Back to Journals » OncoTargets and Therapy » Volume 15
Prognostic Implications of Six Altered Genes in Asian Non-Surgical Esophageal Carcinoma Patients Treated with Chemoradiotherapy
Authors Feng A, Yang N, Yu R , Liu J , Pang J, Wu X, Shao Y, Yang Z, Dai H
Received 24 August 2021
Accepted for publication 26 December 2021
Published 13 January 2022 Volume 2022:15 Pages 41—51
DOI https://doi.org/10.2147/OTT.S334580
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Alei Feng,1,2 Ning Yang,1 Ruoying Yu,3 Jingwen Liu,3 Jiaohui Pang,3 Xue Wu,3 Yang Shao,3,4 Zhe Yang,1 Honghai Dai1
1Tumor Research and Therapy Center, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, 250021, People’s Republic of China; 2Shandong Qidu Pharmaceutical Co. Ltd., Shandong Provincial Key Laboratory of Neuroprotective Drugs, Zibo, 255400, People’s Republic of China; 3Nanjing Geneseeq Technology Inc., Nanjing, Jiangsu, People’s Republic of China; 4School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China
Correspondence: Honghai Dai; Zhe Yang
Tumor Research and Therapy Center, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, 250021, People’s Republic of China
Email [email protected]; [email protected]
Background: Esophageal cancer (EC), especially esophageal squamous cell carcinoma, remained as one of the most aggressive tumors in China with a five-year survival rate of around 40%. Molecular characteristics through next-generation sequencing are becoming an emerging method in identifying prognostic biomarkers for better treatment management for EC patients.
Methods: Targeted next-generation sequencing using a 422-gene pan-cancer panel was performed with tumor tissue samples from a total of 69 Asian non-surgical esophageal carcinoma patients (AEC) treated with chemoradiotherapy. A TCGA cohort of 143 EC patients and another Asian ESCC cohort of 47 patients were employed for validation.
Results: In the AEC cohort, alterations in TP53 (94.2%) and NOTCH1 (55.1%) were the two most frequently observed alterations, whereas in the TCGA cohort, only TP53 alterations were observed at a high ratio (85.3%). Co-amplifications of FGF19 and CCND1 were found at a similar ratio in both cohorts. Multiple alterations in the DNA damage pathway were identified but not associated with overall survival in AEC. Using univariate and multivariate Cox regression analyses, six gene alterations including YAP1 amplification, RB1 alteration, BAP1 mutation, MYC amplification, WRN mutation, and BRIP1 mutation were identified as adverse prognostic factors in the AEC cohort. A Cox proportional hazard model based on the six prognosis-related genes was constructed and showed the ability in distinguishing EC patients with poorer disease outcomes in AEC and two validation cohorts.
Conclusion: Six gene alterations were found to be potential unfavorable prognostic markers that might provide guidance in the treatment management for EC patients.
Keywords: esophageal cancer, YAP1, RB1, BAP1, MYC, BRIP1, WRN, overall survival, chemoradiotherapy
Introduction
Esophageal cancer (EC) is the one of most aggressive tumors in China, with a higher ratio of age-standardized mortality rate in Chinese males compared to Chinese females as well as the UK, the USA and even worldwide.1 Esophageal squamous cell carcinoma (ESCC) is more often observed in the Asian population, while esophageal adenocarcinoma (EAC), on the other hand, occurs mainly in North America and Western Europe.1,2 Smoking and alcohol consumption are two high-risk factors for ESCC,3 whereas obesity is a strong risk factor for EAC.4 The 5-year survival rates of EC depend on several factors, including the stage of cancer at the time of diagnosis. In China, EC patients achieved a 5-year survival rate of around 40%.5,6 In the US, the 5-year survival rate is 47% for EC patients with only localized tumors and 20% for all EC patients combined according to SEER database (cancer.org).
