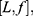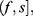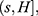Back to Journals » OncoTargets and Therapy » Volume 14
Progesterone Receptor Together with PKCα Expression as Prognostic Factors for Astrocytomas Malignancy
Authors Arcos-Montoya D , Wegman-Ostrosky T, Mejía-Pérez S , De la Fuente-Granada M, Camacho-Arroyo I, García-Carrancá A , Velasco-Velázquez MA , Manjarrez-Marmolejo J, González-Arenas A
Received 11 September 2020
Accepted for publication 20 November 2020
Published 16 June 2021 Volume 2021:14 Pages 3757—3768
DOI https://doi.org/10.2147/OTT.S280314
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Denisse Arcos-Montoya,1 Talia Wegman-Ostrosky,2,3 Sonia Mejía-Pérez,4 Marisol De la Fuente-Granada,1 Ignacio Camacho-Arroyo,5 Alejandro García-Carrancá,6 Marco A Velasco-Velázquez,7 Joaquín Manjarrez-Marmolejo,8 Aliesha González-Arenas1
1Departamento de Medicina Genómica y Toxicología Ambiental, Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México, Ciudad de México, México; 2Dirección de Investigación, Instituto Nacional Cancerología, Ciudad de México, México; 3División de Investigación Básica, Instituto Nacional de Cancerología, Ciudad de México, México; 4Subdirección de Neurocirugía, Instituto Nacional de Neurología y Neurocirugía, Ciudad de México, México; 5Unidad de Investigación en Reproducción Humana, Instituto Nacional de Perinatología-Facultad de Química, Universidad Nacional Autónoma de México, Ciudad de México, México; 6Unidad de Investigación Biomédica en Cáncer, Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México e Instituto Nacional de Cancerología, Ciudad de México, México; 7Departamento de Farmacología, Facultad de Medicina, Universidad Nacional Autónoma de México, Ciudad de México, México; 8Laboratorio de Fisiología de la Formación Reticular, Instituto Nacional de Neurología y Neurocirugía, Ciudad de México, México
Correspondence: Aliesha González-Arenas
Departamento de Medicina Genómica y Toxicología Ambiental, Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México, Ciudad Universitaria, Ciudad de México, 04510, México
Tel +52 5556 22 9209
Email [email protected]
Introduction: Astrocytomas are the most common and aggressive primary brain tumors, and they are classified according to the degree of malignancy on a scale of I to IV, in which grade I is the least malignant and grade IV the highest. Many factors are related to astrocytomas progression as progesterone receptor (PR), whose transcriptional activity could be regulated by phosphorylation by protein kinase C alpha (PKCα) at the residue Ser400. Our aim was to investigate if PR phosphorylation together with PKCα expression could be used as a prognostic factor for astrocytomas malignancy.
Methods: By immunofluorescence, we detected the content of PKCα, PR and its phosphorylation at Ser400 in 46 biopsies from Mexican patients with different astrocytoma malignancy grades; by bioinformatic tools using TCGA data, we evaluated the expression of PR and PKCα mRNA according to astrocytoma malignancy grades. For all statistical analyses, significance was p< 0.05.
Results: We detected a positive correlation between the tumor grade and the content of PKCα, PR and its phosphorylation at Ser400, as well as the intracellular colocalization of these proteins. Interestingly, using an in silico assay, we found that the PR and PKCα expression at mRNA level has an inverse ratio with astrocytomas tumor grade.
Discussion: These results indicate that PR and its phosphorylation at Ser400 site, as well as PKCα and their colocalization, could be considered as possible malignancy biomarkers for astrocytomas grades I–IV.
