Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 14
Regulation of Methylase METTL3 on Fat Deposition
Received 14 October 2021
Accepted for publication 8 December 2021
Published 20 December 2021 Volume 2021:14 Pages 4843—4852
DOI https://doi.org/10.2147/DMSO.S344472
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ming-Hui Zou
Gang Luo, Jialing Chen, Zhanjun Ren
College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi, 712100, People’s Republic of China
Correspondence: Zhanjun Ren
College of Animal Science and Technology, Northwest A&F University, Yangling, Shannxi, 712100, People’s Republic of China
Email [email protected]
Abstract: N6-methyladenosine (m6A) is the most prevalent and abundant type of internal post-transcriptional RNA modification in eukaryotic cells. METTL3 is a methylation modifying enzyme, which can directly or indirectly affect biological processes, such as RNA degradation, translation and splicing. In addition, it was found that 67% of 3’-UTR regions containing m6A sites had at least one miRNA binding site, and the number of m6A at 3’-UTR sites was closely related to the binding sites of miRNA. With the improvement of human living standards, obesity has become a very serious and urgent problem. The essence of obesity is the accumulation of excess fat. Exploring the origin and development mechanisms of adipocyte from the perspective of fat deposition has always been a hotspot in the field of adipocyte research. The aim of the present review is to focus on METTL3 regulating fat deposition through mRNA/adipocyte differentiation axis and pri-miRNA/pre-miRNA/target genes/adipocyte differentiation and to provide a theoretical basis according to the currently available literature for further exploring this association. This review may provide new insights for obesity, fat deposition disease and molecular breeding.
Keywords: METTL3, m6A methylation, miRNA, adipocyte differentiation, intramuscular fat
Introduction
In the early 1970s, a novel RNA epigenetic modification, N6-methyladenosine (m6A), was first discovered and proposed in eukaryotic messenger RNA (mRNA) from Novikoff hepatoma cells.1 N6-methyladenosine (m6A) is one of the most abundant internal modifications in eukaryotic messenger RNA that affects a variety of cellular biological processes, including splicing, processing, nuclear export, stability and decay, translation, cellular differentiation and metabolism.2,3 M6A modification refers to the methylation of the 6th N of adenine on mRNA under the action of methyltransferase complex (MTC), which is a dynamic and reversible process regulated by both methyltransferase and dimethyl transferase,4 such as methyltransferase like 3 (METTL3) and methyltransferase like 4 (METTL4).5,6
METTL3 was discovered and named from Hela cells in 19947 and was conserved from yeast to human, including leading spiral structure LH, nuclear localization signal NLS, Methyltransferase domain MTD containing SAM binding domain and zinc finger motif ZFD.6,8 Studies have shown that zinc finger participates in RNA binding, ZNF1 interacts with RNA electrostatically, whereas ZNF2 interacts with RNA hydrophobically, which suggests that zinc finger is responsible for specifically recognizing RNA and making METTL3 play a role.9 The formation of miRNA requires the cutting of the complex composed of DGCR8 and DROSHA, and METTL3 deletion reduced the binding of DGCR8 to pri-miRNA.10 According to its function, RNA can be divided into two broad categories, including noncoding RNA and encoding protein mRNA. MiRNAs are a group of conservative, small and non-coding RNAs inhibiting translation of or degrading target mRNAs by binding to the complementary sequences in the 3’untranslated region.11 It has been proved that miRNAs play important roles in energy homeostasis,12 sugar and lipid metabolism,13 insulin secretion,14 pancreatic β-cell development,15 and adipocyte differentiation.16 More and more studies have shown that miRNA can interact with transcription factors and important signal molecules related to adipocyte differentiation.17 Adipose tissue deposition is characterized by increased cell size (hypertrophy) and increased cell numbers (hyperplasia) at the cellular level, which indicates that cell differentiation is a necessary process of fat deposition. In addition, studies found the formation of METTL3 and METTL14 heterodimers played an important role in adipocyte differentiation.18 Another study also found METTL3 regulates adipocyte differentiation by regulating genes alone.19 So, METTL3 plays an important role in fat deposition. However, the specific mechanism by which METTL3 regulates fat deposition remains unclear.
Despite recent progress in METTL3 research, the presence and functionality of METTL3 remains largely unknown. Recent studies have reported the emerging roles of METTL3 in the development of fat deposition. The present review focuses on the latest progress in made METTL3 research and provides an up-to-date summary of the association between METTL3 and fat deposition, which may provide insight into METTL3-related molecular biomarkers and increase of fat deposition in animals.
M6A Methylation
Epitranscriptomic m6A modification is dynamically and reversibly regulated by modulators characterized as dedicated demethylases (erasers), m6A binding protein (readers) and methyltransferases (writers), according to their functions,20 Erasers (FTO, ALKBH5) and writers (METTL3, METTL14, WTAP) are responsible for catalyzing and removing m6A, respectively.21–24 In complex METTL3/METTL14/ WTAP, METTL3 and METTL14 form a heterodimer complex and interact with WTAP. METTL3 is identified as a SAM-binding component of the complex and has its own catalytic ability, which is highly conserved in eukaryotes.25 It has been reported that m6A can label pri-miRNAs and identify DGCR8 molecules by METTL3/m6A, participating in the mature process of miRNAs and leading to differential expression of miRNAs in many biological processes.10,26 In addition, it was found that METTL3 knockout decreased the binding activity between DGCR8 and pri-miRNA, leading to decreased expression of mature miRNAs.10 So, understanding the structure of METTL3 and its interaction mechanism with target RNA will help to further understand the post transcriptional regulation level of genetic information.