According to the National Comprehensive Cancer Network (NCCN) guideline, surgery is a major treatment used for locally advanced resectable EC patients with additional preoperative chemoradiation or perioperative chemotherapy to improve survival.7 Targeted treatment strategies have also been explored in EC patients including HER2-targeted therapy, anti-angiogenesis therapy, and immunotherapy. HER2 TKI trastuzumab has been approved by the FDA for HER2-positive advanced ECs.8 Ramucirumab, a VEGFR-2 antibody, has been approved for pre-treated patients with advanced or metastatic EAC initially as monotherapy and subsequently as combination therapy with paclitaxel.9 Pembrolizumab is approved in 2017 by the FDA for EC patients with high microsatellite instability and/or PD-L1 expression.10
With the aid of next-generation sequencing, molecular characteristics are becoming an emerging aspect in multidisciplinary treatment decision-making to create an overall therapeutic plan for cancer patients. The genomic landscape of EC genomes has been studied in both Asian and Western populations.11,12 Highly mutated genes in EC included TP53, NOTCH1, PIK3CA, RB1, CDKN2A have been identified in different ethnic groups. The association between gene alterations and prognosis has been indicated.13,14 Here, we performed targeted panel sequencing of tumor tissues from 69 Asian EC patients. A TCGA cohort of 143 EC patients was also included in this study for comparison. Six gene alterations were shown to be potential prognostic biomarkers for overall survival in EC patients using univariate and multivariate Cox regression analyses. The Cox proportional hazard model built on the six potential prognostic biomarkers demonstrated the ability to select patients with worse disease outcome in the EC patients.
Method
Patient Cohort
A total of 69 patients clinically diagnosed as non-surgical EC was retrospectively recruited from Shandong Provincial Hospital Affiliated to Shandong University according to the NCCN guidelines.7 The inclusion criteria were as follows: 1) all patients had histologically proven primary non-surgical EC and 2) the patients were treated with chemoradiotherapy. The study was approved by the Ethical Review Board of Shandong Provincial Hospital, and informed written consent was obtained from each participant.
DNA Extraction and Library Preparation
As previously described,15 the genomic DNA was extracted from formalin‐fixed and paraffin‐embedded (FFPE) using the QIAamp DNA FFPE Tissue Kit (Qiagen). The extracted DNA quantity was evaluated using a Qubit 3.0 fluorometer and extracted DNA quality was measured using Nanodrop 2000 (Thermo Fisher Scientific). Sequencing libraries were prepared using the KAPA Hyper Prep Kit (KAPA Biosystems) and sequenced with a pan-cancer panel of 422 genes.
Sequencing Data Analysis
Sequencing data analysis was performed as previously described.15 In brief, FASTQ file quality control was performed with trimmomatic16 (below 15 or N bases were removed). Reads were mapped to the reference Human Genome (hg19) using Burrows-Wheeler Aligner (BWA-mem, v0.7.12; https://github.com/lh3/bwa/tree/master/bwakit). VarScan217 was used for somatic mutation detection. Genome Analysis Toolkit was applied to local realignment around the indels and base quality score recalibration (GATK 3.4.0; https://software.broadinstitute.org/gatk/), which was also used for detecting germline mutations. Somatic variants were called if mutant allele frequency (MAF) was at least 0.2%, and at least three supporting-reads from both directions. Common SNPs were filtered out according to dbSNP (v137) and the 1000 Genomes database, and annotated using ANNOVAR.18 Genomic fusions were identified by FACTERA19 with default parameters. Copy-number variations (CNVs) were detected using ADTEx (http://adtex.sourceforge.net) with default parameters. Somatic CNVs were identified with the cut-off of 0.65 for copy-number loss and 1.50 for copy-number gain using paired normal/tumor samples for each exon.