Keywords: astrocytoma, glioblastoma, progesterone receptor, protein kinase C alpha, biomarker
Introduction
Astrocytomas are the most common primary brain tumors which represent about 76% of all gliomas1 and can be found in any part of the brain, especially in the brain cortex, thalamus and basal ganglia.2 The World Health Organization (WHO) proposed a scale from I to IV for their classification, according to the degree of malignancy. Grade I exhibits the lower degree and grade IV or glioblastoma (GBM) exhibits more advanced features of malignancy, including vascular proliferation, mitosis and necrosis, typically associated with rapid disease evolution and with poor prognosis.3 Epidemiological data report that GBM occurs in a greater proportion in men than in women (3:2).2 The average survival of patients with astrocytoma depends on the histological grade, 7 years for patients with astrocytoma grade II, 3 years for grade III, and 12–15 months for patients with glioblastoma, even with the standard treatment (radiotherapy + temozolomide).4 The National Institute of Neurology and Neurosurgery (INNN) reports that in Mexico, 9% of all brain tumors are GBM,5 which present an incidence of 3.5 per 100,000 habitants6 and, particularly for the Mexican population, the average of diagnosis is 49 years old.6
Some factors support the development of astrocytomas, for example, the activated pathways of NFκB, PI3K/AKT and the expression of progesterone receptor (PR).7,8 Regarding PR, immunohistochemical analysis revealed higher detection of this receptor in GBM compared with lower-grade malignant tumors;9 furthermore, blocking this receptor with RU486 antagonist reduces proliferation, migration, and invasion of GBM derived cell lines.10 In in vivo models, the size of tumors resulting from heterotopic and orthotopic xenograft of murine glioma cells (a tumor-like glioblastoma) decreased by 50% after treatment with RU486.11–13
PR receptor belongs to the family of the nuclear transcriptional regulators that can be activated by ligand or without it14,15; PR presents post-translational modifications, such as phosphorylation, sumoylation, acetylation and ubiquitylation.16 The state of phosphorylation of the PR affects its transcriptional activity, cellular localization, specificity and replacement rate.17 At the moment, for the human PR, 15 phosphorylation sites have been recognized.18–21 The residue Ser400 can be basally phosphorylated and in response to mitogens, and this phosphorylation is necessary for the regulation of ligand-independent activity.22 Our group has identified that the protein kinase C (PKC) is able to phosphorylate PR in this residue, in two cell lines derived from GBM.23,24
The protein kinases C (PKC) are a family of isoenzymes that have the activity of Ser/Thr kinases. These proteins participate in many signaling pathways and are implicated in diverse cellular responses.25 The over-expression or hyper-activation of PKC is a characteristic of GBM26; some studies show that the activity of PKC is increased in cell lines derived from gliomas, and the PKC inhibitors reduce their proliferation significantly.27 Isotypes PKCα and PKCδ are expressed in U373 cells (cell line derived from astrocytoma grade III) and its activation induces proliferation, migration and invasiveness,23 interestingly through an interaction between PKCα and PR, which does not exist with the isotype δ.28
In this work, we analyzed 46 biopsies from Mexican patients with diagnosis of astrocytoma (9 for grades I and II, 12 for grade III and 25 for grade IV). These biopsies showed a positive correlation between the tumor grade and the content of PKCα, PR and its phosphorylation at Ser400, as well as the colocalization of these proteins. An in silico assay on Xena browser using the data base of TCGA and GTEX was performed; interestingly, we found that PR and PKCα expression at mRNA level has an inverse ratio with astrocytomas tumor grade.
Materials and Methods
Samples
Pathology slides from 46 astrocytic tumors were obtained from patients (22 females and 24 males). The slides were obtained from the National Institute of Neurology and Neurosurgery Manuel Velasco Suárez (in accordance with the protocol N° 67/12 approved by the Institutional Review Board: Comité Científico y de Bioética del Instituto Nacional de Neurología y Neurocirugía) and were classified according to the WHO histopathological classification (low grade: 9 samples, grade III: 12 samples, and 25 samples for GBM) (Table 1). For this work, six pathology slides of brain epileptic foci (women) were obtained, which were used as non-tumoral tissue.
