Back to Journals » Journal of Inflammation Research » Volume 16
Prediction and Verification of Potential Therapeutic Targets for Non-Responders to Infliximab in Ulcerative Colitis
Authors Lyu X, Zhang Z, Liu X, Geng L, Zhang M, Feng B
Received 3 March 2023
Accepted for publication 9 May 2023
Published 15 May 2023 Volume 2023:16 Pages 2063—2078
DOI https://doi.org/10.2147/JIR.S409290
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Xue Lyu,* Zhe Zhang,* Xia Liu,* Li Geng, Muhan Zhang, Baisui Feng
Department of Gastroenterology, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou, 450014, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Baisui Feng, Email [email protected]
Background: Infliximab (IFX) has been widely used in ulcerative colitis (UC) patients. However, the subsequent effective treatment of IFX non-response in UC patients remains a challenge. This study aims to predict potential therapeutic targets for non-responders by performing a bioinformatic analysis of the data in the Gene Expression Omnibus (GEO) database and validation by biopsies.
Methods: Colonic mucosal biopsies expression profiles of IFX-treated UC patients (GSE73661, GSE16879) were utilized to predict potential therapeutic targets. Bioinformatics analyses were used to explore potential biological mechanisms. CytoHubba was performed to screen hub genes. We used a validation dataset and colonic mucosal biopsies of UC patients to validate hub genes.
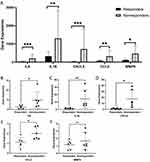
Results: A total of 147 DEGs were identified (119 upregulated genes and 28 downregulated genes). GSEA showed that DEGs in GSE73661 were enriched in the pathways of the cytokine-cytokine receptor, the chemokine, and the adhesion molecules system. Based on the PPI network analysis, we identified four hub genes (and the transcription factor NF-κB). Then, we validate the expression of hub genes by reverse transcription-polymerase chain reaction (RT-PCR). We found higher expression of IL-6, IL1B, CXCL8, and CCL2 in non-responders compared to responders.
Conclusion: In summary, four potential targets (IL-6, IL1B, CXCL8, and CCL2) were finally identified by performing a bioinformatics analysis of the datasets in the GEO database. Their expression was confirmed in colonic mucosal biopsies of patients with UC. These results can help to further explore the mechanism of non-responders to IFX in UC and to provide potential targets for their subsequent treatment.
Keywords: ulcerative colitis, infliximab, GEO dataset, drug targets
Introduction
Ulcerative colitis (UC) is an inflammatory bowel disease (IBD) characterized by continuous and diffuse inflammation of the colonic mucosa, which extends from the proximal rectum. As of now, the pathogenesis of UC is unclear, but risk factors include genetic susceptibility, environmental factors, autoimmunity, and gut microbiota.1,2 The high incidence of UC in developed countries and the substantial rise in the incidence in developing countries have made UC a global health issue.3–5 A first-line drug for mild to moderate UC is 5-amino-salicylic acid (5-ASA).6 Systemic corticosteroids are usually prescribed for moderate to severe UC. Corticosteroids should not be used for a prolonged period and only be used to induce remission. Several types of treatments are available to maintain remission, including azathioprine, anti-TNF, anti-integrins, and Janus kinase inhibitors.7,8
As the first anti-tumor necrosis factor (anti-TNF) drug developed for IBD, infliximab (IFX) can effectively relieve symptoms of the disease.9 Combining IFX with azathioprine can reduce the rate of hospitalization and surgery in patients with UC.10 However, 10–30% of patients with IBD experience primary non-response (PNR) to treatment, and 23–46% will lose response over time (secondary loss of response).11,12 Currently, there are few clinical guidelines on how to manage patients with loss response to therapy.13 Among the clinical methods are switching to another anti-TNF preparation, replacing non-anti-TNF preparations, and surgical treatment, among others.14,15 According to the AGC guidelines, UC patients with IFX failure can be switched to tofacitinib or ustekinumab. However, based on available clinical studies, it was found that none of the switched biologics were as effective as IFX.16,17 Therefore, the screening of novel therapeutic targets is warranted.
In recent years, microarray technology has been extensively used in bioinformatics research. This technology provides valuable information on mRNAs, which can be used to predict disease occurrence and explore the key factors that lead to it.18–20 A bioinformatics-based screening method was used in the current study, and then hub genes were validated using patient datasets and clinical biopsy samples.