METTL3 Promoted the Transformation of pri-miRNA into Mature miRNA (miR-21, miR-25, miR-34a, miR126, miR-143-3p, miR-221/222 and miR-320)
MicroRNAs (miRNAs) are a group of single-stranded, non-coding small RNAs that are broadly present in eukaryotic cells and are highly conserved during evolution with a length of 19–24nt.27 As miRNAs are critical in development, differentiation, and fat deposition, their mature are controlled by multiple ways during their biogenesis cascade. Figure 1 shows the role of METTL3 in miRNA maturation. Alarcon et al demonstrated that m6A modification could mark pri-miRNA for processing by recognizing DGCR8 in a METTL3-dependent manner,10 indicating that altered METTL3 mediated m6A modification might be responsible for the aberrant expression of miRNAs in many biological processes. In addition, it was shown that depletion of METTL3 leads to decreased accumulation of miRNAs and to an overaccumulation of pri-miRNAs due to their impaired processing.10,28 Similar to previous results,1 miR-21 was up-regulated when METTL3 was overexpressed.10,29 METTL3-dependent m6A methylation promoted primary miR-34a (pri-miR34a)30 and miRNA-126 (pri-miR126)31 maturation through DGCR8. Other researchers have demonstrated that upregulation of METTL3/m6A modification promotes pri-miR-25,32 pri-miR-221/22233 and pri-miR-143-3p34 maturation (decreasing the expression of pri-miRNA but increasing the expression of pre-miRNA and miRNA). In addition, pre-miR-320 was much less enriched after METTL3 inhibition, indicating that pre-miR-320 was a target of METTL3.35
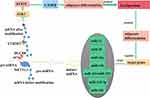 |
Figure 1 METTL3 regulates fat deposition by coding RNA and non-coding RNA. |
MiRNA (miR-21, miR-25, miR-34a, miR126, miR-143-3p, miR-221/222 and miR-320) Regulated Adipocyte Differentiation by Targeting Target Genes
MicroRNAs (miRNAs), a novel class of endogenous, noncoding, single-stranded RNAs, have emerged as a group of important regulators via degradation or translational inhibition of their target mRNAs.36 As shown in Table 1, miRNA regulates adipocyte differentiation by targeting multiple target genes.
 |
Table 1 METTL3 Regulates Adipocyte Differentiation Through Target Genes |
MiR-21 Regulated Adipocyte Differentiation
It was found that nearly 25% of miRNA targets are conserved in the 3’ noncoding region of human, mouse and rabbit.37 In addition, highly conserved miR-21 recognition elements were found in the analysis of PTEN 3’UTR of different species, indicating that PTEN can be combined with mir-21.38 At the same time, the results presented here indicated that the potential signal pathway of miR-21 protection might be achieved by targeting PTEN/AKT signaling pathway. PTEN was the main regulator of the PI3K signaling pathway, which was involved in lipid metabolism and glucose transport in 3 T3-L1 adipocytes.39 Previous studies have also shown that endogenous PTEN expression is down-regulated during 3T3-L1 differentiation,40 and knockdown of PTEN potentiated the increase in insulin-mediated phosphorylation of AKT/ERK and promoted adipogenesis of 3T3-L1 cells.41 The study also indicated that miR-21 directly targets the 3′-UTRs of SMAD7, and negatively regulates mRNA and protein expression levels.42 In addition, SMAD7 regulated 3T3-L1 preadipocyte differentiation and adipogenesis through TGFβ/SMAD and WNT signaling pathway.43
MiR-25 Regulated Adipocyte Differentiation
MiR-25, a member of miR-106b-25 cluster, was significantly downregulated during the differentiation from 3T3-L1 preadipocytes towards mature adipocytes.44 In addition, this study confirmed BTG2,45 FBXW7,46 LATS247 and PTEN48 are targets of miR-25 and had binding sites with miR-25 in the 3’-UTR. Further experiments demonstrated that miR-25 Suppresses 3T3-L1 Adipogenesis by directly targeting KLF4 and C/EBPα.44 FBXW7 inhibits C/EBPα-dependent transcription and inactivation of FBXW7 results in the accumulation of C/EBPα.49 LATS2 regulates the balance between proliferation and differentiation during adipose development. Interestingly, studies provided evidence that LATS2 not only negatively modulates cell proliferation but also positively regulates cell differentiation.50 In addition, a recent study showed that BTG2 downregulates interleukin-6 expression by inhibiting the signal transducer and activator of transcription 3 (STAT3) signaling pathway, which is known to regulate adipocyte differentiation.51 So, miR-25 can regulate adipocyte differentiation through multiple pathways.
MiR-34a Regulated Adipocyte Differentiation
In recent years, reports on miR-34a in human fat have found that miR-34a can target PPARα Gene regulation of human fat deposition in liver,52 which indicated miR-34a played an important role in fat deposition. miR‑34a was revealed to directly target SIRT1 by binding to its 3’‑untranslated region53 and SIRT1 can promote fat mobilization in white adipocytes by repressing PPAR-γ.54 The study provides evidence that miR-34a decreases the mitochondrial content and increases TAG via PPARα and AMPK pathways by targeting the AdipoR2 gene.55 A number of factors regulate the transcriptional activation potential of C/EBP-β in stimulated preadipocytes. DNA binding of C/EBP-β is facilitated by MAPK phosphorylation beginning at 4 h post-stimulation and GSK3β phosphorylation-14 h into differentiation. MiR-34a regulates therapy resistance by targeting HDAC1.56 We have shown that the ability of C/EBP-β to activate C/EBP-β expression in preadipocytes stimulated to differentiate is initially reduced through the interaction of C/EBP-β with an mSin3A/histone deacetylase 1 (HDAC1) complex.57 PPAR-γ and C/EBP-β are marker genes of adipocyte differentiation. So, miR-34a can regulate adipocyte differentiation by targeting target genes.