Data Analysis
The Kaplan–Meier survival curves were performed to estimate OS in different genomic groups. The log‐rank test was performed to analyze differences between groups. The univariate and multivariate analyses were performed to evaluate the prognostic value of clinicopathological characteristics and gene alterations on OS. Overall survival (OS) was measured from the date of pathological diagnosis of EC to the date of death or last follow-up.
Backward stepwise selection with the Akaike information criterion (AIC) was used to identify variables for the multivariate Cox proportional hazards model. Model performance was evaluated by assessing discrimination against the index of concordance (C-index) and plotting Kaplan–Meier curves over the quartiles of prediction by nomogram.20 The optimal cutoff for risk score was selected using X-tile based on the best model as shown in Figure S4. X-tile was a published method for biomarker assessment and out-based cut-point optimization. X-tile can assess the robustness of the relationship between a biomarker and outcome by the construction of a two-dimensional projection of every possible subpopulation, which was also employed in other studies.21,22 Using the optimal cut-off, Kaplan–Meier curve was generated in the test and complete sets to validate the used cutoff and risk score model.
Result
Description of Analytical Cohort
We obtained 69 tumor tissue samples from a total of 69 Asian patients with non-surgical esophageal cancer (AEC cohort, Table 1) was enrolled in this study with a median age of 64 years old (yrs), ranging from 41 to 83 yrs. More than 80% of the patients (81.60%) were male and the rest (17.39%) was female. Almost all patients were squamous cell carcinomas (SCC, 98.55%) except one patient was adenocarcinomas (ADC, 1.45%). There were 17 (24,64%) stage II, 40 (57.97%) stage III, and 12 (17.29%) stage IV patients. Among them, 65.22% of the patients were smokers and more than half of patients (52.17%) had a history of alcohol consumption. All AEC patients were treated with chemoradiotherapy.
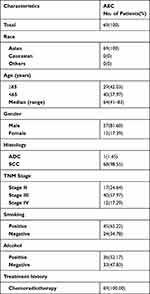 |
Table 1 Demographic Characteristics of Patients in AEC Cohort |
Meanwhile, a TCGA cohort consisting of 143 patients with esophageal cancer was employed in this study (Table S1). The TCGA cohort included 46 (32.17%) Asian cases, 77 (53.84%) Caucasian cases, and 20 (13.99%) cases of other races. The median age was 61 yrs, ranging from 36 to 90 yrs. Similar to our cohort, the majority of the TCGA cohort was male (87.41%). Histology subtypes included 39.86% ADC and 60.13% SCC. The tumor stage included 78 (54.55%) stage II, 56 (39.16%) stage III, and 9 (6/29%) stage IV. In the TCGA cohort, 58.74% of the patients were non-smokers and 55.94% of the patients never had alcohol.
Genomic Landscape of Asian Patients with Esophageal Cancer
The genomic landscape of the AEC cohort and TCGA cohort was shown in Figures 1 and S1, respectively. In the AEC cohort, nearly 95% of patients harbored TP53 alterations and more than half of patients were identified with NOTCH1 alterations. Co-amplifications of FGF19 and CCND1 were found in 36.2% of cases. Other altered genes of high frequencies in the AEC cohort included MCL1(39.1%), MYC (31.9%), PIK3CA (21.7%), and EP300(18.8%). In the TCGA cohort, TP53(85.3%) and PIK3CA (19.6%) were the top two frequently mutated genes. Compared to the AEC cohort, co-amplifications of FGF19 and CCND1 were found at a slightly lower ratio (33.6%). Alterations in multiple DNA damage repair genes were identified in both cohort including ATM (10.1% vs 13.3%), ATR (11.6% vs 4.9%), SMARCA4(7.2% vs 6.3%). Interestingly, considering only targetable mutations, EGFR showed the highest ratio in the AEC cohort and ERBB2 mostly amplification showed the highest ratio in the TCGA cohort.
