Back to Journals » International Journal of General Medicine » Volume 14
Prediction of Survival Outcome in Lower-Grade Glioma Using a Prognostic Signature with 33 Immune-Related Gene Pairs
Authors Chen S , Sun Y, Zhu X , Mo Z
Received 29 September 2021
Accepted for publication 2 November 2021
Published 13 November 2021 Volume 2021:14 Pages 8149—8160
DOI https://doi.org/10.2147/IJGM.S338135
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Shaohua Chen,1,2,* Yongchu Sun,3,4,* Xiaodong Zhu,3– 5 Zengnan Mo1,2
1Department of Urology, The First Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic of China; 2Guangxi Collaborative Innovation Center for Genomic and Personalized Medicine, Nanning, People’s Republic of China; 3Department of Radiation Oncology, Guangxi Medical University Cancer Hospital, Nanning, People’s Republic of China; 4Guangxi Key Laboratory of Early Prevention and Treatment for Regional High Frequency Tumor, Guangxi Medical University, Nanning, People’s Republic of China; 5Department of Oncology, Affiliated Wuming, Hospital of Guangxi Medical University, Nanning, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaodong Zhu
Department of Radiation Oncology, Guangxi Medical University Cancer Hospital, No. 71 He Di Road, Nanning, 530021, People’s Republic of China
Email [email protected]
Zengnan Mo
Institute of Urology and Nephrology, The First Affiliated Hospital of Guangxi Medical University, No. 6 Shuangyong Road, Nanning, 530021, People’s Republic of China
Email [email protected]
Background: Lower-grade glioma (LGG) is one of the prevalent malignancies threatening human health, with considerable intrinsic heterogeneities in their biological behavior. Previous studies have revealed that the immune component is a key factor influencing the formation and development of malignancies. In this study, we aim to use a novel approach to develop a prognostic signature of immune-related gene pairs (IRGPs) to determine the survival outcome of patients with LGG.
Methods: Transcriptomic profiles and clinical data for LGG were obtained from The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) databases, and used as training and validation data sets, respectively. IRGPs influencing the overall survival (OS) of patients with LGG in the training data set were screened by performing univariate Cox regression analysis. Next, a prognostic IRGPs signature was constructed using least absolute shrinkage and selection operator (LASSO) regression. Finally, we cross-validated the two databases to verify the stability of the prognostic signature.
Results: A total of 33 IRGPs influencing prognosis of LGG in the training data set were included in the prognostic signature. Patients with high risk scores (RSs) in the training and validation data sets had a poorer OS than those with low RSs. Moreover, significant differences were observed in tumor-infiltrating immune cells (TICs) between high- and low-RS groups. Functional enrichment analyses results revealed that genes in the high-RS group were enriched in the immune-related activities and developmental processes.
Conclusion: The prognostic signature containing 33 IRGPs has a significant correlation with OS and relative levels of immune cells associated with LGG. The results of the present study provide new insights into the prediction of survival outcome and therapeutic response of LGG.
Keywords: lower-grade glioma, immune-related gene pairs, tumor-infiltrating immune cells, prognostic signature
Introduction
Gliomas are the most prevalent types of primary malignant tumors in the central nervous system.1 Based on the histopathological features outlined by the World Health Organization (WHO), grade II and III gliomas are defined as lower-grade gliomas (LGGs),2 which include astrocytomas, oligodendrogliomas, and oligoastrocytomas.3 Currently, the standard care for LGG comprises surgical resection combined with postoperative radiotherapy and chemotherapy. LGG exhibits considerable intrinsic heterogeneity in biological behavior, with overall survival (OS) ranging from 1 to 15 years.4 Nevertheless, there are some patients who develop drug-resistant high-grade gliomas, resulting in adverse long-term outcomes. Despite advances in the diagnosis and treatment modalities of LGG in recent years, the prognosis of patients with LGG has not improved substantially over the last few decades.4,5 Therefore, identification of novel targets or biomarkers to facilitate early diagnosis and treatment response of LGG is imperative. Risk classification of glioma has been frequently applied in clinical decision-making, and WHO established specific molecular markers for brain tumor classification in 2016, such as chromosome 1p and 19q (chr1p/19q) co-deletions, isocitrate dehydrogenase, and histone H3K27M mutations.6 This classification is superior to the previous histopathological classification. However, the widespread clinical dissemination of such a classification system remains a huge challenge due to the level of molecular diagnostic technology in each medical center.
