantimicag.2020.105882
17. Büchler AC, Gehringer C, Widmer AF, Egli A, Tschudin-Sutter S. Risk factors for colistin-resistant Enterobacteriaceae in a low-endemicity setting for carbapenem resistance - a matched case-control study. Euro Surveill. 2018;23(30):1700777. doi:10.2807/1560-7917.ES.2018.23.30.1700777
18. Kazlauskas E, Gegieckaite G, Eimontas J, Zelviene P, Maercker A. A brief measure of the international classification of diseases-11 adjustment disorder: investigation of psychometric properties in an adult help-seeking sample. Psychopathology. 2018;51(1):10–15. doi:10.1159/000484415
19. European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version. 2023;13(1):1.
20. Giamarellou H. Epidemiology of infections caused by polymyxin-resistant pathogens. Int J Antimicrob Agents. 2016;48(6):614–621. doi:10.1016/j.ijantimicag.2016.09.025
21. Arjun R, Gopalakrishnan R, Nambi PS, Kumar DS, Madhumitha R, Ramasubramanian V. A study of 24 patients with colistin-resistant gram-negative isolates in a tertiary care hospital in South India. Indian J Crit Care Med. 2017;21(5):317–321. doi:10.4103/ijccm.IJCCM_454_16
22. de Maio Carrillho CM, Gaudereto JJ, Martins RC, et al. Colistin-resistant Enterobacteriaceae infections: clinical and molecular characterization and analysis of in vitro synergy. Diagn Microbiol Infect Dis. 2017;87(3):253–257. doi:10.1016/j.diagmicrobio.2016.11.007
23. Balkhair A, Saadi KA, Adawi BA. Epidemiology and mortality outcome of carbapenem- and colistin-resistant Klebsiella pneumoniae, Escherichia coli, Acinetobacter baumannii, and Pseudomonas aeruginosa bloodstream infections. IJID Reg. 2023;7:1–5. doi:10.1016/j.ijregi.2023.01.002
24. Huang PH, Cheng YH, Chen WY, et al. Risk factors and mechanisms of in vivo emergence of colistin resistance in carbapenem-resistant Klebsiella pneumoniae. Int J Antimicrob Agents. 2021;57(6):106342. doi:10.1016/j.ijantimicag.2021.106342
25. Gundogdu A, Ulu-Kilic A, Kilic H, et al. Could frequent carbapenem use be a risk factor for colistin resistance? Microb Drug Resist. 2018;24(6):774–781. doi:10.1089/mdr.2016.0321
26. Giacobbe DR, Del Bono V, Trecarichi EM, et al. Risk factors for bloodstream infections due to colistin-resistant KPC-producing Klebsiella pneumoniae: results from a multicenter case-control-control study. Clin Microbiol Infect. 2015;21(12):
27. Zhang HL, Han JH, Lapp Z, et al. Risk factors for colistin-resistant carbapenem-resistant Klebsiella pneumoniae in the postacute care setting. Open Forum Infect Dis. 2022;9(9):ofac452. doi:10.1093/ofid/ofac452
28. Xu X, Zhu R, Lian S, et al. Risk factors and molecular mechanism of polymyxin B resistance in carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary hospital in Fujian, China. Infect Drug Resist. 2022;15:7485–7494. doi:10.2147/IDR.S391674
29. Wang Y, Tian GB, Zhang R, et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. 2017;17(4):390–399. doi:10.1016/S1473-3099(16)30527-8
30. Mills JP, Rojas LJ, Marshall SH, et al. Risk factors for and mechanisms of COlistin resistance among Enterobacterales: getting at the CORE of the issue. Open Forum Infect Dis. 2021;8(7):ofab145. doi:10.1093/ofid/ofab145
31. Papadimitriou-Olivgeris M, Bartzavali C, Spyropoulou A, et al. Molecular epidemiology and risk factors for colistin- or tigecycline-resistant carbapenemase-producing Klebsiella pneumoniae bloodstream infection in critically ill patients during a 7-year period. Diagn Microbiol Infect Dis. 2018;92(3):235–240. doi:10.1016/j.diagmicrobio.2018.06.001
32. Balkan II, Alkan M, Aygün G, et al. Colistin resistance increases 28-day mortality in bloodstream infections due to carbapenem-resistant Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 2021;40(10):2161–2170. doi:10.1007/s10096-020-04124-y
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.