MiR-126 Regulated Adipocyte Differentiation
MiR-126 is a single stranded small RNA molecule with a length of 23 nucleotides encoded by endogenous genes,58 which can widely mediate the regulation of physiological reactions such as cell differentiation, proliferation and migration.59 Functional analysis of miR-126 demonstrated that its overexpression conveys neurotoxicity by impairing IGF-1/PI3K/AKT signaling, and that its inhibition increases the trophic effects of IGF-1.60 Studies also confirmed that miR-126 exerted these pivotal functions by down-regulating the expression of CRK.61 During 3T3-Ll cell differentiation induction, C-CRK is phosphorylated on tyrosine by IGF-1 receptor kinase and dephosphorylated by PTPase.62 In addition, over-expression of miR126 down-regulated IRS-1 expression, suppressed AKT and ERK1/2 activation. Decreased expression of IRS-1 in embryonic fibroblast cells severely decreased the expression of C/EBPα and PPARγ.63 The inhibitory effect of mir-126 on VEFG expression was investigated and indicated that VEGF is a target of miR-126.64 Retrovirus-mediated restoration of VEGF expression in mutant cells reduced adipocyte differentiation to the levels exhibited by control cells.65 In a word, miR-126 played an important role in adipocyte differentiation.
MiR–143-3p Regulated Adipocyte Differentiation
miR-143 was identified to promote adipocyte differentiation by using antisense oligonucleotides.66 There are many target genes of miR-143-3p that play a regulatory role in adipocyte differentiation, such as MAPK7,67 MAP3K7,68 AKT,69 KLF5,70 PI3K71 and EZH2.72 Firstly, MAPK7 inhibited adipocyte differentiation73 and MAP3K7 induces adipocyte differentiation through PPARγ signaling.74 Secondly, AKT/PKB may play a role in suppression of apoptosis and negatively regulate preadipocyte differentiation.75 KLF5 is also induced by C/EBPβ/δ, and that it then acts in concert with C/EBPβ/δ to regulate PPARγ2 expression76 and EZH2-induced H3K27me3 of WNT gene promoters facilitated adipogenic differentiation of murine preadipocytes.77 Finally, IRSs/PI3K signal pathway may play an important role in the differentiation of 3T3-L1 preadipocytes by regulating the expression of C/EBPα and PPARγ.78 These results suggest that miR-143-3p can regulate adipocyte differentiation.
MiR–221/222 Regulated Adipocyte Differentiation
miR-221/222, located in a cluster on chromosome Xp11.3, are considered part of the same family. They share the same’seed’ sequence, short regions at their 5’ ends through which they bind their target sites in mRNA 3’-UTRs. Studies showed that miR-221 and 222, by targeting PTEN and TIMP3 tumor suppressors, induce TRAIL resistance and activate the AKT pathway.79,80 SH2-B is a key regulator of adipogenesis both in vivo and in vitro by regulating the insulin/IGF-I receptor-AKT-FOXO1-PPARγ pathway,81 which indicates PTEN, TIMP3 and AKT genes play an important role in adipogenesis. In addition, miR-221 and 222 inhibited the expression of p27Kip182 and Genetic ablation of p27Kip1 in mice leads to adipocyte hyperplasia.83 In a word, miR–221/222 can regulate adipocyte differentiation by multiple pathways.
MiR-320 Regulated Adipocyte Differentiation
MiR-320 is involved in a variety of pathological processes, including cell proliferation and differentiation.114 The present results provided evidence that the miR-320/ELF3 axis regulated tumor progression via the PI3K/AKT signaling pathway.111 Activated form of PI3K, a critical target of IRS1 downstream, led to phosphorylation of phosphatidyl inositides and then activated the downstream main target AKT, which is pivotal in regulating 3T3-L1 preadipocyte differentiation.115,116 In addition, Data study indicates that miR-320 negatively regulates expression of ET-1, VEGF, and FN through ERK1/2.109 The adipocyte-specific transcription factor PPARγ can be phosphorylated by ERK1/2 to decrease its transcriptional activity and inhibit adipocyte differentiation.117 Finally, A luciferase assay confirmed that miR-320 binds to the 3’-untranslated regions of AdipoR1, which indicated AdipoR1 is a target gene of miR-320.112 CTRP6 regulates proliferation and differentiation of intramuscular and subcutaneous adipocytes through the AdipoR1 (Adiponectin Receptor 1)/MAPK pathway.118 So miR-320 can regulate adipocyte differentiation by targeting ERK1/2, PI3K and adipoR1.
METTL3 Regulated Adipocyte Differentiation by Directly Modifying Key Genes
Methyltransferase-like 3 (METTL3), a key RNA methyltransferase, has been demonstrated to regulate neurogenesis,119 spermatogenesis,120,121 early embryonic development,122 stem cell pluripotency in mice,122,123 and white fat cell differentiation in vitro.18 Recently, Yao et al found that METTL3 plays an important role in BMSCs differentiation and adipogenesis and there was a negative correlation between METTL3 expression and porcine BMSCs (pBMSCs) adipogenesis.124 It was demonstrated that the deletion of METTL3 significantly promoted the pBMSCs adipogenesis process and janus kinase 1 (JAK1) protein expression via an m6A-dependent way.124 Specifically, METTL3 inhibited pBMSCs adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6A-YTHDF2–dependent manner.124 C/EBPβ is a marker gene of adipocyte differentiation, which indicates METTL3 plays an important role in regulating adipocyte differentiation.
Effect of Adipocyte Differentiation and on Fat Deposition
Fat deposition is the main means of energy storage in animals. Mammalian adipose tissue mainly exists in four forms: subcutaneous, visceral, intermuscular and intramuscular fat. Generally, the differentiation of adipocytes refers to the process of preadipocytes differentiating into multi compartment adipocytes.125,126 After 8 days of culture in vitro, precursor adipocytes were induced to differentiate into mature adipocytes by PPARγ, CEBP/a and FABP4.127 The number and volume of lipid droplets in mature adipocytes increased. At the same time, the volume of mature adipocytes also increased significantly, which also increased the content of adipose tissue. So, adipocyte differentiation promoted fat deposition.