 |
Figure 1 Genomic landscape of AEC cohort. The type of alterations was indicated by color. Each column represented one patient. |
Univariate and Multivariate Cox Regression Analyses of Prognostic Parameters
Clinicopathological features and genetic alterations are all potential predictors of prognosis in cancer treatment. Next, we examined the association of these possible prognostic features with patients’ overall survival (OS) using the univariate Cox regression model in the AEC cohort. As shown in Table 2, clinicopathological features including gender, age, smoking status, and alcohol consumption were not predictors of the OS in the AEC cohort. TNM stage showed some association with OS, with poorer outcome in stage III–IV patients compared to stage II (Figure S2). The most frequently observed alterations TP53 mutation was not able to predict OS (Figure S3), which might due to limited TP53 wild-type patients.
 |
Table 2 Univariate Cox Regression Analyses of Prognostic Parameters |
Interestingly, seven gene alterations were found to be independent markers of OS (Table 2) in the univariate model including BAP1 mutation, BRIP1 mutation, KDR mutation, MYC amplification, RB1 variant (mutation and deletion), WRN mutation, YAP1 amplification. In the AEC cohort, the frequencies of these seven gene alterations varied from 5.8% to 31.88% (Table 2). Next, we performed a multivariate cox regression analysis to predict OS using seven potential prognosis-related genes identified in univariate analysis (Table 3). Except for KDR with an insignificant p-value of 0.91, the rest six genes all showed as independent factors in multivariate analysis. Compared to wild type, patients with alterations in these genes were associated with poorer OS (Figure 2). Meanwhile, we also look into the association of POLE gene alterations and DDR pathway gene alterations with OS, however, the results were insignificant (Figure S4).
 |
Table 3 Multivariate Cox Regression Analyses of Prognostic Parameters. |
The Construction of Proportional Hazards Model with Six Prognosis-Related Genes and Evaluation
To distinguish EC patients with poor disease outcomes, a Cox proportional hazards model has been constructed using identified prognosis-related genes. The model was evaluated using a stepwise selection approach and the best model was then chosen with the combination of six prognosis-related genes, which minimized the Akaike Information Criterion (AIC) and maximized index of concordance (C-index). The comparison of the C-index and AIC was shown in Table 4. The six-gene model achieved the lowest AIC of 264.09 and the highest C-index of 0.75 compared to models using single-gene alterations or other gene alteration combinations. The optimal cutoff point for risk score was set at 18 according to X-tile22 (maximum χ2, p<0.0001, Figure S5). Based on this cutoff, the Kaplan–Meier survival curves showed AEC patients with a risk score >18 (15 patients, median OS:10.40 months) displayed a poorer OS than a risk score˂18 group (54 patients, median OS:41.86 months) (Figure 3A). We further examine the performance of this model using the TCGA cohort. The frequency of six prognosis-related genes in TCGA was shown in Table S2. With a cutoff at 18, this model was able to distinguish six EC patients with poor OS (median OS 14.31 vs 28.09 months, p=0.0008) (Figure 3B). We also validated our model using an independent Asian cohort of 47 ESCC patients treated with dCRT.23 As shown in Figure S6, the model identified 22 ESCC patients with a risk score higher than 18, which displayed significantly worse OS (p=0.0022) than patients with a risk score lower than 18.