 |
Table 1 Clinic and Pathological Characteristics from 46 Astrocytic Tumor Samples |
Immunofluorescence
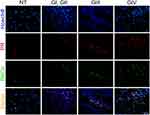
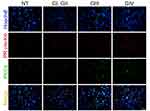
The slides were dewaxed in a dry bath at 60°C for 50 minutes; after that, they were passed for the following gradients: xylol, xylol:ethanol (1:1), ethanol (100%), ethanol (96%) and ethanol (90%), for 5 minutes each. For the identification of antigens, the epitopes were exposed to a solution of sodium citrate (10 mM, pH 6.0), boiling for an hour. The tissue was blocked in 5% normal goat serum/TBS-Tween 0.01% (blocking buffer) overnight at 4°C; the sections were washed and then incubated with the primary antibodies: rabbit anti-PR (2 μg/mL) (sc-7208, Santa Cruz Biotechnology, Dallas, TX) and mouse anti-PKCα (2 μg/mL) (sc-8393, Santa Cruz Biotechnology, Dallas, TX) and rabbit anti-PR pSer400 (1.3 μg/mL) (ab60954, Abcam) at 4°C overnight. The antibodies were removed and the sections were washed three times with TBS-Tween 0.1% for 5 minutes and then incubated with secondary antibodies at room temperature for 1 hour: Alexa Fluor 488 anti-mouse (1:1000) (A11001, Life Technologies, Carlsbad, CA) and Alexa Fluor 594 anti-rabbit (2 drops/mL according with the guide) (R37117, Molecular Probes by Life Technologies, Carlsbad, CA). Nuclei were stained with Hoechst 3342 (Thermo Scientific, Waltham, MA). Sections were covered from light, washed, mounted with Fluoro Care Anti-Fade Mountant (Biocare Medical, Concord, CA) and visualized in an Olympus Bx43 fluorescence microscope. The images were analyzed using the program Image-Pro Plus 7.0 Media Cybernetics (Rockville, MD). Twenty-five fields were taken for each sample; subsequently, the fluorescence intensity for each field was evaluated, considered as a proportional measure of the density of positive cells. Finally, the values were averaged to have a unique value for each patient. An immunofluorescence control was done using samples and secondary antibody, non-signal was detected. Two persons determined fluorescence intensity for each sample independently and blinded (no one knew the malignancy grade of samples).
Results Classification
In order to define three intervals for the fluorescence intensity as low, medium, or high for PR or PKCα based on the average of 25 fields per sample, we used the formula modified from Villegas–Pineda et al.29 I= (H-L)/3, where I is the intensity, H is the highest intensity value of all samples and L is the lowest intensity value of non-tumoral tissue. With this formula, we can define the intervals as follows:
where f = L+Iand 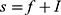 . Each sample was categorized in one of these intervals.
. Each sample was categorized in one of these intervals.
PGR and PRKCA Genes Evaluation
For the determination of PGR and PRKCA mRNA we used the platform of the California University, Santa Cruz: UCSC Xena (XenaBrowser.net) and the database from TCGA and GTEX for tumor tissue and non-tumor tissue, respectively. For the non-tumor tissue a total of 283 samples from brain cortex of GTEX database were used. For the tumor tissue, we used the TCGA database and obtained 258 samples for grade II (GII), 271 for grade III (GIII) and 166 for grade IV (GIV or GBM). The levels of mRNA were obtained and plotted on Graph Pad Prism 5.0. For this analysis, we used EGFR (Supplementary Figure 4) as positive control expression for GBM. The data are summarized in Table 2.
 |
Table 2 Clinical Pathological Features of the Samples from Data Bases TCGA and GTEX |
Spearman Correlations
To obtain the correlation between the degree of expression of PGR and PRKCA, the gene expression database called “TCGA Glioblastoma (GBM)” were used, the samples that did not have data for both genes were eliminated; 172 samples were used and the Spearman coefficient was determined using the GraphPad Prism 8.0.2 program.