Materials and Methods
Datasets Selection and Identification of DEGs
The analysis process of this study is described in Figure 1. Two datasets of mucosal array profiles in IFX-treated UC patients were employed in this study. The raw data of the dataset we used (GSE73661, GSE16879) (Table 1) were downloaded from the Gene expression omnibus (GEO) website (https://www.ncbi.nlm.nih.gov/geo/). GSE73661 is the training dataset, and GSE16879 is the validation dataset.21,22 The raw data were processed and standardized using the R software (version 4.1.0). Responders to IFX and non-responders to IFX were analyzed using the limma package, and statistically significant altered and DEGs were identified. The significant DEGs were identified with the threshold: adjust P value <0.05 and |log2 (Fold Change) |>1.5.
 |
Table 1 The Information of Datasets |
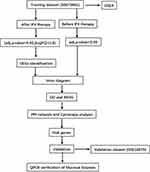 |
Figure 1 Flow chart of the study. |
Biological Function and Pathway Enrichment Analyses
The Gene Set Enrichment Analysis (GSEA) tool was used for exploring different pathways between responders and non-responders.23 Significance criteria were nominal P-value <0.05 and false positive rate (FDR) <0.25. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the DEGs were performed using the Cluster Profile package in the R software.24 The GO enrichment analysis can be divided into three parts: biological process (BP), cell component (CC), and molecular function (MF).25 The criterion for the DEGs enrichment analysis was P <0.05.
PPI Network Analysis
STRING database (https://string-db.org/) was used to integrate and build the Protein-Protein Interaction (PPI) network.26 Removing isolated nodes and filtering out low-confidence nodes, resulting in 98 genes remaining. The plugin CytoHubba in Cytoscape (version 3.9.1) was used to discover key nodes in the PPI network.27,28 The other plugin molecular complex detection (MCODE) in Cytoscape was used to identify the important functional modules.29 Hub genes were identified by CytoHubba and sorted by degree scores.
Patients
Patients with moderate to severe active UC were selected using the Mayo score, which was defined as a Mayo score of 6–12 and an endoscopic score of at least 2 points. Response to IFX was defined as a minimum of 3 points decrease in the total Mayo score and a 30% decrease in the baseline score, with the rectal bleeding score decreased by 1 point or the absolute score of rectal bleeding was 0 or 1 point, and the Mayo endoscopic subscore decreased by at least 1 point.30 12 post-IFX treatment colonic mucosal biopsies from 11 UC patients were obtained (Two colonic mucosal biopsies were obtained during different periods of colonoscopy in the same patient after the fourth infliximab treatment), who received a loading dose of IFX (5 or 10 mg/kg) for refractory UC (Table 2).
 |
Table 2 Baseline Clinical Characteristics of Ulcerative Colitis Patients |
RNA Extraction and RT-PCR
To validate target genes and determine gene expression in biopsy samples, RT-PCR was performed for IL1B, IL-6, CXCL8, CCL2, MMP9, and GAPDH, which was used as the endogenous reference gene. Total RNA was extracted from mucosal biopsies using the Trizol reagent (Invitrogen) and cDNA was synthesized by reverse transcription kit (TAKARA). Primer sequences for target genes were designed by Sangon Biotech (https://www.sangon.com/) and constructed by Sigma-Aldrich (Table 3). RT-PCR was carried out using the ABI STEPONE PLUS system and the PowerUp SYBR Green Master Mix (Thermo Fisher Scientific). The relative expression level of the target genes was calculated as the expression level of GAPDH. Relative expression was determined using the 2-ΔΔCt method. The final results were visualized by GraphPad Prism 9.2.0.
 |
Table 3 Primers for RT-PCR |
Drug-Gene Interaction
To explore drug-gene interaction, we used the DrugBank31 (https://go.drugbank.com/drugs) database to identify existing or/and potentially relevant drug substances. Moreover, the Cytoscape software was utilized for data visualization.
Statistical Analyses
Data were analyzed using the R software. Differences between the two groups were analyzed using Student’s t-test, and Mann–Whitney U-test. Statistical significance was judged when the p-value< 0.05 (two-sided). GraphPad Prism 9.2.0 was used for statistical analysis and image construction.
Results
Identification of DEGs
Based on dataset GSE73661, we identified 155 DEGs, of which 29 of them were downregulated and 126 upregulated in non-responders. We observed that most DEGs were upregulated in non-responders. These differentially expressed genes were visualized by a volcano plot (Figure 2A). We also performed a differential analysis between patients with responders and non-responders before using IFX, and a total of 470 genes were statistically different (Figure 2B).
As is shown in Figure 2C, to exclude differences between groups due to individual differences, we removed genes that differed both before and after using IFX. A total of 147 DEGs were identified (119 upregulated genes and 28 downregulated genes). Differential genes before the use of IFX can be used to predict whether IFX will respond or not, and differential genes after the use of IFX can be used to find therapeutic targets.