Immune components of the tumor microenvironment (TME) that influence the formation and development of malignancies have been elucidated. Consequently, several promising preclinical and clinical immunotherapeutic approaches, including immune checkpoint inhibitors (ICIs), immunotherapy, and gene therapy have been applied in the management of gliomas with satisfactory results.7–9 Strikingly, tumor development is thought to be dependent on the infiltration of immune cells,10 while immune-related genes (IRGs) are thought to be indicative of patient prognosis, which has been reported in various types of cancers, such as esophageal cancer,11 bladder cancer,12 lung cancer,13 and colon cancer.14 Therefore, the expression profiles of IRGs present great potential in predicting the prognosis and therapeutic response of LGG, which merits further investigation to identify a sophisticated prognostic signature that can predict survival outcome and guide appropriate individualized treatment strategies.
However, traditional methods of data normalization and scaling in the processing of gene expression data have certain limitations, making it difficult to draw reliable conclusions. In the present study, we used a novel method that involved ranking of relative gene expression levels,15,16 thereby establishing a prognostic signature for IRGPs. In addition, two separate datasets, including LGG cases from the Cancer Genome Atlas (TCGA) and The Chinese Glioma Genome Atlas (CGGA) databases, were cross-validated to verify its reliability and authenticity as a prognostic marker for LGG.
Materials and Methods
Acquisition and Processing of Transcriptomic and Clinical Data
Transcriptomic data for LGG, which included 529 tumor samples, and corresponding clinical data were obtained from TCGA database (https://portal.gdc.cancer.gov/) and used as the training data set. Other dataset containing 443 LGG samples from CGGA database (http://www.cgga.org.cn/) were used as the validation data set.
Construction of a Prognostic IRGPs Signature for LGG
Next, we obtained IRGs from the ImmPort database (https://www.immport.org/). Subsequently, the transcriptome data of TCGA-LGG cohort were analyzed using limma package of R, intersected with IRGs from the ImmPort database, and retained the IRGs. The relevant IRGs were screened further based on a median absolute deviation (MAD) of >0.5 to attain a comparatively high variation. Afterwards, the transcriptome data for LGG cases obtained from the CGGA database were screened based on MAD value of >0.5. Gene lists obtained from the two databases were intersected to identify a shared list of significant IRGs and their respective expression matrices from the two databases. LGG cases obtained from TCGA and CGGA databases were used as training and validation data sets, respectively. Thereafter, we constructed IRGPs for each patient in two databases: in the pairwise comparison of each sample, the expression level of the first gene was regarded as zero if the expression level of the gene was lower than the second gene; otherwise, it was regarded as one. Samples with ratios of zero and one that were less than 20% and greater than 80% were removed to retain gene pairs that were associated with LGG prognosis. Subsequently, univariate Cox regression analysis of IRGPs from TCGA was performed using the survival package in R based on p-value <0.001 as a filtering condition. Least absolute shrinkage and selection operator (LASSO) regression was performed using the glmnet package in R, and we finally obtained prognostic signatures containing 33 IRGPs. A time-dependent receiver operating characteristic (ROC) curve with three-year survival rate for the training data set was constructed using survROC package. LGG cases in the training and validation data sets were divided into high and low risk score (RS) groups based on the optimal cutoff value of the ROC curve for three-year survival rate.
Cross-Validation of Prognostic IRGPs Signatures
The high- and low-RS groups in the training and validation data sets were compared by performing Kaplan–Meier survival analyses using survival and survminer packages in R. Furthermore, univariate and multivariate Cox regression analyses were used to assess the efficacy of the signatures and the prognostic value of other clinical data.