Survival Curves
To evaluate the relationship between gene expression of PGR and PRKCA with patients survival, Kaplan–Meier type curves were performed using the XenaBrowser.net platform, the database “TCGA low grade astrocytoma and glioblastoma,” we obtained the following data: for PGR: grade II: 257 samples, grade III: 270 samples, grade IV: 172 samples, for PRKCA: Grade II: 270 samples, Grade III: 270 samples, and Grade IV: 172 samples.
Statistical Analysis
The results were expressed as the mean ± SD for % of positive cells and for their overlap. Statistical analysis between groups was performed with an ANOVA followed by a Dunnet’s post-test. A value P < 0.05 was considered statistically significant as stated in figure legends. All these analyses were performed in Graph Pad Prism 5.0 (Graph Pad Software, San Diego, CA).
Results
PR, PKCα Localization and Their Merge Increase According to the Tumor Grade
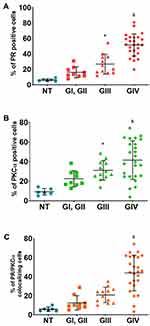
Immunofluorescence assays were performed to evaluate the presence of PR, PKCα, and their colocalization in astrocytoma biopsies (Figure 1). We observed that the number of PR and PKCα positive cells increased in accordance with the tumor malignancy grade: the percentage for PR positive cells in the non-tumor tissue (NT) was 6.2%, GI–II 16.1%, GIII 26.7% and GIV (GBM) 51.6% (Figure 2A). For PKCα, the data were as follows: NT 9.2%, GI-II 22.5%, GIII31.1% and GIV 41.5% (Figure 2B), and for colocalization: 5.9% for NT, 12.6% for GI-II, 20.8% for GIII and 43.9% for GIV (Figure 2C).
To know if a correlation exists between the tumor grade and the PR/PKCα detection, we performed a Spearman Correlation. We found a positive statistical correlation between PR (Figure 3A), PKCα (Figure 3B), and their colocalization with tumor malignancy grade (Figure 3C).
To determine if some features as sex or age could influence PR and PKCα detection, the samples were separated according to the sex in two groups (F: feminine and M: masculine), but no significant differences were found between sexes (Supplementary Figures 1a-c). No correlation was observed between astrocytomas grades and the age of patients according to detection of PR, PKCα and PR/PKCα colocalization (Supplementary Figures 2a-c).
Fluorescence Intensity from Each Protein in Immunofluorescence Assays
We sorted the fluorescence intensity for each protein in three intervals (low, medium, and high) according to the formula described in the Materials and Methods section,28 then the samples were classified in accordance with their intensity (Table 3): 66.6% of the samples from grade I-II showed a low intensity for PR, and 70% for PKCα, in the case of astrocytomas grade III, 66.66% for PR and 61.54% for PKCα of the samples showed a medium intensity, and for grade IV or GBM 72% for PR and 64% for PKCα presented high intensity.
 |
Table 3 Fluorescence Intensity from Each Protein and Their Classification in Intervals |
PR pSer400 and PKCα Detection, and Their Colocalization Increased According to the Tumor Grade
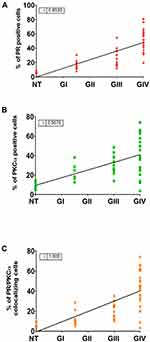
For the analysis of phosphorylation, we randomly took six samples for each group. We found that the percentage of positive cells for pSer400 and their merge with PKCα increased according to the tumor grade (Figure 4). The mean of cells positive to PR pSer400 in the NT tissue was 4.2%, 8.4% for GI-II, 17.9% for GIII, and 42.4% for GIV (Figure 5A), for PKCα this mean was 4.1% for NT, 8.0% for GI-II, 20.63% for GIII and 41.3% for GIV (Figure 5B). The colocalization of these two proteins provided a mean of positive cells of 3.3% for NT, 6.7% for GI-II, 16.1% for GIII, and 38.9% for GIV (Figure 5C). We performed a Spearman correlation between colocalization of PR pSer400/PKCα with tumor grade, a positive statistical correlation was found (Figure 5D). In Supplementary Figure 3, representative images of PR pSer400 and PKCα colocalization for each patient are shown. These results suggest an astrocytoma grade-dependent correlation with the detection of PKC- α and PR pSer400.