 |
Table 4 Comparison of C-Index and Akaike Information Criterion (AIC) Between Models Using Different Variables |
 |
Figure 3 Kaplan–Meier curves for overall survival according to optimal cut-off point of six gene alterations in AEC (A) and TCGA (B) cohorts. |
Discussion
Here, we employed a pan-cancer NGS panel of 422 genes to study the association between clinical characteristics, gene alterations, and overall survival in the Asian EC population. The prognostic value of six potential biomarkers was further evaluated in an EC cohort from TCGA. Compared to previous studies, the genomic landscape of this AEC displayed a higher ratio of NOTCH1, MCL1, PIK3CA alterations, which is likely due to the difference in the tumor stage as well as the sequencing method used.11 In this AEC cohort, clinical characteristics including gender, age, smoking status, alcohol consumption were not associated with OS. Stage II EC patients showed slightly better OS than stage III–IV patients. Meanwhile, despite their high prevalence in EC, alterations in TP53, PIK3CA, NOTCH1 as well as FGF19 /CCND1 co-amplification were not associated with OS, which is consistent with previous studies.13,23
Several gene alterations were identified as adverse prognostic factors in the AEC cohort, some of which belonged to different oncogenic signaling pathways in cancers such as YAP1 in the Hippo pathway, MYC in the MYC pathway, RB1 in the cell cycle pathway.24 YAP1 amplification has been reported to be a prognostic factor of chemoradiotherapy in nonsurgical esophageal squamous cell carcinoma and is associated with shorter local recurrent‐free survival and OS.23 MYC amplification was also reported to be associated with poorer OS in ESCC patients.25 The associations between the rest alterations and OS in EC were First reported in this study with some indications of their prognostic value in other cancer types. BAP1, WRN, and RBIP1 are homologous recombination pathway-related genes, which can be targeted for DNA repair-targeted therapy.26,27 BAP1 is deubiquitylase associated with many cancer pathways and BAP1 mutation has been reported as prognostic factors in predicting metastatic risk.28,29 BRIP1 and WRN encode RecQ DNA helicases and are important in the normal double-strand break repair.30,31 Altered BRIP1 is a targetable alteration in ovarian cancers and breast cancer patients with overexpression of BRIP1 displayed a poor survival rate.32,33 KDR, also known as VEGFR-2, plays an important role in tumor angiogenesis and potential therapeutic target for esophageal carcinoma.34
The use of genetic biomarkers in clinical settings is increasing, and many studies have been carried out on the characterization of prognostic biomarkers in EC patients to predict disease outcomes.35–37 The identification of these markers provides a basis for detecting potential therapeutic strategies for specific molecular subtypes in clinical trials and will ultimately contribute to the personalized treatment plan for EC patients. For instance, some patients in AEC and TCGA cohort were identified as high-risk for poor disease outcomes using our model (a risk score higher than 18), different strategies could be considered for these patients: firstly, targeted therapy including trastuzumab (HER2 positive), ramucirumab (VEGFR); secondly, immunotherapy can be applied if the patient had MSI-H or PD-L1 expression according to NCCN guidelines;7 Thirdly, as shown in Figure 1, altered DDR genes especially homologous recombination pathway genes such as BRCA2, CHEK2, ATM/ATR were also identified. Preliminary studies showed that PARP inhibitors may enhance the radiosensitivity in ESCC patients.38,39 Therefore, PARP inhibitor together with radiotherapy may also be an option for high-risk populations. More studies are needed to validate our observation.
Major limitations of this study were the small cohort size and the lack of a large-scale Asian EC cohort to validate the six candidates for OS prediction. Most Asian EC studies were either RNA and gene expression level or the survival information of the cohort was unavailable. Here, we used a TCGA cohort, which consisted of 32.17% of Asian cases and 53.84% of Caucasian cases, whereas the AEC cohort was all Asian. Meanwhile, the distribution of histology subtypes varied in both cohorts. ESCC accounted for 60.13% of the TCGA and 97.1% of the AEC. Most of the treatment history from the TCGA cohort is unavailable, which may have some impact on the interpretation of the result. In conclusion, our study identified potential prognostic biomarkers for Asian EC patients. Further studies and validations of the prognostic value of these biomarkers in larger Asian clinical cohorts are warranted.
Abbreviations
EC, esophageal cancer; YAP1, yes-associated protein 1; RB1, retinoblastoma protein 1; BAP1, BRCA1-associated protein 1; MYC, MYC Proto-Oncogene; BRIP1, BRCA1 interacting protein C-terminal helicase 1; WRN, WRN RecQ Like Helicase; KDR, kinase insert domain receptor; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; NOTCH1, Notch homolog 1; FGF19, fibroblast growth factor 19; CCND1, cyclinD1 gene; ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3 related; SMARCA4, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 4.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Availability of Supporting Data
All data that support the findings of this study are available from the corresponding authors upon a reasonable request.