Tumor-Infiltrating Immune Cell Analysis
To evaluate the relative levels of tumor-infiltrating immune cells (TICs) associated with LGG, we used the CIBERSORT algorithm to calculate the relative levels of 22 TICs in each tumor sample in the training and validation data sets. CIBERSORT is an analytical tool developed by Newman et al, and it requires an input matrix of transcriptomic data, thereby providing an estimation of the relative level of immune cell subtype based on linear support vector regression.17,18 Thereafter, cancer samples with P > 0.05 were excluded, and differential analyses of the relative levels of 22 TIC subtypes in the high- and low-RS groups were performed using Wilcoxon test. The results were then visualized using vioplot package in R.
Gene Set Enrichment Analysis of High- and Low-RS Groups
We performed gene set enrichment analysis (GSEA) of differentially expressed genes in high- and low-RS groups using the fgsea package in R. Log2 fold change was made between the two groups, and Padj < 0.05 was considered statistically significant. Finally, the results were visualized using the ggplot2 package in R.
Statistical Analysis
Statistical analyses and visualization of data were performed using R software (version 4.0.3). A p-value less than 0.05 (two-sided) was considered statistically significant.
Results
Screening of IRGs
A list of 2483 IRGs was obtained from the ImmPort database, which intersected the transcriptome data for LGG obtained from TCGA database, and finally obtained 1692 IRGs. Subsequently, the transcriptome data for glioma in the CGGA database was downloaded, and the LGG cases were retained. Gene expression matrices obtained from the two databases were filtered based on MAD > 0.5, while the IRGs that were shared by the two databases were obtained by intersection analysis for subsequent analysis. A total of 659 IRGs were identified. In the present study, TCGA-LGG and CGGA-LGG cohorts were used as the training and validation data sets, respectively.
Establishment of Prognostic Signature for IRGPs
IRGPs were constructed based on a one-on-one matching of 659 IRGs in the training data set. Univariate Cox regression and Kaplan–Meier survival analyses were used to screen for key IRGPs influencing prognosis of LGG, and 10469 key IRGPs were identified. Finally, LASSO regression was used to further screen the IRGPs, and 33 IRGPs used to establish the prognostic signature were obtained, which included a total of 49 unique IRGs. The names and categories of the IRGs and their coefficients in the prognostic signature are presented in Table 1.
 |
Table 1 The Name and Category of Each Immune-Related Gene and Coefficient in the Prognostic Signature |
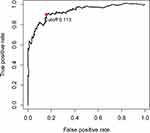
Subsequently, the time-dependent ROC curve analysis results for three-year OS revealed that the optimal cutoff value was 0.113 (Figure 1). The RSs of the cases in the training and validation data sets were calculated and used to classify patients into high- and low-RS groups based on the optimal cutoff value, RSs above the optimal cut-off were assigned to the high-RS group, while RSs below the optimal cut-off were classified into the low-RS group. The heatmap displays the distribution of the 33 IRGPs in the training data set (Figure 2A). Risk curve (Figure 2B) as well as the survival status (Figure 2C) of the LGG cases in the training data set indicated that the proportion of deaths in the high-RS group was significantly higher than in the low-RS group, similar results were obtained in the same analyses of validation data set (Figure 2D–F). Further Kaplan–Meier survival analysis of the training data set demonstrated that the prognosis of patients with LGG in the high-RS group was poorer than that in the low-RS group (Figure 3A). Kaplan–Meier survival analysis of the LGG cases in the validation data set was conducted for cross-validation, confirming that the adverse prognosis of LGG cases in the high-RS group was also presented in the validation data set (Figure 3B).