Conclusions
In summary, although the correlation between m6A modification and fat deposition, as a hotspot in the field of genetics, has been extensively explored, most studies concentrated on gene sequencing analysis, differential expression analysis, and modification site analysis. There are few studies on the functional phenotypes and mechanisms of action at the cell level, but studies in this field are likely to be key to revealing the origin of fat deposition, especially the origin and development of obesity. With a deep understanding of mechanism of fat deposition and the targeted study for m6A modification, m6A modification then provides a new perspective for elucidating the occurrence and development of related obesity diseases, providing a new direction for guiding the diagnosis and treatment of obesity diseases.
Data Sharing Statement
All of the data used in this research appears in the manuscript and is available at request from corresponding author.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Yangling Demonstration Area Industry University research and application collaborative innovation major project(1017cxy-15), Agricultural science and technology innovation and tackling key projects in Shaanxi Province (2016NY-108) and Integration and demonstration of rabbit breeding and factory breeding technology (2018ZDXM-NY-041).
Disclosure
The authors declare no conflict of interest.
References
1. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71(10):3971–3975. doi:10.1073/pnas.71.10.3971
2. Shi H, Wang X, Lu Z, et al. Ythdf3 facilitates translation and decay of n6-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–328. doi:10.1038/cr.2017.15
3. Wang X, He C. Dynamic RNA modifications in posttranscriptional regulation. Mol Cell. 2014;56(1):5–12. doi:10.1016/j.molcel.2014.09.001
4. Luo J, Xu T, Sun K. N6-methyladenosine RNA modification in inflammation: roles, mechanisms, and applications. Front Cell Dev Biol. 2021;9:670711. doi:10.3389/fcell.2021.670711
5. Zhao W, Cui Y, Liu L, Ma X, Wu J. Mettl3 facilitates oral squamous cell carcinoma tumorigenesis by enhancing c-myc stability via ythdf1-mediated m6a modification. Mol Ther Nucleic Acids. 2020;20:1–12. doi:10.1016/j.omtn.2020.01.033
6. Schller E, Weichmann F, Treiber T, Ringle S, Meister G. Interactions, localization, and phosphorylation of the m6A generating METTL3–METTL14–WTAP complex. RNA. 2018;24(4):499–512. doi:10.1261/rna.064063.117
7. Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from hela cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269(26):17697–17704. doi:10.1016/S0021-9258(17)32497-3
8. Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of mettl3 and mettl14 methyltransferases. Mol Cell. 2016;36(2):306–317. doi:10.1016/j.molcel.2016.05.041
9. Huang J, Dong X, Gong Z, et al. Solution structure of the RNA recognition domain of mettl3-mettl14 n6-methyladenosine methyltransferase. Protein Cell. 2018;10(4):272–284. doi:10.1007/s13238-018-0518-7
10. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi:10.1038/nature14281
11. Bartel DP. microRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
12. Teleman AA. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Develop. 2006;20(4):417–422. doi:10.1101/gad.374406
13. Poy MN, Spranger M, Stoffel M. microRNAs and the regulation of glucose and lipid metabolism. Diabetes Obes Metab. 2010;9(s2):67–73. doi:10.1111/j.1463-1326.2007.00775.x
14. Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi:10.1038/nature03076
15. Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk R. Targeted inhibition of miRNA maturation with morpholinos reveals a role for mir-375 in pancreatic islet development. PLoS Biol. 2007;5(8):e203. doi:10.1371/journal.pbio.0050203
16. Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3t3-l1 adipogenesis. Mol Endocrinol. 2009;23(6):925–931. doi:10.1210/me.2008-0298
17. You HS, Ka S, Kim AY, Kim JB. Regulation of adipocyte differentiation via microRNAs. Endocrinol Metab. 2014;29(2):122–135. doi:10.3803/EnM.2014.29.2.122
18. Masatoshi K, Mitsuru O, Takayoshi S, et al. The RNA methyltransferase complex of wtap, mettl3, and mettl14 regulates mitotic clonal expansion in adipogenesis. Mol Cell Biol. 2018;38:116–118.
19. Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. doi:10.1038/nchembio.1432
20. Roignant JY, Soller M. M6a in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33(6):380–390. doi:10.1016/j.tig.2017.04.003
21. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated fto. Nat Chem Biol. 2011;1(12):885–887. doi:10.1038/nchembio.687
22. Qi JY, Sang L, Lin M, et al. Mettl3–mettl14 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells. Cell Res. 2018;28(9):952–954. doi:10.1038/s41422-018-0062-2
23. Zheng G, Dahl JA, Niu Y, et al. Alkbh5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi:10.1016/j.molcel.2012.10.015
24. Ping XL, Sun BF, Xiao W, et al. Mammalian wtap is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. doi:10.1038/cr.2014.3
25. Shuibin L, Junho C, Du P, et al. The m(6)a methyltransferase mettl3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. doi:10.1016/j.molcel.2016.03.021
26. Jin-Zhao M, Fu Y, Chuan-Chuan Z, Feng L, Ji-Hang Y. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary microRNA processing. Hepatology. 2017;65(2):529–543. doi:10.1002/hep.28885
27. Xin Z, Hou Y, Tuo Z, Wei F. Application values of mir-194 and mir-29 in the diagnosis and prognosis of gastric cancer. Exp Ther Med. 2018;15:4179–4184.
28. Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6) a-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–1308. doi:10.1016/j.cell.2015.08.011
29. Diao LT, Xie SJ, Lei H, et al. Mettl3 regulates skeletal muscle specific miRNAs at both transcriptional and post-transcriptional levels. Biochem Biophys Res Commun. 2021;552:52–58. doi:10.1016/j.bbrc.2021.03.035
30. Lintao Zhong M, Xiang H, Song H, et al. Mettl3 induces abdominal aortic aneurysm development and progression by modulating n6-methyladenosine-dependent primary mir34a processing. Mol Ther Nucleic Acids. 2020;21:394–411.