Functional Enrichment Analysis
GO and KEGG pathway analyses were then performed based on these DEGs. In the GO functional enrichment, the top eight pathways were selected. In the BP category, the DEGs were mainly enriched in leukocyte and granulocyte chemotaxis and migration. DEGs were markedly enriched in the external side of the plasma membrane, the apical part of the cell, and the secretory granule lumen in the CC category. Enrichment of the MF category included cytokine activity, cytokine receptor binding, and immune receptor activity (Figure 3A). The KEGG pathway enrichment analysis indicated that the DEGs were mainly enriched in the cytokine-cytokine receptor interaction pathway, IL-17 signaling pathway, TNF signaling pathway, and chemokine signaling pathway (Figure 3B). Meanwhile, we also performed the enrichment analysis of the original matrix of the dataset by GSEA and found that the inflammatory and immune-related pathways were significantly activated in the non-responders, which coincides with the analysis results of GO and KEGG (Figure 4).
 |
Figure 3 Functional enrichment analysis of the DEGs. (A) Gene Ontology (GO) analysis of DEGs. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for DEGs. |
PPI Network Construction
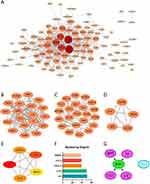
PPI network analysis was performed using the STRING database, where 147 DEGs removed the isolated nodes and filtered out low-confidence nodes, finally remaining with 98 genes (Figure 5A). The PPI network consists of 98 nodes with 566 edges. Visualization was performed using the Cytoscape software. The MCODE plugin was used to identify important functional modules. Three cluster modules were identified based on the screening criteria (Figures 5B–D). Cluster 1 has a high score (score: 12,111, 19 nodes and 109 edges), followed by cluster 2 (score: 7714, 22 nodes and 81 edges), and cluster 3 (score: 4500, 5 nodes and 9 edges). Hub genes were filtered according to the Degree values using the Cytohubba plugin. We found that the top five genes with Degree values greater than 35 (IL-6, IL1B, CXCL8, CCL2, and MMP9) were also within the first module (Figure 5E and F). Transcription factors bind to non-transcribed regions of genes to regulate gene expression. The TF binding motifs of five hub genes associated with non-responders were tested using the iRegulon plugin. We found that four genes except CXCL8 received NF-κB1 regulation (Figure 5G).
Verification of Hub Genes
We performed a differential expression analysis of five hub genes in the validation dataset. In agreement with our predictions, five hub genes specifically expressed in non-responders were significantly upregulated in their mRNA expression levels compared to responders (Figure 6A). Finally, the expression of these five hub genes was validated in colonic mucosal biopsies of UC patients using IFX. A total of 12 colonic mucosal biopsies with moderate to severe UC were selected to determine the mRNA expression levels of hub genes by RT-PCR. We found that IL-6, IL1B, CXCL8, and CCL2 were significantly upregulated in non-responders (Figures 6B–E). However, there was no significant difference in the expression of MMP9 in the biopsies of the 12 patients (Figure 6F).
Drug-Gene Interaction
The development of therapeutic agents targeting the hub genes in non-responders is a novel therapeutic strategy. The drug-gene interaction network for target genes is shown in Figure 7. Thirty potential therapeutic agents for four target genes have been identified (Five of these drugs can target both IL-6 and IL1B), including 11 for market approval (Table 4). Among them, antagonists and inhibitors can be used in non-responders.
 |
Table 4 The Drugs Approved to Interact with Target Genes |
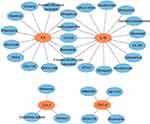 |
Figure 7 Drug-gene interaction network of target genes. Drugs are in blue, and target genes are in Orange. |
Discussion
UC is an idiopathic chronic inflammatory disease characterized by repeated periods of activity and remission. A subset of patients with UC experienced a primary non-response or secondary loss of response to IFX therapy despite its effectiveness.17 AGA guidelines recommend switching to tofacitinib or ustekinumab in patients with UC unresponsive to IFX. With its excellent safety profile, vedolizumab may be a viable option for patients with mild disease.14 However, there are few reports in the literature on the effectiveness of biologics after switching. In the published study, the authors used machine learning techniques to predict the effect of PNR on IFX in UC patients.18 Some authors have also identified predictors of IFX non-response in IBD patients by a cell-centered meta-analysis.32 Follow-up studies after IFX failure are scarce. Based on the lack of response to IFX in UC patients, we searched microarray data from the GEO database for potential new therapeutic targets.
This study compared the colonic mucosal expression profiles of UC patients who responded to IFX therapy with those who did not. 147 DEGs were identified for gene function analysis. GO and KEGG showed that DEGs are mainly involved in the chemotaxis and migration of leukocytes as well as the inflammatory response. The PPI network analysis of DEGs eliminated the isolated and low-confidence nodes. Hub gene screening and modular analysis revealed five hub genes, IL-6, IL1B, CXCL8, CCL2, and MMP9 (all upregulated in non-response patients). We validated these five hub genes in another dataset, consistent with the conclusions we obtained. The expression of five hub genes was then validated in clinical biopsy samples, demonstrating that IL-6, IL1B, CXCL8, and CCL2 were upregulated in non-responders. They are common inflammatory cytokines and chemokines but have not been previously reported to associate with IFX non-response.