Furthermore, univariate and multivariate Cox regression analyses were performed on the training data set to analyze prognostic IRGPs signature and clinical features associated with LGG. Overall, age and RS of signature were identified as the independent prognostic factors for predicting the survival outcome of patients with LGG (Figure 4A and B). Univariate and multivariate Cox regression analyses of the validation data set also indicated that RS of signature was a detrimental prognostic factor for LGG (Figure 4C and D). In conclusion, the prognostic IRGPs signature exhibited superior stability and could assess the prognosis of patients with LGG accurately.
Analysis of TICs
To analyze variations in the profiles of TICs, we used the CIBERSORT algorithm to calculate the relative levels of TICs in the TCGA-LGG and CGGA-LGG cohorts, thereby constructing 22 kinds of TICs profiles and their correlation analysis of training data set (Figure 5A and B) and validation data set (Figure S1A and B). Violin plots were used to visualize comparative analyses of TICs between high- and low-RS groups in the training data set (Figure 6). Notably, naïve B cells, activated memory CD4+ T cells, CD8+ T cells, M0 macrophages, M1 macrophages, M2 macrophages, and regulatory T cells (Treg) were highly expressed in the high-RS group. In contrast, eosinophils, activated mast cells, monocytes, and activated NK cells were highly expressed in the low-RS group. Nevertheless, TICs profiles of CGGA-LGG cohort indicated that monocytes were the only TIC subtype that exhibited statistically significant differences between the two groups (Figure 7). The above conclusions shed light on the fact that the relative levels of specific TIC subtypes in the TME of LGG could be associated with the formation and development of LGG.
 |
Figure 6 Violin plots representing comparative analysis of TICs between the high- and low-RS groups of the training data set. |
 |
Figure 7 Violin plots representing comparative analysis of TICs between the high- and low-RS groups of validation data set. |
Functional Enrichment Analysis of Prognostic IRGPs Signature
Next, GSEA was performed to elucidate variations in the gene expression profiles between high- and low-RS groups in the training data set. The results revealed that enriched terms of GSEA analysis in the high-RS group were mostly immune-related activities, including immune effector process, cell activation, and defense response to other organisms. Intriguingly, development-related processes were also involved, including regulation of multicellular organismal development and epithelium development. The enriched terms ranked based on p-value were visualized using bubble plots (Figure 8), and top five enriched terms in GSEA analysis of training data set were immune effector process, cell activation, defense response to other organisms, response to cytokine, and chromatin (Figure 9), which were consistently up-regulated in the high-RS group.
 |
Figure 8 Bubble plots showing the enriched terms based on GSEA analysis ranked by p-value in the high-RS group of the training data set. |
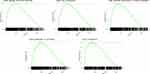 |
Figure 9 Top five enriched terms based on GSEA analysis in the high-RS group of the training data set. |
Similar results were acquired in the functional enrichment analysis of validation data set (Figure S2), and top five enriched terms in GSEA analysis of training data set were molecular transducer activity, animal organ morphogenesis, positive regulation of immune system process, regulation of immune system process, and tube development (Figure S3), explaining underlying reasons for the variations in the prognosis of the two groups of patients.
Discussion
Glioma is the most widespread malignancy that influences the nervous system, accounting for approximately 80% of malignant brain tumors.19 The overall estimated incidence of gliomas is 4.7–5.7 cases per 100,000 people.20 LGG is one of the common pathological types of gliomas that is characterized by gradual growth. Nevertheless, LGG is associated with an extremely high recurrence rate, which frequently leads to severe disability and death due to its aggressive and invasive nature.21,22 ICIs have received increasing attention because of their promising results in the treatment of tumors.23,24 Similar results have been obtained in clinical trials of glioma,25–27 with a certain therapeutic value and response rate. However, the efficacy of glioma has not been outstanding when compared to other cancers, such as melanoma and lung cancer,28,29 while drug efficacy exhibits considerable individual variations and has been associated with varying degrees of treatment-related adverse events.30 In the present study, we established a prognostic IRGPs signature for predicting the prognosis of LGG with an unbiased method for data analysis based on ranking and pairwise comparisons of relative gene expressions. A similar approach has been used in numerous studies, with remarkable results.31 Furthermore, the stability and reliability of the prognostic signature were verified by cross-validating the two databases.