Expression of mRNA for PGR andPRKCA
In silico analysis using TCGA data showed that mRNA levels for both genes are proportionally inverse to the tumor grade (Figure 6A and B). We observed that no correlation exists at mRNA level between PGR and PRKCA expression (Figure 6C), suggesting that the expression of one of these factors does not depend on the other. For this analysis, we used the gene EGFR (Supplementary Figure 4) as a positive control because many reports indicate that the mRNA level of this gene correlates positively with tumor astrocytoma grade.30 From the in silico data, we separated the data according to the patient’s sex to determine if a difference exists between mRNA for PGR and PRKCA. No difference was observed between sexes or tumor grade (Supplementary Figures 5a and 5b).
Expression of PGR and PRKCA Correlation with Patient Survival
In order to evaluate the relationship between PGR and PRKCA expression (mRNA) with patient survival, Kaplan–Meier graphs using TCGA data were done (Figure 7). Interestingly, patients with astrocytoma grade III and IV with less expression of both factors have a large survival than those with high expression (Figure 6C–F). Patients with astrocytoma grade II survival are independent of the expression of these factors (Figure 6A and B). The results suggest that the expression level of both factors in high-grade astrocytomas correlates with a poor prognosis.
Discussion
In this work, we detected the presence of PR, PKCα, and the colocalization of both proteins in 46 biopsies from Mexican patients with a diagnosis of astrocytomas grades I–IV. Interestingly, we observed that the number of positive cells for total PR and its phosphorylation at the Ser400 site, as well as PKCα and their respective colocalization, positively correlates with the tumor grade. Previously, a positive relationship between the percentage of PR positive cells with respect to tumor grade had been observed in patient biopsies; however, it had not been compared with respect to non-tumor tissue.9,31
The activity of transcription factors is regulated by post-translational modifications such as phosphorylation; PR can be activated by phosphorylation at Ser400 by PKCα in human GBM cell lines.28 In human glioblastoma cell lines, PKCα can be activated by lysophosphatidic acid receptor 1 (LPA1) which activates Gαq, which in turn activates PKCα;24 the activation of this kinase causes an increase in the transcriptional activity of PR23 that regulates expression of genes involved in proliferation, migration, and invasiveness.32,33 In this work, we detected that PR phosphorylated at Ser400 and their colocalization with PKCα increased according to the tumor grade, which suggests that the PKCα induces PR phosphorylation, which afterward could induce receptor transcriptional activity.23,28,34 Phosphorylation sites have been proposed as possible tumor biomarkers; the phosphorylation at Ser294 of PR in breast cancer is associated with bad prognostic. This phosphorylation induces stem cell phenotype and increases signaling pathways activated by growth factors; in relation to this, a use of therapies with anti-estrogen activity has been suggested to avoid the recurrence of patients with a high content of pSer294 PR.35
Interestingly, we observed that the expression of PGR and PRKCA genes decreased in relation to the malignancy grade of astrocytomas, opposite effect observed at the protein level. The abundance of mRNA and the protein levels are unrelated in many cases36; in lung adenocarcinoma, 29 genes with at least two isoforms, show a different correlation coefficient mRNA/protein, for example, for OP18 (oncoprotein 18), three of the four isoforms showed a statistically significant correlation between the mRNA abundance and the protein, the fourth isoform showed no correlation between protein and mRNA expression.37 On patients with lung adenocarcinoma, some mechanisms that can modify the transcription at mRNA levels have been suggested; the Kozak sequences alterations, the codons polarization, the nonsense lectures, or even these alterations can be attributed to the quantification methods, such as temperature and lifetime of mRNA .36 Differences between the transcriptome and proteome studies from samples of the same patient with GBM have been found; in genes as synapsin 1 (SYN1, related to cellular communication), mRNA levels are under-expressed, meanwhile protein is over-expressed.38 In another study, a profile of proteins was performed in eight patients with GBM and compared to their respective non-tumor tissue; afterward, they analyzed the relation between mRNA/protein, they observed that only 2% of total proteins correlated with their respective mRNA.39
Gene mutations could be related with differences between mRNA and protein levels. In chordoid gliomas PRKCAD463H transcript is overexpressed compared to PRKCAWT mRNA transcripts.40 We determined the presence of punctual mutations in the PGR and PRKCA genes in glioblastoma tissue using the TCGA data bank in the Xena browser platform (Supplementary Figure 6), we found the presence of mutations that provides changes in protein amino acids at the sites: 709, 813 and 836 for PR and 506 for PKCα, it would be necessary to study if these transcripts are abundant in GBM compares to NT tissue. Interestingly, Kaplan–Meier curves point out that the expression of mRNA of PGR and PRKCA in different astrocytoma grades correlates with patient survival, indicating the relevance of these factors for this pathology.