31. Li X, Xiong W, Long X, et al. Inhibition of mettl3/m6a/mir126 promotes the migration and invasion of endometrial stromal cells in endometriosis. Biol Reprod. 2021;104(1):1–39. doi:10.1093/biolre/ioaa162
32. Zhang J, Bai R, Li M. Excessive mir-25-3p maturation via n6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10(1):1858. doi:10.1038/s41467-019-09712-x
33. Han J, Wang JZ, Yang X, Yu H, Yang H. Mettl3 promote tumor proliferation of bladder cancer by accelerating pri-mir221/222 maturation in m6a-dependent manner. Mol Cancer. 2019;18(1):2–15. doi:10.1186/s12943-019-1036-9
34. Wang H, Deng Q, Lv Z, Ling Y, Chen L. N6-methyladenosine induced mir-143-3p promotes the brain metastasis of lung cancer via regulation of vash1. Mol Cancer. 2019;18(1):181. doi:10.1186/s12943-019-1108-x
35. Yan G, Yuan Y, He M, et al. M 6 a methylation of precursor-mir-320/runx2 controls osteogenic potential of bone marrow-derived mesenchymal stem cells. Mol Ther Nucleic Acids. 2020;19:421–435. doi:10.1016/j.omtn.2019.12.001
36. Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–676. doi:10.1016/S0092-8674(03)00428-8
37. Meng F, Henson R, Wehbe–Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. doi:10.1053/j.gastro.2007.05.022
38. Dey N, Das F, Mariappan MM, Mandal CC, Choudhury GG. MicroRNA-21 orchestrates high glucose-induced signals to torc1 for renal cell pathology in diabetes. J Biol Chem. 2011;286(29):25586–25603. doi:10.1074/jbc.M110.208066
39. Nakashima N. The tumor suppressor PTEN negatively regulates insulin signaling in 3t3-l1 adipocytes. J Biol Chem. 2000;275(17):12889–12895. doi:10.1074/jbc.275.17.12889
40. Li YX, Meng JX, Cai XZ, Li DF, Yu XY. [Induced differentiation and signaling factor PTEN expression of 3t3-l1 adipocytes]. Nan Fang Yi Ke Da Xue Xue Bao. 2007;27(3):259–263. Chinese.
41. Lee SK, Lee JO, Ji HK, et al. Metformin sensitizes insulin signaling through AMPK-mediated PTEN down-regulation in preadipocyte 3t3-l1 cells. J Cell Biochem. 2011;112(5):1259–1267. doi:10.1002/jcb.23000
42. Luo M, Tan X, Mu L, et al. MiRNA-21 mediates the antiangiogenic activity of metformin through targeting PTEN and SMAD7 expression and PI3K/AKT pathway. Sci Rep. 2017;7(1):43427. doi:10.1038/srep43427
43. Ouyang D, Xu L, Zhang L, et al. MiR-181a-5p regulates 3T3-L1 cell adipogenesis by targeting Smad7 and Tcf7l2. Acta Biochim Biophys Sin (Shanghai). 2016;48(11):1034–1041. doi:10.1093/abbs/gmw100
44. Liang WC, Wang Y, Liang PP, et al. Mir-25 suppresses 3t3-l1 adipogenesis by directly targeting klf4 and c/ebpα. J Cell Biochem. 2015;116(11):2658–2666. doi:10.1002/jcb.25214
45. He Z, Liu Y, Xiao B, Qian X. Mir-25 modulates NSCLC cell radio-sensitivity through directly inhibiting btg2 expression. Biochem Biophys Res Commun. 2015;457(3):235–241. doi:10.1016/j.bbrc.2014.12.094
46. Jie X, Hang JB, Che JM, Li HC. Mir-25 is up-regulated in non-small cell lung cancer and promotes cell proliferation and motility by targeting fbxw7. Int J Clin Exp Pathol. 2015;8:9147–9153.
47. Wu T, Hu H, Zhang T, Jiang L, Lu Z. Mir-25 promotes cell proliferation, migration, and invasion of non-small-cell lung cancer by targeting the LATS2/YAP signaling pathway. Oxid Med Cell Longev. 2019;2019:1–14. doi:10.1155/2019/3832648
48. Feng X, Jiang J, Shi S, Xie H, Zhou L, Zheng S. Knockdown of mir-25 increases the sensitivity of liver cancer stem cells to trail-induced apoptosis via PTEN/PI3K/AKT/BAD signaling pathway. Int J Oncol. 2016;49(6):2600–2610. doi:10.3892/ijo.2016.3751
49. Bengoechea-Alonso MT, Ericsson J. The ubiquitin ligase fbxw7 controls adipocyte differentiation by targeting c/ebpα for degradation. Proc Natl Acad Sci USA. 2010;107(26):11817–11822. doi:10.1073/pnas.0913367107
50. Yang A, Kang Q, Zhao Y, Hu X, Li N, Hong W. Lats2 modulates adipocyte proliferation and differentiation via hippo signaling. PLoS One. 2013;8(8):e72042. doi:10.1371/journal.pone.0072042
51. Wang D, Zhou Y, Lei W, et al. Signal transducer and activator of transcription 3 (stat3) regulates adipocyte differentiation via peroxisomeâ-proliferatorâ-activated receptor-r. Biol Cell. 2010;102(1):1–12. doi:10.1042/BC20090070
52. Ding J, Li M, Wan X, et al. Effect of mir-34a in regulating steatosis by targeting pparα expression in nonalcoholic fatty liver disease. Sci Rep. 2015;5:13729.