Interleukin 6 (IL-6) belongs to a family of pro-inflammatory cytokines. IL-6, transmembrane IL-6 receptor (mIL-6R) or soluble IL-6 receptor (sIL-6R), binds to gp130 signaling subunit molecules to mediate IL-6 Signal Transduction.33,34 The classical IL-6 signaling pathway is initiated by the IL-6/IL-6R complex and two molecules of the signal transducer gp130.35 Prolonged activation of this pathway leads to protein cleavage of the mIL-6R, causing the release of sIL-6R. Additionally, sIL-6R can form complexes with IL-6, resulting in IL-6 trans-signaling,36 which is mainly responsible for chronic inflammation. Many drugs that target the IL-6 signaling pathway lack specificity and inhibit both classical and trans-signaling pathways. IL-6 and IL-6R inhibitors are effective in treating rheumatoid arthritis, and their applications also extend to other acute and chronic inflammation.37,38 IL-6 is a pleiotropic cytokine in IBD patients. On the one hand, IL-6 may be involved in the regulation of intestinal epithelial barrier integrity. On the other hand, IL-6 induces chronic colitis by inhibiting T-cell apoptosis.39,40 Therefore, the use of IL-6 inhibitors may also be accompanied by other side effects. PF-04236921, a human anti-IL-6 antibody, is in Phase I clinical trials in Crohn’s Disease (CD) patients who had had an inadequate response to anti-TNF therapy. The study concluded that treatment with PF-04236921 50 mg produced clinical and symptomatic remission in patients with moderately to severely refractory CD who had failed TNF therapy. Side effects such as gastrointestinal abscesses and perforation can also occur.41 Sgp130Fc (olamkicept) is an IL-6 trans-signaling inhibitor that has been tested in clinical trials for patients with UC. In a double-blind placebo-controlled phase IIb clinical study, it was administered to treat moderate-to-severe UC patients who did not respond adequately to conventional therapy. It is noteworthy that the majority of patients (94.5%) had not been treated with biologics before. After 12 weeks of treatment, clinical remission was observed in 0% (placebo), 6.7% (olamkicept 300 mg/injection), and 20.7% (olamkicept 600 mg/injection) of patients. It is important to note that the incidence of adverse events (AEs), such as upper respiratory tract infections, elevated AST levels, and elevated urinary bilirubin levels, were similar in all groups. These AEs were mostly mild to moderate and transient. No deaths or serious adverse events were associated with pan-IL-6 inhibitors.42 It may be more reasonable to selectively block the IL-6 trans-signaling pathway rather than fully blocking IL-6 with neutralizing antibodies.
Interleukin 1 beta (IL1B) is a potent pro-inflammatory cytokine that plays a role in both innate and adaptive immune responses. IL1B is involved in the development of intestinal inflammation in the absence of IL10R.43 Blocking IL1B inhibited DSS-induced colitis in mice.44 Canakinumab, a human anti-IL-1β monoclonal antibody, has been FDA-approved for treating Periodic Fever Syndromes and Still’s disease. It effectively controls patient progression, with the most commonly reported AE being infection but most events being mild to moderate. Injection site reactions are mild or moderate.45 Anakinra, a recombinant human IL-1 receptor antagonist (IL-1RA), has shown efficacy in treating various diseases including rheumatoid arthritis (RA), gout, and idiopathic pericarditis, to rare hereditary diseases. The safety profile of anakinra has been well-established since its introduction in the United States in 2001. The most common adverse drug reactions associated with anakinra treatment are injection site reactions, which are typically mild to moderate in severity.46 One of the known reasons for IFX failure in IBD is the formation of anti-drug antibodies (ADA).47 Previous studies have shown that the formation of IFX-TNF complexes is positively correlated with the formation of ADA and that IFX-TNF complexes induce innate immune activation, leading to increased expression of IL1B.48 Therefore, IL1B levels may be elevated in non-responders. In our study, IL1B expression levels were significantly increased in the colonic mucosa of non-responders compared with responders. Reduction of intestinal IL1B expression by IL1B inhibitors reduces intestinal inflammation in non-responders.
CXCL8 (Interleukin-8, IL8) is one of the earliest identified chemokines, and its main function is to guide the transport of leukocytes to the sites of inflammation.49 The expression of CXCL8 was increased in the colonic mucosa of patients with active UC.50 By binding to its receptor, CXC chemokine receptor type 1 (CXCR1) and CXC chemokine receptor type 2 (CXCR2), CXCL8 recruits neutrophils to sites of inflammation. CXCR1/2 is widely co-expressed in immune cells, and inhibition of CXCR1/2 reduces neutrophil recruitment in vivo.51 In animal trials, the CXCR2 antagonist (SB225002) has significantly alleviated the DSS-induced colon inflammation in mice.52 Several studies have also demonstrated that the CXCR2 gene deficiency is protective against DSS-induced colitis in mice.53 While the role of CXCR2 antagonists in UC has not been extensively studied in clinical trials, early lung trials have shown promising results. Specifically, a clinical trial involving the CXCR2 antagonist (SCH527123) reported that it was safe, well tolerated, and effective in reducing sputum neutrophil counts in patients with severe asthma.54 In our study, non-responders had significantly increased CXCR1/2 expression in the colonic mucosa compared with responders (Supplementary Figure 1A and B). Therefore, IFX failure in UC patients could be treated by inhibiting the CXCL8-CXCR1/2 axis.