Conclusion
Our results indicate that PR and its phosphorylation at Ser400 site, as well as PKCα and their colocalization, could be considered as possible malignancy biomarkers for astrocytomas grades I–IV and that the analysis of mRNA expression in GBM is not enough to determine the role of a gene in tumor malignancy.
Ethical Statement
The pathology slides were obtained from the National Institute of Neurology and Neurosurgery Manuel Velasco Suárez in accordance with the protocol N° 67/12 approved by the Institutional Review Board. All patients signed the informed consent and the guidelines outlined in the Declaration of Helsinki were followed.
Acknowledgments
We thank Carlos Humberto Martínez Álvarez (Universidad Autónoma del Estado de México), MSc. Ángel Jonathan Ruiz Moreno (Facultad de Medicina, Universidad Nacional Autónoma de México) and MSc. Gabriela Piña-Medina (Facultad de Química, Universidad Nacional Autónoma de México) for their technical support.
Funding
This work was supported by Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT) IA201120 and IN219719. Denisse Arcos-Montoya is a PhD student from Programa de Maestría y Doctorado en Ciencias Bioquímicas, Universidad Nacional Autónoma de México (UNAM) and received fellowship 894557 from CONACyT.
Disclosure
The authors reported no conflicts of interest for this work and declare that they have not known competing financial interests or personal relationships that could have appeared to influence this work.
References
1. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20(suppl_4):iv1–iv86. doi:10.1093/neuonc/noy131
2. Ohgaki H. Genetic pathways to glioblastomas. Neuropathology. 2005;25(1):1–7. doi:10.1111/j.1440-1789.2004.00600.x
3. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System; a summary. Acta Neuropatho. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1.
4. Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi:10.1101/gad.1596707
5. Magaña-Maldonado R, Chávez-Cortez EG, Olascoaga-Arellano NK, et al. Immunological Evasion in Glioblastoma. Biomed Res Int. 2016;2016:7487313. doi:10.1155/2016/7487313
6. Wegman-Ostrosky T, Reynoso-Noverón N, Mejía-Pérez SI, et al. Clinical prognostic factors in adults with astrocytoma: historic cohort. Clin Neurol Neurosurg. 2016;146:116–122. doi:10.1016/j.clineuro.2016.05.002
7. Soubannier V, Stifani S. NF-κB signalling in glioblastoma. Biomedicines. 2017.
8. González-Agüero G, Gutiérrez AA, González-Espinosa D, et al. Progesterone effects on cell growth of U373 and D54 human astrocytoma cell lines. Endocrine. 2007;32(2):129–135. doi:10.1007/s12020-007-9023-0
9. Khalid H, Shibata S, Kishikawa M, Yasunaga A, Iseki M, Hiura T. Immunohistochemical analysis of progesterone receptor and Ki-67 labeling index in astrocytic tumors. Cancer. 1997;80(11):2133–2140.