53. Li T, Pang Q, Liu Y, Bai M, Zhang Z. Sulforaphane protects human umbilical vein endothelial cells from oxidative stress via the miR34a/SIRT1 axis by upregulating nuclear factor erythroid-2-related factor 2. Exp Ther Med. 2021;21(1):1–9. doi:10.3892/etm.2020.9451
54. Picard F, Kurtev M, Chung NJ, Topark-Ngarm A, Senawong T, Oliveira D. Sirt1 promotes fat mobilization in white adipocytes by repressing ppar-gamma. Nature. 2004;429(6993):771–776. doi:10.1038/nature02583
55. Wen F, An C, Wu X, Yi Y, Yang Z. Mir-34a regulates mitochondrial content and fat ectopic deposition induced by resistin through the AMPK/PPARα pathway in HEPG2 cells. Int J Biochem Cell Biol. 2018;94:133–145. doi:10.1016/j.biocel.2017.11.008
56. Myw A, Jf B, Xx C, Jw D, Rcw A. Mir-34a regulates therapy resistance by targeting hdac1 and hdac7 in breast cancer. Cancer Lett. 2014;354(2):311–319. doi:10.1016/j.canlet.2014.08.031
57. Abdou HS, Atlas E, Hache R. Liver-enriched inhibitory protein (LIP) actively inhibits preadipocyte differentiation through histone deacetylase 1 (HDAC1). J Biol Chem. 2011;286(24):21488–21499. doi:10.1074/jbc.M110.211540
58. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight. Nat Rev Genet. 2008;9(2):102–114. doi:10.1038/nrg2290
59. Zhou J, Li YS, Nguyen P, Wang KC, Shu C. Regulation of vascular smooth muscle cell turnover by endothelial cell-secreted microRNA-126: role of shear stress. Circ Res. 2013;113(1):40–51. doi:10.1161/CIRCRESAHA.113.280883
60. Kim W, Lee Y, Mckenna ND, et al. Mir-126 contributes to Parkinson’s disease by dysregulating the insulin-like growth factor/phosphoinositide 3-kinase signaling. Neurobiol Aging. 2014;35(7):1712–1721. doi:10.1016/j.neurobiolaging.2014.01.021
61. Feng R, Chen X, Yu Y, et al. Mir-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298(1):50–63. doi:10.1016/j.canlet.2010.06.004
62. Liao K, Zhai B, Jin S. Role of proto-oncogene crk in the differentiation induction of 3t3-l1 preadipocyte. Biochem Soc Trans. 2000;28(5):
63. Miki H, Yamauchi T, Suzuki R, et al. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Mol Cell Biol. 2001;21(7):2521–2532. doi:10.1128/MCB.21.7.2521-2532.2001
64. Liu B, Peng XC, Zheng XL, Wang J, Qin YW. Mir-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66(2):169–175. doi:10.1016/j.lungcan.2009.01.010
65. Liu Y, Berendsen AD, Jia S, et al. Intracellular VEGF regulates the balance between osteoblast and adipocyte differentiation. J Clin Invest. 2012;122(9):3101–3113. doi:10.1172/JCI61209
66. Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279(50):52361. doi:10.1074/jbc.C400438200
67. Xia C, Yang Y, Kong F, Kong Q, Shan C. Mir-143-3p inhibits the proliferation, cell migration and invasion of human breast cancer cells by modulating the expression of mapk7. Biochimie. 2018;147:98–104. doi:10.1016/j.biochi.2018.01.003
68. Fan H, Ge Y, Ma X, Li Z, Xu Z. Long non-coding RNA CCDC144NL-AS1 sponges miR-143-3p and regulates MAP3K7 by acting as a competing endogenous RNA in gastric cancer. Cell Death Dis. 2020;11(7):1–12. doi:10.1038/s41419-020-02740-2
69. Dong Y, Feng S, Dong F. Maternally-expressed gene 3 (meg3)/mir-143-3p regulates injury to periodontal ligament cells by mediating the akt/inhibitory κb kinase (ikk) pathway. Med Sci Monit. 2020;26:e922486. doi:10.12659/MSM.922486
70. Wangzhou K, Lai Z, Lu Z, Fu W, Hao C. Mir-143-3p inhibits osteogenic differentiation of human periodontal ligament cells by targeting klf5 and inactivating the wnt/β-catenin pathway. Front Physiol. 2021;11:606967. doi:10.3389/fphys.2020.606967
71. Jin YP, Hu YP, Wu XS, et al. Mir-143-3p targeting of itga6 suppresses tumour growth and angiogenesis by downregulating plgf expression via the pi3k/akt pathway in gallbladder carcinoma. Cell Death Dis. 2018;9:1–15.
72. Feng Y, Zhang C, Zhang S, Hou L, Luo H. miR-143-3p regulates proliferation, migration and invasion of colon cancer RKO cells via targeting EZH2. Chin J Cancer Biother. 2020;27:735–741.
73. Peiwen Z, Du J, Linghui W, Lili N, Zhao Y. MicroRNA-143a-3p modulates preadipocyte proliferation and differentiation by targeting mapk7. Biomed Pharmacother. 2018;108:531–539.
74. Zhang Y, O’Keefe RJ, Jonason JH. Bmp-tak1 (map3k7) induces adipocyte differentiation through pparγ signaling. J Cell Biochem. 2017;118:204–210.
75. Wang ZX, Jiang CS, Liu L, et al. The role of akt on arsenic trioxide suppression of 3t3-l1 preadipocyte differentiation. Cell Res. 2005;15(5):379–386. doi:10.1038/sj.cr.7290305
76. Oishi Y, Manabe I, Nagai R. [krüppel-like family of transcription factor 5 (klf5). Klf5 is a key regulator of adipocyte differentiation]. Nihon Rinsho. 2011;69(Suppl 1):264–268. Japanese.
77. Wang L, Jin Q, Lee J-E, Su I-H, Ge K. Histone h3k27 methyltransferase ezh2 represses wnt genes to facilitate adipogenesis. Proc Nat Acad Sci. 2010;107(16):7317–7322. doi:10.1073/pnas.1000031107
78. Gao JZ, Zheng RD, Wang CB, Ying YQ, Luo XP. Effects of pi3k inhibitor ly294002 on the differentiation of mouse preadipocytes and the expression of c/ebpα and pparγ. Zhongguo Dang Dai Er Ke Za Zhi. 2011;13(10):823–826.