The prognostic signature comprised 33 IRGPs, including 49 independent IRGs, which predominantly consist of cytokines and cytokine receptor-related functions. Notably, cytokines have been reported to play an essential role in the communication between different components of TME in glioblastoma (GBM), while cytokine networks constituted by transforming growth factor β (TGF-β), interleukin (IL)-6, and IL-10 can impede T cell proliferation and response, thereby suppressing anti-tumor immunity, and promoting tumor progression and metastasis.32 The characteristics of low immunogenicity of glioma and the complex cytokine network of immunosuppression in TME could contribute to the poor efficacy of immunotherapy. Similarly, chemokines also have a primary function among the IRGs in the prognostic signature. Chemokines influence the regulation of inflammation, infection, immune response, tissue damage response, and apoptosis by providing optimal host defense against infection with pathogens.33 Strikingly, chemokines and their receptors also play a decisive role in tumor progression. Chemokines can indirectly regulate tumor growth by recruiting leukocytes, in turn, influencing the immune status of TME.34 Previous studies have revealed that chemokines can influence tumor progression in a multifaceted manner in the TME of GBM. CXCL8 is an inflammation-associated chemokine with tumorigenic and pro-angiogenic properties, which has been demonstrated to be upregulated in tumor cells of gliomas, and its expression level is consistent with the pathological grades of gliomas.35 Studies have revealed that CXCL8 is involved in the cellular process of epithelial–mesenchymal transition (EMT) by activating the JAK/STAT1/HIF-1α/Snail signaling pathway in GBM cells, thus promoting progression of cancer cells.36 Therefore, targeting cytokines and chemokines could be an alternative solution to the improvement of immunotherapy efficacy in the treatment of glioma.
TICs have been identified as crucial components in tumor biology, and they influence tumor development and growth.37,38 The roles of TICs in gliomas have also been reported in previous studies.38 Several studies have revealed that Tregs are key factors influencing the immunosuppressive state of glioma TME. Tregs inhibit the secretion of pro-inflammatory factors (eg, IL-2 and interferon gamma [IFN]-γ) while promoting the excretion of immunosuppressive factors (eg, TGF-β), thereby suppressing antitumor response in various lymphocytes.39 In addition, Tregs bind to CD80 or CD86 through CTLA-4, which can substantially inhibit activation of effector T cells.40 Similarly, macrophages are a canonical subtype of TICs that stably accumulate through the development of gliomas. Macrophages with pro-angiogenic factors have been found to be significantly upregulated in macrophages isolated from tumors, which interact with tumor vessels and are associated with vascular remodeling.41 In the present study, we observed that the relative levels of Tregs and macrophages in patients with LGG in the high-RS group were relatively higher, suggesting that the two subtypes of TICs are associated with progression and metastasis of LGG. Conversely, the relative levels of activated NK cells and monocytes were relatively higher in the low-RS group and were associated with better prognosis of LGG. The findings of the present study suggest that TICs play a pivotal role in the TME, which provide a novel avenue for the improvement of the immunosuppressive state of TME and guides clinical treatment responses of LGG.
However, the present study had certain limitations. First, we established a prognostic IRGPs signature of LGG based on data obtained from TCGA database and cross-validated the signature with LGG cases in the CGGA database, which achieved satisfactory test efficiency and stability. Nevertheless, future studies involving larger samples are required to validate the reliability of the IRGPs signature. Second, gene expression profiling is still dependent on RNA-seq and microarray data; therefore, extensive promotion remains a huge challenge, which requires further optimization of testing programs to enhance their clinical application.
Conclusion
In general, we established a prognostic IRGPs signature that can accurately evaluate the prognosis of patients with LGG. The risk stratification results based on the signature revealed that the relative levels of TICs and gene expression profiles of patients with low- and high-RSs were significantly different. Therefore, the IRGPs signature is a robust and stable approach for predicting the survival outcome and treatment responses of LGG immunotherapy.