10. Piña-Medina AG, Hansberg-Pastor V, González-Arenas A, Cerbón M, Camacho-Arroyo I. Progesterone promotes cell migration, invasion and cofilin activation in human astrocytoma cells. Steroids. 2016;105:19–25. doi:10.1016/j.steroids.2015.11.008
11. Germán-Castelán L, Manjarrez-Marmolejo J, González-Arenas A, González-Morán MG, Camacho-Arroyo I. Progesterone induces the growth and infiltration of human astrocytoma cells implanted in the cerebral cortex of the rat. Biomed Res Int. 2014;2014:1–8. doi:10.1155/2014/393174
12. Llaguno-Munive M, Medina LA, Jurado R, Romero-Piña M, Garcia-Lopez P. Mifepristone improves chemo-radiation response in glioblastoma xenografts. Cancer Cell Int. 2013;13(1):29. doi:10.1186/1475-2867-13-29
13. Llaguno-Munive M, Romero-Piña M, Serrano-Bello J, Medina LA, Uribe-Uribe N, Ana Salazar AM. Mifepristone overcomes tumor resistance to temozolomide associated with DNA damage repair and apoptosis in an orthotopic model of glioblastoma. Cancers (Basel). 2018;11(16):1–15. doi:10.3390/cancers11010016
14. Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi:10.1042/bse0400011
15. McEwan IJ. Nuclear receptors: one big family. MethodsMol Biol. 2009;505:3–18.
16. Hagan CR, Daniel AR, Dressing GE, Lange CA. Role of phosphorylation in progesterone receptor signaling and specificity. Mol Cell Endocrinol. 2012;357(1–2):43–49. doi:10.1016/j.mce.2011.09.017
17. Qiu M, Lange CA. MAP kinases couple multiple functions of human progesterone receptors: degradation, transcriptional synergy, and nuclear association. J Steroid Biochem Mol Biol. 2003;85(2–5):147–157. doi:10.1016/S0960-0760(03)00221-8
18. Zhang Y, Beck CA, Poletti A, Edwards DP, Weigel NL. Identification of phosphorylation sites unique to the B form of human progesterone receptor. In vitro phosphorylation by casein kinase II. J Biol Chem. 1994;269(49):31034–31040. doi:10.1016/S0021-9258(18)47386-3
19. Zhang Y, Beck CA, Poletti A, et al. Phosphorylation of human progesterone receptor by cyclin-dependent kinase 2 on three sites that are authentic basal phosphorylation sites in Vivo. Mol Endocrinol. 1997;11(6):823–32. doi:10.1210/mend.11.6.0006
20. Knotts TA, Orkiszewski RS, Cook RG, Edwards DP, Weigel NL. Identification of a phosphorylation site in the hinge region of the human progesterone receptor and additional amino-terminal phosphorylation sites. J Biol Chem. 2001;276(11):8475–8483. doi:10.1074/jbc.M009805200
21. Lange CA, Shen T, Horwitz KB. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc Natl Acad Sci U S A. 2000;97(3):1032–7. doi:10.1073/pnas.97.3.1032
22. Pierson-Mullany LK, Lange CA. Phosphorylation of progesterone receptor serine 400 mediates ligand-independent transcriptional activity in response to activation of cyclin-dependent protein kinase 2. Mol Cell Biol. 2004;24(24):10542–10557. doi:10.1128/MCB.24.24.10542-10557.2004
23. Marquina-Sánchez B, González-Jorge J, Hansberg-Pastor V, et al. The interplay between intracellular progesterone receptor and PKC plays a key role in migration and invasion of human glioblastoma cells. J Steroid Biochem Mol Biol. 2017;172:198–206. doi:10.1016/j.jsbmb.2016.10.001
24. Valdés-Rives SA, de la Fuente-granada M, Velasco-Velázquez MA, González-Flores O, González-Arenas A. LPA 1 receptor activation induces PKCα nuclear translocation in glioblastoma cells. Int J Biochem Cell Biol. 2019;110:91–102. doi:10.1016/j.biocel.2019.03.003
25. Steinberg SF. Structural basis of protein kinase C isoform function. Physiol Rev. 2008;88(4):1341–1378. doi:10.1152/physrev.00034.2007
26. Cameron AJ, Procyk KJ, Leitges M, Parker PJ. PKC alpha protein but not kinase activity is critical for glioma cell proliferation and survival. Int J Cancer. 2008;123(4):769–779. doi:10.1002/ijc.23560
27. Mandil R, E Ashkenazi, M Blass, et al. Protein kinase Cα and protein kinase Cδ play opposite roles in the proliferation and apoptosis of glioma cells. Cancer Res. 2001;61(11):4612–4619.