79. Garofalo M, Leva GD, Romano G, et al. Mir-221&222 regulate trail resistance and enhance tumorigenicity through PTEN and timp3 downregulation. Cancer Cell. 2009;16(6):498–509. doi:10.1016/j.ccr.2009.10.014
80. Zhang J, Han L, Ge Y, et al. Mir-221/222 promote malignant progression of glioma through activation of the akt pathway. Int J Oncol. 2010;36:913–920.
81. Daigo Y, Naoichi S, Takehiro T, et al. Adaptor protein sh2-b linking receptor-tyrosine kinase and akt promotes adipocyte differentiation by regulating peroxisome proliferator-activated receptor gamma messenger ribonucleic acid levels. Mol Endocrinol. 2007;21:1120–1131.
82. Galardi S, Mercatelli N, Giorda E, et al. Mir-221 and mir-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27kip1. J Biol Chem. 2007;282:23716–23724.
83. Naaz A, Holsberger DR, Iwamoto GA, Nelson A, Cooke PS. Loss of cyclin-dependent kinase inhibitors produces adipocyte hyperplasia and obesity. FASEB J. 2004;18(15):1925–1927. doi:10.1096/fj.04-2631fje
84. Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J. Mir-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology. 2013;306:162–168. doi:10.1016/j.tox.2013.02.014
85. He H, Cai M, Zhu J, et al. Mir-148a-3p promotes rabbit preadipocyte differentiation by targeting PTEN. In Vitro Cell Dev Biol Anim. 2018;54(3):241–249. doi:10.1007/s11626-018-0232-z
86. He Z, Long J, Yang C, Gong B, Tang J. Lncrna dgcr5 plays a tumor-suppressive role in glioma via the miR-21/SMAD7 and miR-23a/PTEN axes. Aging. 2020;12(20):20285–20307. doi:10.18632/aging.103800
87. Chen H, Pan H, Qian Y, Zhou W, Liu X. Mir-25-3p promotes the proliferation of triple negative breast cancer by targeting btg2. Mol Cancer. 2018;17(1):4. doi:10.1186/s12943-017-0754-0
88. Kim S, Hong JW, Park KW. B cell translocation gene 2 (btg2) is regulated by stat3 signaling and inhibits adipocyte differentiation. Mol Cell Biochem. 2016;413(1–2):145–153. doi:10.1007/s11010-015-2648-z
89. Yao Y, Sun F, Ming L. miR-25 inhibits sepsis-induced cardiomyocyte apoptosis by targeting PTEN. Biosci Rep. 2018;38(2):1–9. doi:10.1042/BSR20171511
90. Lu D, Davis M, Abreu-Goodger C, Wang W, Liu P. MiR-25 regulates Wwp2 and Fbxw7 and promotes reprogramming of mouse fibroblast cells to iPSCs. PLoS One. 2012;7(8):e40938. doi:10.1371/journal.pone.0040938
91. Lee HW, Rhee DK, Kim BO, Pyo S. Inhibitory effect of sinigrin on adipocyte differentiation in 3t3-l1 cells: involvement of AMPK and MAPK pathways. Biomed Pharmacother. 2018;102:670–680. doi:10.1016/j.biopha.2018.03.124
92. Choi Y, Um SJ, Park T. Indole-3-carbinol directly targets sirt1 to inhibit adipocyte differentiation. Int J Obes. 2013;37(6):881–884. doi:10.1038/ijo.2012.158
93. Guohua L, Yanning L, Shanshan W, Jihua X, Fan Y. The p53/mir-34a/sirt1 positive feedback loop in quercetin-induced apoptosis. Cell Physiol Biochem. 2015;35(6):2192–2202. doi:10.1159/000374024
94. Claire K, Houssein-Salem A, Rj H, Atlas E. Inactivation of histone deacetylase 1 (hdac1) but not hdac2 is required for the glucocorticoid-dependent ccaat/enhancer-binding protein α (c/ebpα) expression and preadipocyte differentiation. Endocrinology. 2014;155(12):4762–4773. doi:10.1210/en.2014-1565
95. Yu J, Fan Q, Li L. The MCM3AP-AS1/miR-126/VEGF axis regulates cancer cell invasion and migration in endometrioid carcinoma. World J Surg Oncol. 2021;19(1):1–8. doi:10.1186/s12957-021-02316-0
96. Kim W, Lee Y, Mckenna ND, et al. Mir-126 contributes to Parkinson disease by dysregulating igf-1/pi3k signaling. Neurobiol Aging. 2014;35:1712–1721.
97. Di Z, Shi S, Wang H, Kan L. Growth arrest induces primary-cilium formation and sensitizes igf-1-receptor signaling during differentiation induction of 3t3-l1 preadipocytes. J Excell. 2009;122:2760–2768.