CC chemokine ligand-2 (CCL2) is one of the chemokines secreted by immune system cells and plays an important role in the migration of monocytes.55,56 CCL2 can participate in the inflammatory response. When tissues are infected and damaged, large amounts of CCL2 are secreted to recruit CCR2-expressing cells to the site of injury.57 Studies have shown that CCL2 expression is increased in IBD patients.58 Tenascin C (TNC) and CCL2 expression were reduced in IFX responders with UC. TNC may be a regulator of CCL2 expression, suggesting downregulation of CCL2.59 Therefore, the use of CCL2 inhibitors may prevent monocytes from entering the site of inflammation, thereby reducing inflammation.
GSEA enrichment results showed that, in addition to the activation of the cytokine-cytokine receptor and chemokine pathways, cell adhesion molecule pathways were also significantly activated in non-responders. Ranked by logFC value, E-selectin (SELE) was the largest gene in the cell adhesion molecule pathway. In the GSE16879 validation dataset, SELE was upregulated in non-responders. Gene expression levels measured by RT-PCR in intestinal mucosal biopsies indicated upregulation of SELE in non-responders (Supplementary Figure 1C). Selectins include L-, P-, and E-selectins. Selectins and their receptors mediate the initial contact between neutrophils and endothelial cells and are involved in rolling neutrophils along endothelial cells.60 SELE expression is absent on the surface of resting endothelial cells but is upregulated when endothelial cells are exposed to pro-inflammatory cytokines such as TNF-α and interleukins. SELE regulates intestinal inflammation by participating in leukocyte trafficking, and by preventing leukocyte homing with selectin inhibitors, it helps reduce the exaggerated immune response in colitis.61 SELE regulates intestinal inflammation by participating in leukocyte trafficking, and by preventing leukocyte homing with selectin inhibitors, it helps reduce the exaggerated immune response in colitis.
The current study has several limitations. First, the number of patients in the database is relatively small. Furthermore, this study lacked follow-up functional testing and mechanistic exploration. However, we think this study is meaningful. Bioinformatics analysis screened out the hub genes between responders and non-responders, providing evidence for subsequent clinical treatment and switching to biological agents.
Biological agents clinically used in patients with UC can be divided into two categories: (1) agents that antagonize pro-inflammatory cytokines and (2) agents that block lymphocyte homing. Integrins, selectins, and chemokines jointly regulate leukocyte trafficking.62 Due to the high potency of anti-TNF biologics, anti-TNF drugs are usually the biologics of choice for patients with moderate to severe UC. In the case of ineffective anti-TNF drugs, which drug to choose as an alternative treatment has become the focus of clinicians’ research. We found that the expression of IL-6, IL1B, CXCL8, CCL2, and SELE was significantly increased in IFX non-responders of UC patients. The use of antagonists of these hub genes can reduce intestinal inflammation in IFX non-responders of UC patients, providing a basis for the selection of subsequent biologics.
Conclusions
In summary, four potential targets (IL-6, IL1B, CXCL8, and CCL2) were finally identified by performing a bioinformatics analysis of the datasets in the GEO database. Their expression was confirmed in colonic mucosal biopsies of patients with UC. Some biologics for these targets have been used in clinical trials, and olamkicept has been used in the treatment of patients with moderate-to-severe UC. These results can help to further explore the mechanism of non-responders to IFX in UC and to provide potential targets for their subsequent treatment.
Abbreviations
IFX, Infliximab; UC, ulcerative colitis; GEO, Gene expression omnibus; DEGs, Differentially expressed genes; GSEA, Gene Set Enrichment Analysis; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction; MCODE, molecular complex detection; RT-PCR, reverse transcription-polymerase chain reaction; IBD, inflammatory bowel disease; 5-ASA, 5-amino-salicylic acid; TNF, tumor necrosis factor; PNR, primary non-response; FC, fold change BP, biological process; CC, cell component; MF, molecular function; FDR, false discovery rate; NES, normalized enrichment score; IL-6, Interleukin-6; mIL-6R, transmembrane IL-6 receptor; sIL-6R, soluble IL-6 receptor; CD, Crohn’s Disease; AEs, adverse events; IL1B, Interleukin 1beta; IL-1RA, IL-1 receptor antagonist; RA, rheumatoid arthritis; ADA, anti-drug antibodies; CXCL8/IL8, Interleukin-8; CXCR1, CXC chemokine receptor type 1; CXCR2, CXC chemokine receptor type 2; CCL2, CC chemokine ligand-2; TNC, Tenascin C; SELE, E-selectin.