Ethical Approval and Consent to Participate
Patient consent form was obtained from each patient following the guideline of Institutional Review Board requirements and the Declaration of Helsinki.
Consent for Publication
No individual data were used in this study.
Acknowledgments
We would like to thank the patients who participated in this study and their family, as well as the investigators and research staff involved.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Natural Science Foundation of Shandong (Grant No. ZR2020MH229) and the project of Shandong University (Grant No. 199/2019 heng).
Disclosure
Ruoying Yu, Jingwen Liu, Jiaohui Pang, Xue Wu and Yang Shao are shareholders or employees of Nanjing Geneseeq Technology Inc. The remaining authors have no conflicts of interest to declare.
References
1. Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun. 2019;39:22. doi:10.1186/s40880-019-0368-6
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
3. Torre L, Siegel R, Ward E. Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol Biomarkers Prevent. 2016;25:16–27. doi:10.1158/1055-9965.EPI-15-0578
4. Turati F, Tramacere I, La Vecchia C. A meta-analysis of body mass index and esophageal and gastric cardia adenocarcinoma. Ann Oncol. 2013;24:609–617. doi:10.1093/annonc/mds244
5. Hou H, Meng Z, Zhao X, et al. Survival of esophageal cancer in china: a pooled analysis on hospital-based studies from 2000 to 2018. Front Oncol. 2019;9:548. doi:10.3389/fonc.2019.00548
6. He Y, Liang D, Du L, et al. Clinical characteristics and survival of 5283 esophageal cancer patients: a multiCenter study from eighteen hospitals across six regions in China. Cancer Commun. 2020;40:531–544. doi:10.1002/cac2.12087
7. Ajani JA, D’Amico TA, Bentrem DJ, et al. Esophageal and Esophagogastric Junction Cancers, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Nat Comprehen Cancer Net. 2019;17:855–883. doi:10.6004/jnccn.2019.0033
8. Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a Phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi:10.1016/S0140-6736(10)61121-X
9. Food US, Administration D. Ramucirumab in combination with paclitaxel; 2014.
10. Food US, Administration D. FDA grants accelerated approval to pembrolizumab for first tissue/site agnostic indication; 2017.
11. Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–95. doi:10.1038/nature13176
12. Agrawal N, Jiao Y, Bettegowda C, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899–905. doi:10.1158/2159-8290.CD-12-0189
13. Zhang N, Shi J, Shi X, Chen W, Liu J. Mutational characterization and potential prognostic biomarkers of Chinese patients with esophageal squamous cell carcinoma. Onco Targets Ther. 2020;13:12797–12809. doi:10.2147/OTT.S275688
14. Wang Y, Liang N, Xue Z, Xue X. Identifying an eight-gene signature to optimize overall survival prediction of esophageal adenocarcinoma using bioinformatics analysis of ceRNA network. Onco Targets Ther. 2020;13:13041–13054. doi:10.2147/OTT.S287084
15. Yang Z, Yang N, Ou Q, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res. 2018;24:3097–3107. doi:10.1158/1078-0432.CCR-17-2310
16. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi:10.1093/bioinformatics/btu170
17. Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi:10.1101/gr.129684.111
18. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi:10.1093/nar/gkq603
19. Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30:3390–3393. doi:10.1093/bioinformatics/btu549
20. Wang J, Ma Z, Wang Q, et al. Prognostic utility of six mutated genes for older patients with acute myeloid leukemia. Int j Cancer. 2018;142:1664–1670. doi:10.1002/ijc.31178
21. Li Y, Liang L, Dai W, et al. Prognostic impact of programed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor infiltrating lymphocytes in colorectal cancer. Mol Cancer. 2016;15:55. doi:10.1186/s12943-016-0539-x
22. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi:10.1158/1078-0432.CCR-04-0713
23. Dai H, Shao YW, Tong X, et al. YAP1 amplification as a prognostic factor of definitive chemoradiotherapy in nonsurgical esophageal squamous cell carcinoma. Cancer Med. 2020;9:1628–1637. doi:10.1002/cam4.2761
24. Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173:321–37 e10. doi:10.1016/j.cell.2018.03.035
25. Zhang X, Xu Y, He C, et al. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol. 2015;111:834–839. doi:10.1002/jso.23888
26. Heeke AL, Pishvaian MJ, Lynce F, et al. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precision Oncol. 2018;2018. doi:10.1200/PO.17.00286