98. Lu W, Tao X, Fan Y, et al. Linc00888 promoted tumorigenicity of melanoma via mir-126/crk signaling axis. Onco Targets Ther. 2018;11:4431–4442. doi:10.2147/OTT.S164711
99. Hu L, Xu H, Lu J, Zhou Y, Chu F. MicroRNA-126 deficiency affects the development of thymus CD4+ single-positive cells through elevating IRS-1. Int Arch Allergy Immunol. 2018;177(3):207–218. doi:10.1159/000490710
100. Kim HL, Lee HJ, Choi BK, Park SB, Lee DR. Roots extract of adenophora triphylla var. Japonica inhibits adipogenesis in 3t3-l1 cells through the downregulation of irs1. J Physiol Pathol Korean. 2020;34(3):136–141. doi:10.15188/kjopp.2020.06.34.3.136
101. Chen JJ, Zhou SH. Mesenchymal stem cells overexpressing mir-126 enhance ischemic angiogenesis via the akt/erk-related pathway. Cardiol J. 2011;18(6):675–681. doi:10.5603/CJ.2011.0032
102. Kim H, Sakamoto K. (-)-epigallocatechin gallate suppresses adipocyte differentiation through the mek/erk and pi3k/akt pathways. Cell Biol Int. 2013;36(2):147–153. doi:10.1042/CBI20110047
103. Gao H, Peng C, L. W, et al. Yiqi-huoxue granule promotes angiogenesis of ischemic myocardium through mir-126/pi3k/akt axis in endothelial cells. Phytomedicine. 2021;92:153713. doi:10.1016/j.phymed.2021.153713
104. Wu X, Li J, Chang K, Yang F, Xu Y. Histone h3 methyltransferase ezh2 promotes white adipocytes but inhibits brown and beige adipocyte differentiation in mice. Biochim Biophys Acta Mol Cell Biol Lipids. 2021;1866:158901. doi:10.1016/j.bbalip.2021.158901
105. Li L, Wang Z, Hu X, et al. Human aortic smooth muscle cell-derived exosomal mir-221/222 inhibits autophagy via a PTEN/akt signaling pathway in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2016;479(2):343–350. doi:10.1016/j.bbrc.2016.09.078
106. Bernot D, Barruet E, Poggi M, Bonardo B, Alessi MC, Peiretti F. Down-regulation of tissue inhibitor of metalloproteinase-3 (timp-3) expression is necessary for adipocyte differentiation. J Biol Chem. 2010;285(9):6508–6514. doi:10.1074/jbc.M109.078444
107. Zhang C, Zhang A, Hao J, Zhang J, Kang C. Abstract 3962: knocked down mir-221/222 inhibits akt pathway to suppress malignant glioma cell growth independent of PTEN status. Cancer Res. 2011;71:3962.
108. Okada M, Sakai T, Nakamura T, et al. Skp2 promotes adipocyte differentiation via a p27kip1-independent mechanism in primary mouse embryonic fibroblasts. Biochem Biophys Res Commun. 2009;379(2):249–254. doi:10.1016/j.bbrc.2008.12.069
109. Feng B, Chakrabarti S. Mir-320 regulates glucose-induced gene expression in diabetes. ISRN Endocrinol. 2012;2012:549875. doi:10.5402/2012/549875
110. Turpin E, Muscat A, Vatier C, et al. Carbamazepine directly inhibits adipocyte differentiation through activation of the erk 1/2 pathway. Br J Pharmacol. 2013;168(1):139–150. doi:10.1111/j.1476-5381.2012.02140.x
111. Zhang Z, Zhang J, Li J, et al. Mir320/elf3 axis inhibits the progression of breast cancer via the pi3k/akt pathway. Oncol Lett. 2020;19(4):3239–3248. doi:10.3892/ol.2020.11440
112. Guo W, Hu S. Mir-320 mediates diabetes amelioration after duodenal-jejunal bypass via targeting adipor1. Surg Obes Relat Dis. 2018;14(7):960–971. doi:10.1016/j.soard.2018.03.007
113. Liu HF, Gui MX, Dong H, Wang X, Li XW. Differential expression of adipor1, igfbp3, pparγ and correlative genes during porcine preadipocyte differentiation. In Vitro Cell Dev Biol Anim. 2012;48(1):54–60. doi:10.1007/s11626-011-9468-6
114. Li H, Fan J, Zhao Y, Zhang X, Wang DW. Nuclear mir-320 mediates diabetes-induced cardiac dysfunction by activating transcription of fatty acid metabolic genes to cause lipotoxicity in the heart. Circ Res. 2019;125(12):1106–1120. doi:10.1161/CIRCRESAHA.119.314898
115. Hirsch E, Katanaev VL, Garlanda C, et al. Central role for g protein-coupled phosphoinositide 3-kinase γ in inflammation. Science. 2000;287(5455):1049–1053. doi:10.1126/science.287.5455.1049
116. Xu J, Liao K. Protein kinase b/akt 1 plays a pivotal role in insulin-like growth factor-1 receptor signaling induced 3t3-l1 adipocyte differentiation. J Biol Chem. 2004;279(34):35914–35922. doi:10.1074/jbc.M402297200
117. Hu E, Kim J, Sarraf P, Spiegelman BM. Inhibition of adipogenesis through map kinase-mediated phosphorylation of ppargamma. Science. 1996;274(5295):2100–2103. doi:10.1126/science.274.5295.2100
118. Wu W, Zhang J, Zhao C, Sun Y, Wei-Jun P, Yang G. Ctrp6 regulates porcine adipocyte proliferation and differentiation by the adipor1/MAPK signaling pathway. J Agric Food Chem. 2017;65(27):5512–5522. doi:10.1021/acs.jafc.7b00594
119. Mac C, Chang M, Hongyi L, et al. RNA m(6)a methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018;19(1):68. doi:10.1186/s13059-018-1435-z
120. Zea L. Mettl3-/mettl14-mediated mRNA n6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017;27(10):1216–1230. doi:10.1038/cr.2017.117
121. Xu K, Yang Y, Feng GH, et al. Mettl3-mediated m6a regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27(9):1100–1114. doi:10.1038/cr.2017.100
122. Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. doi:10.1126/science.1261417
123. Xiao-Li P, Tong C, Meng W, Miao-Miao Y, Xu Z. M(6)a RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:389–401.
124. Yao Y, Bi Z, Wu R, et al. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6 A-YTHDF2–dependent manner. FASEB J. 2019;33(6):7529–7544. doi:10.1096/fj.201802644R
125. Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14(11):1293–1307. doi:10.1101/gad.14.11.1293
126. Rodríguez C, Acosta C, Badía P, Cejas JR, Santamaría F, Lorenzo A. Assessment of lipid and essential fatty acids requirements of Black Seabream (spondyliosoma cantharus) by comparison of lipid composition in muscle and liver of wild and captive adult fish. Comp Biochem Physiol B Biochem Mol Biol. 2004;139(4):619–629. doi:10.1016/j.cbpc.2004.07.013
127. Fang D, Qing-qing L, Le L, Chao G, Xin Y. Isolation, culture and differentiation of duck (anas platyrhynchos) preadipocytes. Cytotechnology. 2015;67(5):773–781. doi:10.1007/s10616-014-9715-2
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