Data Sharing Statement
The datasets analyzed during the current study are available in the Gene Expression Omnibus (GEO) datasets (https://www.ncbi.nlm.nih.gov/geo/). And the authors confirm that the data supporting the findings of this study are available within the article.
Ethics Statement
The study involving human participants were reviewed and approved by the Ethical Review Board of the Second Affiliated Hospital of Zhengzhou University. The patients/participants provided their written informed consent to participate in this study. This study complies with the Declaration of Helsinki.
Acknowledgments
The authors would like to acknowledge the excellent assistance all study doctors and nurses provide.
Funding
This project was supported by the Science and Technology Benefiting the People Program of Zhengzhou, 2021 (2021KJHM0018), the Science and Technology Breakthrough Project of Henan Province (222102310531), the Medical Science and Technology Breakthrough Project of Henan Province, China (LHGJ20210409, LHGJ20200396, and LHGJ20210415).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389(10080):1756–1770. doi:10.1016/S0140-6736(16)32126-2
2. Kobayashi T, Siegmund B, Le Berre C, et al. Ulcerative colitis. Nat Rev Dis Primers. 2020;6(1):74. doi:10.1038/s41572-020-0205-x
3. Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390(10114):2769–2778. doi:10.1016/S0140-6736(17)32448-0
4. Coward S, Clement F, Benchimol EI, et al. Past and future burden of inflammatory bowel diseases based on modeling of population-based data. Gastroenterology. 2019;156(5):1345–1353 e1344. doi:10.1053/j.gastro.2019.01.002
5. Ng SC, Kaplan GG, Tang W, et al. Population density and risk of inflammatory bowel disease: a prospective population-based study in 13 countries or regions in Asia-pacific. Am J Gastroenterol. 2019;114(1):107–115. doi:10.1038/s41395-018-0233-2
6. Harbord M, Eliakim R, Bettenworth D, et al. Third European evidence-based consensus on diagnosis and management of ulcerative colitis. Part 2: current management. J Crohns Colitis. 2017;11(7):769–784. doi:10.1093/ecco-jcc/jjx009
7. Neurath MF, Leppkes M. Resolution of ulcerative colitis. Semin Immunopathol. 2019;41(6):747–756. doi:10.1007/s00281-019-00751-6
8. Feuerstein JD, Moss AC, Farraye FA. Ulcerative Colitis. Mayo Clin Proc. 2019;94(7):1357–1373. doi:10.1016/j.mayocp.2019.01.018
9. Levin AD, Wildenberg ME, van den Brink GR. Mechanism of action of anti-TNF therapy in inflammatory bowel disease. J Crohns Colitis. 2016;10(8):989–997. doi:10.1093/ecco-jcc/jjw053
10. Panaccione R, Ghosh S, Middleton S, et al. Combination therapy with infliximab and azathioprine is superior to monotherapy with either agent in ulcerative colitis. Gastroenterology. 2014;146(2):392–400 e393. doi:10.1053/j.gastro.2013.10.052
11. Pouillon L, Travis S, Bossuyt P, Danese S, Peyrin-Biroulet L. Head-to-head trials in inflammatory bowel disease: past, present and future. Nat Rev Gastroenterol Hepatol. 2020;17(6):365–376. doi:10.1038/s41575-020-0293-9
12. Sprakes MB, Ford AC, Warren L, Greer D, Hamlin J. Efficacy, tolerability, and predictors of response to infliximab therapy for Crohn’s disease: a large single centre experience. J Crohns Colitis. 2012;6(2):143–153. doi:10.1016/j.crohns.2011.07.011
13. Roda G, Jharap B, Neeraj N, Colombel JF. Loss of response to anti-TNFs: definition, epidemiology, and management. Clin Transl Gastroenterol. 2016;7:e135. doi:10.1038/ctg.2015.63
14. Feuerstein JD, Isaacs KL, Schneider Y, et al. AGA clinical practice guidelines on the management of moderate to severe ulcerative colitis. Gastroenterology. 2020;158(5):1450–1461. doi:10.1053/j.gastro.2020.01.006
15. van Rheenen PF, Aloi M, Assa A, et al. The medical management of paediatric crohn’s disease: an ECCO-ESPGHAN guideline update. J Crohns Colitis. 2020. doi:10.1093/ecco-jcc/jjaa161
16. Singh S, George J, Boland BS, Vande Casteele N, Sandborn WJ. Primary non-response to tumor necrosis factor antagonists is associated with inferior response to second-line biologics in patients with inflammatory bowel diseases: a systematic review and meta-analysis. J Crohns Colitis. 2018;12(6):635–643. doi:10.1093/ecco-jcc/jjy004
17. Fine S, Papamichael K, Cheifetz AS. Etiology and management of lack or loss of response to anti-tumor necrosis factor therapy in patients with inflammatory bowel disease. Gastroenterol Hepatol. 2019;15(12):656–665.