27. Huselid E, Bunting SF. The regulation of homologous recombination by helicases. Genes. 2020;1;11.
28. Zhang H, Kalirai H, Acha-Sagredo A, Yang X, Zheng Y, Coupland SE. Piloting a deep learning model for predicting nuclear BAP1 immunohistochemical expression of uveal melanoma from hematoxylin-and-eosin sections. Transl Vis Sci Technol. 2020;9:50. doi:10.1167/tvst.9.2.50
29. Jensen DE, Proctor M, Marquis ST, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi:10.1038/sj.onc.1201861
30. Litman R, Peng M, Jin Z, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi:10.1016/j.ccr.2005.08.004
31. Cheng WH, Kusumoto R, Opresko PL, et al. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res. 2006;34:2751–2760. doi:10.1093/nar/gkl362
32. Gupta I, Ouhtit A, Al-Ajmi A, et al. BRIP1 overexpression is correlated with clinical features and survival outcome of luminal breast cancer subtypes. Endocr Connect. 2018;7:65–77. doi:10.1530/EC-17-0173
33. Ciccone MA, Adams CL, Bowen C, et al. Inhibition of poly (ADP-ribose) polymerase induces synthetic lethality in BRIP1 deficient ovarian epithelial cells. Gynecol Oncol. 2020;159:869–876. doi:10.1016/j.ygyno.2020.09.040
34. Zhang L, Niu X, Bi Y, Cui H, Li H, Cheng X. Potential role of targeting KDR and proteasome inhibitors in the therapy of esophageal squamous cell carcinoma. Technol Cancer Res Treat. 2020;19:1533033820948060. doi:10.1177/1533033820948060
35. Liu M, An H, Zhang Y, et al. Molecular analysis of Chinese oesophageal squamous cell carcinoma identifies novel subtypes associated with distinct clinical outcomes. EBioMedicine. 2020;57:102831. doi:10.1016/j.ebiom.2020.102831
36. Weng NQ, Chi J, Wen J, et al. The prognostic value of a seven-lncRNA signature in patients with esophageal squamous cell carcinoma: a lncRNA expression analysis. J Transl Med. 2020;18:47. doi:10.1186/s12967-020-02224-z
37. Yu Y, Cao J, Wu W, et al. Genome-wide copy number variation analysis identified ANO1 as a novel oncogene and prognostic biomarker in esophageal squamous cell cancer. Carcinogenesis. 2019;40:1198–1208. doi:10.1093/carcin/bgz077
38. Zhan L, Qin Q, Lu J, et al. Novel poly (ADP-ribose) polymerase inhibitor, AZD2281, enhances radiosensitivity of both normoxic and hypoxic esophageal squamous cancer cells. Dis Esophagus. 2016;29:215–223. doi:10.1111/dote.12299
39. Miyamoto K, Minegaki T, Tanahashi M, et al. Synergistic effects of olaparib and DNA-damaging agents in oesophageal squamous cell carcinoma cell lines. Anticancer Res. 2019;39:1813–1820. doi:10.21873/anticanres.13288
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