Ethical Statement
This study was approved by the Guangxi Medical University Cancer Hospital (LW2021070).
Acknowledgments
We acknowledge TCGA and CGGA databases for providing their platforms and contributors for uploading their meaningful datasets.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Forst DA, Nahed BV, Loeffler JS, Batchelor TT. Low-grade gliomas. Oncologist. 2014;19(4):403–413. doi:10.1634/theoncologist.2013-0345
2. Gittleman H, Sloan AE, Barnholtz-Sloan JS. An independently validated survival nomogram for lower-grade glioma. Neuro Oncol. 2020;22(5):665–674. doi:10.1093/neuonc/noz191
3. Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl 2):ii1–ii56. doi:10.1093/neuonc/not151
4. Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498.
5. Claus EB, Walsh KM, Wiencke JK, et al. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus. 2015;38(1):E6. doi:10.3171/2014.10.FOCUS12367
6. Wesseling P, Capper DW. WHO 2016 Classification of gliomas. Neuropathol Appl Neurobiol. 2018;44(2):139–150. doi:10.1111/nan.12432
7. Sadik A, Somarribas Patterson LF, Öztürk S, et al. IL4I1 is a metabolic immune checkpoint that activates the ahr and promotes tumor progression. Cell. 2020;182(5):1252–1270.e1234. doi:10.1016/j.cell.2020.07.038
8. Xu S, Tang L, Li X, Fan F, Liu Z. Immunotherapy for glioma: current management and future application. Cancer Lett. 2020;476:1–12. doi:10.1016/j.canlet.2020.02.002
9. Tamura R, Miyoshi H, Yoshida K, Okano H, Toda M. Recent progress in the research of suicide gene therapy for malignant glioma. Neurosurg Rev. 2021;44(1):29–49. doi:10.1007/s10143-019-01203-3
10. Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32(19–20):1267–1284. doi:10.1101/gad.314617.118
11. Guo X, Wang Y, Zhang H, et al. Identification of the prognostic value of immune-related genes in esophageal cancer. Front Genet. 2020;11:989. doi:10.3389/fgene.2020.00989
12. Qiu H, Hu X, He C, Yu B, Li Y, Li J. Identification and validation of an individualized prognostic signature of bladder cancer based on seven immune related genes. Front Genet. 2020;11:12. doi:10.3389/fgene.2020.00012
13. Guo D, Wang M, Shen Z, Zhu J. A new immune signature for survival prediction and immune checkpoint molecules in lung adenocarcinoma. J Transl Med. 2020;18(1):123. doi:10.1186/s12967-020-02286-z
14. Wang J, Yu S, Chen G, et al. A novel prognostic signature of immune-related genes for patients with colorectal cancer. J Cell Mol Med. 2020;24(15):8491–8504. doi:10.1111/jcmm.15443
15. Popovici V, Budinska E, Tejpar S, et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J Clin Oncol. 2012;30(12):1288–1295. doi:10.1200/JCO.2011.39.5814
16. Ren L, Xu Y, Liu C, Wang S, Qin G. IL-17RB enhances thyroid cancer cell invasion and metastasis via ERK1/2 pathway-mediated MMP-9 expression. Mol Immunol. 2017;90:126–135. doi:10.1016/j.molimm.2017.06.034
17. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
18. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259.
19. Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205(12):613–621. doi:10.1016/j.cancergen.2012.10.009
20. Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. doi:10.1093/neuonc/nou087
21. Shields LB, Choucair AK. Management of low-grade gliomas: a review of patient-perceived quality of life and neurocognitive outcome. World Neurosurg. 2014;82(1–2):e299–e309. doi:10.1016/j.wneu.2014.02.033
22. Su X, Li H, Chen S, Qin C. Study on the prognostic values of dynactin genes in low-grade glioma. Technol Cancer Res Treat. 2021;20:15330338211010143. doi:10.1177/15330338211010143
23. Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi:10.1056/NEJMoa1200690
24. Rizvi NA, Mazières J, Planchard D, et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a Phase 2, single-arm trial. Lancet Oncol. 2015;16(3):257–265. doi:10.1016/S1470-2045(15)70054-9
25. Carter T, Shaw H, Cohn-Brown D, Chester K, Mulholland P. Ipilimumab and bevacizumab in glioblastoma. Clin Oncol. 2016;28(10):622–626. doi:10.1016/j.clon.2016.04.042
26. Ott PA, Bang YJ, Piha-Paul SA, et al. T-cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J Clin Oncol. 2019;37(4):318–327. doi:10.1200/JCO.2018.78.2276
27. Cloughesy TF, Mochizuki AY, Orpilla JR, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med. 2019;25(3):477–486. doi:10.1038/s41591-018-0337-7
28. McCall NS, Dicker AP, Lu B. Beyond concurrent chemoradiation: the emerging role of PD-1/PD-L1 inhibitors in stage III lung cancer. Clin Cancer Res. 2018;24(6):1271–1276. doi:10.1158/1078-0432.CCR-17-3269
29. Chae YK, Arya A, Iams W, et al. Current landscape and future of dual anti-CTLA4 and PD-1/PD-L1 blockade immunotherapy in cancer; lessons learned from clinical trials with melanoma and non-small cell lung cancer (NSCLC). J Immunother Cancer. 2018;6(1):39. doi:10.1186/s40425-018-0349-3
30. Reiss SN, Yerram P, Modelevsky L, Grommes C. Retrospective review of safety and efficacy of programmed cell death-1 inhibitors in refractory high grade gliomas. J Immunother Cancer. 2017;5(1):99. doi:10.1186/s40425-017-0302-x
31. Shen S, Wang G, Zhang R, et al. Development and validation of an immune gene-set based prognostic signature in ovarian cancer. EBioMedicine. 2019;40:318–326. doi:10.1016/j.ebiom.2018.12.054
32. Bagley SJ, Desai AS, Linette GP, June CH, O’Rourke DM. CAR T-cell therapy for glioblastoma: recent clinical advances and future challenges. Neuro Oncol. 2018;20(11):1429–1438. doi:10.1093/neuonc/noy032
33. Grayson MH, Holtzman MJ. Chemokine signaling regulates apoptosis as well as immune cell traffic in host defense. Cell Cycle. 2006;5(4):380–383. doi:10.4161/cc.5.4.2427
34. Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17(9):559–572.
35. Brat DJ, Bellail AC, Van Meir EG. The role of interleukin-8 and its receptors in gliomagenesis and tumoral angiogenesis. Neuro Oncol. 2005;7(2):122–133. doi:10.1215/S1152851704001061
36. Bronte V. Myeloid-derived suppressor cells in inflammation: uncovering cell subsets with enhanced immunosuppressive functions. Eur J Immunol. 2009;39(10):2670–2672. doi:10.1002/eji.200939892
37. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi:10.1038/s41591-018-0014-x
38. Clara JA, Monge C, Yang Y, Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells - A clinical update. Nat Rev Clin Oncol. 2020;17(4):204–232. doi:10.1038/s41571-019-0293-2
39. Reardon DA, Freeman G, Wu C, et al. Immunotherapy advances for glioblastoma. Neuro Oncol. 2014;16(11):1441–1458. doi:10.1093/neuonc/nou212
40. Brusko TM, Wasserfall CH, Agarwal A, Kapturczak MH, Atkinson MA. An integral role for heme oxygenase-1 and carbon monoxide in maintaining peripheral tolerance by CD4+CD25+ regulatory T cells. J Immunol. 2005;174(9):5181–5186. doi:10.4049/jimmunol.174.9.5181
41. Brandenburg S, Müller A, Turkowski K, et al. Resident microglia rather than peripheral macrophages promote vascularization in brain tumors and are source of alternative pro-angiogenic factors. Acta Neuropathol. 2016;131(3):365–378. doi:10.1007/s00401-015-1529-6
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.