18. Feng J, Chen Y, Feng Q, Ran Z, Shen J. Novel gene signatures predicting primary non-response to infliximab in ulcerative colitis: development and validation combining random forest with artificial neural network. Front Med. 2021;8:678424. doi:10.3389/fmed.2021.678424
19. Xu M, Kong Y, Chen N, et al. Identification of immune-related gene signature and prediction of CeRNA network in active ulcerative colitis. Front Immunol. 2022;13:855645. doi:10.3389/fimmu.2022.855645
20. Cortes-Ciriano I, Gulhan DC, Lee JJ, Melloni GEM, Park PJ. Computational analysis of cancer genome sequencing data. Nat Rev Genet. 2022;23(5):298–314. doi:10.1038/s41576-021-00431-y
21. Arijs I, De Hertogh G, Lemmens B, et al. Effect of vedolizumab (anti-alpha4beta7-integrin) therapy on histological healing and mucosal gene expression in patients with UC. Gut. 2018;67(1):43–52. doi:10.1136/gutjnl-2016-312293
22. Arijs I, De Hertogh G, Lemaire K, et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One. 2009;4(11):e7984. doi:10.1371/journal.pone.0007984
23. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
24. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
25. Dalmer TRA, Clugston RD. Gene ontology enrichment analysis of congenital diaphragmatic hernia-associated genes. Pediatr Res. 2019;85(1):13–19. doi:10.1038/s41390-018-0192-8
26. Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi:10.1093/nar/gkaa1074
27. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11
28. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
29. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi:10.1186/1471-2105-4-2
30. D’Haens G, Sandborn WJ, Feagan BG, et al. A review of activity indices and efficacy end points for clinical trials of medical therapy in adults with ulcerative colitis. Gastroenterology. 2007;132(2):763–786. doi:10.1053/j.gastro.2006.12.038
31. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi:10.1093/nar/gkx1037
32. Gaujoux R, Starosvetsky E, Maimon N, et al. Cell-centred meta-analysis reveals baseline predictors of anti-TNFalpha non-response in biopsy and blood of patients with IBD. Gut. 2019;68(4):604–614. doi:10.1136/gutjnl-2017-315494
33. Ghandadi M, Sahebkar A. Interleukin-6: a critical cytokine in cancer multidrug resistance. Curr Pharm Des. 2016;22(5):518–526. doi:10.2174/1381612822666151124234417
34. Uciechowski P, Dempke WCM. Interleukin-6: a masterplayer in the cytokine network. Oncology. 2020;98(3):131–137. doi:10.1159/000505099
35. Schreiber S, Aden K, Bernardes JP, et al. Therapeutic interleukin-6 trans-signaling inhibition by olamkicept (sgp130Fc) in patients with active inflammatory bowel disease. Gastroenterology. 2021;160(7):2354–2366 e2311. doi:10.1053/j.gastro.2021.02.062
36. Kaur S, Bansal Y, Kumar R, Bansal G. A panoramic review of IL-6: structure, pathophysiological roles and inhibitors. Bioorg Med Chem. 2020;28(5):115327. doi:10.1016/j.bmc.2020.115327
37. Kaneko Y, Atsumi T, Tanaka Y, et al. Comparison of adding tocilizumab to methotrexate with switching to tocilizumab in patients with rheumatoid arthritis with inadequate response to methotrexate: 52-week results from a prospective, randomised, controlled study (Surprise study). Ann Rheum Dis. 2016;75(11):1917–1923. doi:10.1136/annrheumdis-2015-208426
38. Kang S, Tanaka T, Narazaki M, Kishimoto T. Targeting interleukin-6 signaling in clinic. Immunity. 2019;50(4):1007–1023. doi:10.1016/j.immuni.2019.03.026
39. Shahini A, Shahini A. Role of interleukin-6-mediated inflammation in the pathogenesis of inflammatory bowel disease: focus on the available therapeutic approaches and gut microbiome. J Cell Commun Signal. 2022;17(1):55–74. doi:10.1007/s12079-022-00695-x
40. Zhou L, Sonnenberg GF. Essential immunologic orchestrators of intestinal homeostasis. Sci Immunol. 2018;3(20). doi:10.1126/sciimmunol.aao1605
41. Danese S, Vermeire S, Hellstern P, et al. Randomised trial and open-label extension study of an anti-interleukin-6 antibody in Crohn’s disease (ANDANTE I and II). Gut. 2019;68(1):40–48. doi:10.1136/gutjnl-2017-314562
42. Chen B, Zhang S, Wang B, et al. 775b olamkicept, an IL-6 trans-signaling inhibitor, is effective for induction of response and remission in a randomized, placebo-controlled trial in moderate to severe ulcerative colitis. Gastroenterology. 2021;161(2):e28–e29. doi:10.1053/j.gastro.2021.06.038