28. González-Arenas A, Peña-Ortiz MÁ, Hansberg-Pastor V, et al. Pkcα and pkcδ activation regulates transcriptional activity and degradation of progesterone receptor in human astrocytoma cells. Endocrinology. 2015;156(3):1010–1022. doi:10.1210/en.2014-1137
29. Villegas-Pineda JC, Garibay-Cerdenares OL, Hernández-Ramírez VI, et al. Integrins and haptoglobin: molecules overexpressed in ovarian cancer. Pathol Res Pract. 2015;211(12):973–981. doi:10.1016/j.prp.2015.10.002
30. Puliyappadamba VT, Hatanpaa KJ, Chakraborty S, Habib AA. The role of NF-κB in the pathogenesis of glioma. Mol Cell Oncol. 2014;1(3):e963478. doi:10.4161/23723548.2014.963478
31. González-Agüero G, Ondarza R, Gamboa-Domínguez A, Cerbón MA, Camacho-Arroyo I. Progesterone receptor isoforms expression pattern in human astrocytomas. Brain Res Bull. 2001;56(1):43–48. doi:10.1016/S0361-9230(01)00590-1
32. González-Arenas A, Valadez-Cosmes P, Jiménez-Arellano C, López-Sánchez M, Camacho-Arroyo I. Progesterone-induced blocking factor is hormonally regulated in human astrocytoma cells, and increases their growth through the IL-4R/JAK1/STAT6 pathway. J Steroid Biochem Mol Biol. 2014;144(PB):463–470. doi:10.1016/j.jsbmb.2014.09.007
33. Hernández-Hernández OT, Camacho-Arroyo I. Regulation of gene expression by progesterone in cancer cells: effects on cyclin D1, EGFR and VEGF. Mini Rev Med Chem. 2013;13(5):635–42. doi:10.2174/1389557511313050002
34. Hernández-Hernández OT, González-García TK, Camacho-Arroyo I. Progesterone receptor and SRC-1 participate in the regulation of VEGF, EGFR and Cyclin D1 expression in human astrocytoma cell lines. J Steroid Biochem Mol Biol. 2012;132(1–2):127–134. doi:10.1016/j.jsbmb.2012.04.005
35. Knutson TP, Truong TH, Ma S, et al. Posttranslationally modified progesterone receptors direct ligand-specific expression of breast cancer stem cell-associated gene programs. J Hematol Oncol. 2017;10(1). doi:10.1186/s13045-017-0462-7.
36. Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966–3973. doi:10.1016/j.febslet.2009.10.036
37. Chen G, Gharib TG, Huang C-C, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1(4):304–313. doi:10.1074/mcp.M200008-MCP200
38. Lemée J-M, Clavreul A, Aubry M, et al. Integration of transcriptome and proteome profiles in glioblastoma: looking for the missing link. BMC Mol Biol. 2018;19(1):13. doi:10.1186/s12867-018-0115-6
39. Song YC, Lu G-X, Zhang H-W, et al. Proteogenomic characterization and integrative analysis of glioblastoma multiforme. Oncotarget. 2017;8(57):97304–97312. doi:10.18632/oncotarget.21937
40. Rosenberg S, Simeonova I, Bielle F, et al. A recurrent point mutation in PRKCA is a hallmark of chordoid gliomas. Nat Commun. 2018;9(1). doi:10.1038/s41467-018-04622-w
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.