43. Shouval DS, Biswas A, Kang YH, et al. Interleukin 1beta mediates intestinal inflammation in mice and patients with interleukin 10 receptor deficiency. Gastroenterology. 2016;151(6):1100–1104. doi:10.1053/j.gastro.2016.08.055
44. Yin Q, Pi X, Jiang Y, et al. An immuno-blocking agent targeting IL-1beta and IL-17A reduces the lesion of DSS-induced ulcerative colitis in mice. Inflammation. 2021;44(5):1724–1736. doi:10.1007/s10753-021-01449-4
45. De Benedetti F, Gattorno M, Anton J, et al. Canakinumab for the treatment of autoinflammatory recurrent fever syndromes. N Engl J Med. 2018;378(20):1908–1919. doi:10.1056/NEJMoa1706314
46. Vastert SJ, Jamilloux Y, Quartier P, et al. Anakinra in children and adults with Still’s disease. Rheumatology. 2019;58(Suppl6):vi9–vi22. doi:10.1093/rheumatology/kez350
47. Ben-Horin S, Chowers Y. Tailoring anti-TNF therapy in IBD: drug levels and disease activity. Nat Rev Gastroenterol Hepatol. 2014;11(4):243–255. doi:10.1038/nrgastro.2013.253
48. Bar-Yoseph H, Pressman S, Blatt A, et al. Infliximab-tumor necrosis factor complexes elicit formation of anti-drug antibodies. Gastroenterology. 2019;157(5):1338–1351 e1338. doi:10.1053/j.gastro.2019.08.009
49. Ha H, Debnath B, Neamati N. Role of the CXCL8-CXCR1/2 axis in cancer and inflammatory diseases. Theranostics. 2017;7(6):1543–1588. doi:10.7150/thno.15625
50. Bruno ME, Rogier EW, Arsenescu RI, et al. Correlation of biomarker expression in colonic mucosa with disease phenotype in crohn’s disease and ulcerative colitis. Dig Dis Sci. 2015;60(10):2976–2984. doi:10.1007/s10620-015-3700-2
51. Chapman RW, Phillips JE, Hipkin RW, Curran AK, Lundell D, Fine JS. CXCR2 antagonists for the treatment of pulmonary disease. Pharmacol Ther. 2009;121(1):55–68. doi:10.1016/j.pharmthera.2008.10.005
52. Zhu F, He H, Fan L, et al. Blockade of CXCR2 suppresses proinflammatory activities of neutrophils in ulcerative colitis. Am J Transl Res. 2020;12(9):5237–5251.
53. Ranganathan P, Jayakumar C, Manicassamy S, Ramesh G. CXCR2 knockout mice are protected against DSS-colitis-induced acute kidney injury and inflammation. Am J Physiol Renal Physiol. 2013;305(10):F1422–F1427. doi:10.1152/ajprenal.00319.2013
54. Nair P, Gaga M, Zervas E, et al. Safety and efficacy of a CXCR2 antagonist in patients with severe asthma and sputum neutrophils: a randomized, placebo-controlled clinical trial. Clin Exp Allergy. 2012;42(7):1097–1103. doi:10.1111/j.1365-2222.2012.04014.x
55. Yoshimura T. The production of monocyte chemoattractant protein-1 (MCP-1)/CCL2 in tumor microenvironments. Cytokine. 2017;98:71–78. doi:10.1016/j.cyto.2017.02.001
56. Behfar S, Hassanshahi G, Nazari A, Khorramdelazad H. A brief look at the role of monocyte chemoattractant protein-1 (CCL2) in the pathophysiology of psoriasis. Cytokine. 2018;110:226–231. doi:10.1016/j.cyto.2017.12.010
57. Raghu H, Lepus CM, Wang Q, et al. CCL2/CCR2, but not CCL5/CCR5, mediates monocyte recruitment, inflammation and cartilage destruction in osteoarthritis. Ann Rheum Dis. 2017;76(5):914–922. doi:10.1136/annrheumdis-2016-210426
58. Singh UP, Singh NP, Murphy EA, et al. Chemokine and cytokine levels in inflammatory bowel disease patients. Cytokine. 2016;77:44–49. doi:10.1016/j.cyto.2015.10.008
59. Magnusson MK, Strid H, Isaksson S, et al. Response to infliximab therapy in ulcerative colitis is associated with decreased monocyte activation, reduced CCL2 expression and downregulation of Tenascin C. J Crohns Colitis. 2015;9(1):56–65. doi:10.1093/ecco-jcc/jju008
60. Cappenberg A, Kardell M, Zarbock A. Selectin-mediated signaling-shedding light on the regulation of integrin activity in neutrophils. Cells. 2022;11(8):1310. doi:10.3390/cells11081310
61. Vestweber D, Blanks JE. Mechanisms that regulate the function of the selectins and their ligands. Physiol Rev. 1999;79(1):181–213. doi:10.1152/physrev.1999.79.1.181
62. Song Y, Yuan M, Xu Y, Xu H. Tackling inflammatory bowel diseases: targeting proinflammatory cytokines and lymphocyte homing. Pharmaceuticals. 2022;15(9):1080. doi:10.3390/ph15091080
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.